Risk of Fat Mass- and Obesity-Associated Gene-Dependent Obesogenic Programming by Formula Feeding Compared to Breastfeeding
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Breastfeeding in Comparison to Infant Formula Feeding Modifies DNA Methylation
3.2. Variations in DNA CpG Methylation Modify FTO and CUX1 Gene Expression
3.3. The Epigenetic Impact of RNA N6-Methyladenosine Modifications on Adipogenesis
3.4. Formula Feeding Compared to Breastfeeding Increases FTO Expression
3.5. High Protein Formula Intake and the Risk of FTO and mTORC1/S6K1 Overactivation
3.6. The Potential Adipogenic Impact of Exosomal Milk miRNA Deficiency in Infant Formula
3.7. Hypothalamic FTO Expression Enhances Energy Intake
3.8. FTO Expression, Circadian Rhythm, and Obesity
3.9. Potential Impact of Exosomal Milk miRNAs in FTO-Circadian Clock Regulation
3.10. Zinc Finger Protein 217 Promotes FTO Expression and Adipogenesis
3.11. FTO Deficiency and Long Non-Coding RNA H19 Promote Brown Adipose Tissue
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AA | amino acid |
| AARS | amino-acyl tRNA synthetases |
| ADIPOQ | adiponectin |
| ATF4 | activating transcription factor 4 |
| BAT | brown adipose tissue |
| BF | breastfeeding |
| BCAA | branched-chain amino acids |
| BMAL1 | brain and muscle ARNT-like protein 1 |
| BMI | body mass index |
| CCNA2 | cyclin A2 |
| CDK2 | cyclin-dependent kinase 2 |
| CEBPA | CCAAT/enhancer-binding protein α |
| CEBPB | CCAAT/enhancer-binding protein β |
| CEBPD | CCAAT/enhancer-binding protein δ |
| CLOCK | circadian locomotor output cycles kaput |
| CRY1 | cryptochrome 1 |
| CRY2 | cryptochrome 2 |
| CTCF | CCCTC-binding factor |
| CUX1 | cut-like homeobox 1 |
| DOHaD | developmental origins of health and disease |
| DNMT1 | DNA methyltransferase 1 |
| EZH2 | enhancer of zeste homolog 2 |
| FF | formula feeding |
| FOXO1A | forkhead box O1A |
| FTO | fat mass and obesity-associated gene |
| GHRL | ghrelin |
| G6PD | glucose-6-phosphate dehydrogenase |
| H19 | long non-coding RNA H19 |
| HFD | high-fat diet |
| HIF1α | hypoxia-inducible factor 1α |
| HIF1AN | hypoxia-inducible factor 1-α inhibitor |
| KLF2 | Krüppel-like factor 2 |
| KYN | kynurenine |
| LARS | leucyl-tRNA synthase |
| LEU | leucine |
| m6A | N6-methyladenosine |
| METTL3 | methyltransferase 3, N6-adenosine-methyltransferase complex catalytic subunit 3 |
| METTL14 | methyltransferase 14, N6-adenosine-methyltransferase complex catalytic subunit 14 |
| MCE | mitotic clonal expansion |
| miRNA | micro ribonucleic acid |
| MSC | mesenchymal stem cell |
| mTORC1 | mechanistic target of rapamycin complex 1 |
| MYC | MYC protooncogene |
| NAD | nicotinamide adenosine dinucleotide |
| NADP | nicotinamide adenosine dinucleotide phosphate |
| NAMPT | nicotinamide phosphoribosyltransferase |
| NRIP1 | nuclear receptor-interacting protein 1 |
| PPAR-α | peroxisome proliferator-activated receptor-α |
| PPAR-γ | peroxisome proliferator-activated receptor-γ |
| PPARGC1A | peroxisome proliferator-activated receptor-γ co-activator-1α |
| RAGD | Ras-related GTP-binding protein D |
| RPGRIP1L | retinitis pigmentosa GTPase regulator interacting protein 1-like |
| RUNX1T1 | runt-related transcription factor 1, translocated, 1 |
| S6K1 | S6 kinase 1 |
| SNP | single nucleotide polymorphism |
| SREBF1 | sterol regulatory element-binding transcription factor 1 |
| TSC1 | tuberous sclerosis complex 1 |
| TRP | tryptophan |
| UCP1 | uncoupling protein 1 |
| UTR | untranslated region |
| WAT | white adipose tissue |
| WNT10a | wingless-type MMTV integration site family, member 10A |
| YTHDF2 | YTH N6-methyladenosine RNA binding protein 2 |
| ZNF217 | zinc finger protein gene 217 |
| ZFP217 | zinc finger protein 217 |
References
- Rajamoorthi, A.; LeDuc, C.A.; Thaker, V.V. The metabolic conditioning of obesity: A review of the pathogenesis of obesity and the epigenetic pathways that “program” obesity from conception. Front. Endocrinol. 2022, 13, 1032491. [Google Scholar] [CrossRef]
- Şahin, Ö.N. The part breast milk plays in epigenetic programming. In Breastfeeding and Metabolic Programming; Şahin, Ö.N., Briana, D.D., Di Renzo, G.C., Eds.; Springer: Cham, Switzerland, 2023. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s epigenetic regulators. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442. [Google Scholar] [CrossRef]
- Melnik, B.C.; Stremmel, W.; Weiskirchen, R.; John, S.M.; Schmitz, G. Exosome-derived microRNAs of human milk and their effects on infant health and development. Biomolecules 2021, 11, 851. [Google Scholar] [CrossRef]
- Röszer, T. Co-evolution of breast milk lipid signaling and thermogenic adipose tissue. Biomolecules 2021, 11, 1705. [Google Scholar] [CrossRef]
- Röszer, T. Mother-to-child signaling through breast milk biomolecules. Biomolecules 2021, 11, 1743. [Google Scholar] [CrossRef]
- Ozkan, H.; Tuzun, F.; Taheri, S.; Korhan, P.; Akokay, P.; Yılmaz, O.; Duman, N.; Özer, E.; Tufan, E.; Kumral, A.; et al. Epigenetic programming through breast milk and its impact on milk-siblings mating. Front. Genet. 2020, 11, 569232. [Google Scholar] [CrossRef]
- Yan, J.; Liu, L.; Zhu, Y.; Huang, G.; Wang, P.P. The association between breastfeeding and childhood obesity: A meta-analysis. BMC Public Health 2014, 14, 1267. [Google Scholar] [CrossRef]
- Horta, B.L.; Loret de Mola, C.; Victora, C.G. Long-term consequences of breastfeeding on cholesterol, obesity, systolic blood pressure and type 2 diabetes: A systematic review and meta-analysis. Acta Paediatr. 2015, 104, 30–37. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Y. Nutrition and metabolism in the first 1000 days of life. Nutrients 2023, 15, 2554. [Google Scholar] [CrossRef]
- Mannan, H. Early infant feeding of formula or solid foods and risk of childhood overweight or obesity in a socioeconomically disadvantaged region of Australia: A longitudinal cohort analysis. Int. J. Environ. Res. Public Health 2018, 15, 1685. [Google Scholar] [CrossRef]
- Rito, A.I.; Buoncristiano, M.; Spinelli, A.; Salanave, B.; Kunešová, M.; Hejgaard, T.; García Solano, M.; Fijałkowska, A.; Sturua, L.; Hyska, J.; et al. Association between characteristics at birth, breastfeeding and obesity in 22 countries: The WHO European Childhood Obesity Surveillance Initiative-COSI 2015/2017. Obes. Facts 2019, 12, 226–243. [Google Scholar] [CrossRef] [PubMed]
- Usheva, N.; Lateva, M.; Galcheva, S.; Koletzko, B.V.; Cardon, G.; De Craemer, M.; Androutsos, O.; Kotowska, A.; Socha, P.; Moreno, L.A.; et al. Breastfeeding and overweight in European preschoolers: The ToyBox Study. Nutrients 2021, 13, 2880. [Google Scholar] [CrossRef]
- Liu, F.; Lv, D.; Wang, L.; Feng, X.; Zhang, R.; Liu, W.; Han, W. Breastfeeding and overweight/obesity among children and adolescents: A cross-sectional study. BMC Pediatr. 2022, 22, 347. [Google Scholar] [CrossRef]
- Mantzorou, M.; Papandreou, D.; Vasios, G.K.; Pavlidou, E.; Antasouras, G.; Psara, E.; Taha, Z.; Poulios, E.; Giaginis, C. Exclusive breastfeeding for at least four months is associated with a lower prevalence of overweight and obesity in mothers and their children after 2–5 years from delivery. Nutrients 2022, 14, 3599. [Google Scholar] [CrossRef]
- Ma, J.; Qiao, Y.; Zhao, P.; Li, W.; Katzmarzyk, P.T.; Chaput, J.P.; Fogelholm, M.; Kuriyan, R.; Lambert, E.V.; Maher, C.; et al. Breastfeeding and childhood obesity: A 12-country study. Matern. Child Nutr. 2020, 16, e12984. [Google Scholar] [CrossRef]
- Qiao, J.; Dai, L.J.; Zhang, Q.; Ouyang, Y.Q. A meta-analysis of the association between breastfeeding and early childhood obesity. J. Pediatr. Nurs. 2020, 53, 57–66. [Google Scholar] [CrossRef]
- Wagner, K.J.P.; Rossi, C.E.; Hinnig, P.F.; Alves, M.A.; Retondario, A.; Vasconcelos, F.A.G. Association between breastfeeding and overweight/obesity in schoolchildren aged 7–14 years. Rev. Paul. Pediatr. 2021, 39, e2020076. [Google Scholar] [CrossRef]
- Cieśla, E.; Stochmal, E.; Głuszek, S.; Suliga, E. Breastfeeding history and the risk of overweight and obesity in middle-aged women. BMC Womens Health 2021, 21, 196, Erratum in BMC Womens Health 2021, 21, 245. [Google Scholar]
- Yuan, M.; Lu, M.; Guo, Y.; Lam, K.B.H.; Lu, J.; He, J.; Shen, S.; Wei, D.; Thomas, G.N.; Cheng, K.K.; et al. Timing of infant formula introduction in relation to BMI and overweight at ages 1 and 3 years: The Born in Guangzhou Cohort Study (BIGCS). Br. J. Nutr. 2023, 129, 166–174. [Google Scholar] [CrossRef]
- World Health Organization. e-Library of Evidence for Nutrition Actions (eLENA). Exclusive Breastfeeding to Reduce the Risk of Childhood Overweight and Obesity. Available online: https://www.who.int/tools/elena/interventions/breastfeeding-childhood-obesity (accessed on 5 July 2024).
- Owen, C.G.; Martin, R.M.; Whincup, P.H.; Smith, G.D.; Cook, D.G. Does breastfeeding influence risk of type 2 diabetes in later life? A quantitative analysis of published evidence. Am. J. Clin. Nutr. 2006, 84, 1043–1054, Erratum in Am. J. Clin. Nutr. 2012, 95, 779. [Google Scholar] [CrossRef]
- Horta, B.L.; de Lima, N.P. Breastfeeding and type 2 diabetes: Systematic review and meta-Analysis. Curr. Diab. Rep. 2019, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Victora, C.G.; Bahl, R.; Barros, A.J.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Lampousi, A.M.; Carlsson, S.; Löfvenborg, J.E. Dietary factors and risk of islet autoimmunity and type 1 diabetes: A systematic review and meta-analysis. EBioMedicine 2021, 72, 103633. [Google Scholar] [CrossRef] [PubMed]
- Kadayifci, F.Z.; Zheng, S.; Pan, Y.X. Molecular mechanisms underlying the link between diet and DNA methylation. Int. J. Mol. Sci. 2018, 19, 4055. [Google Scholar] [CrossRef]
- Reizel, Y.; Sabag, O.; Skversky, Y.; Spiro, A.; Steinberg, B.; Bernstein, D.; Wang, A.; Kieckhaefer, J.; Li, C.; Pikarsky, E.; et al. Postnatal DNA demethylation and its role in tissue maturation. Nat. Commun. 2018, 9, 2040. [Google Scholar] [CrossRef]
- Plagemann, A.; Harder, T.; Schellong, K.; Schulz, S.; Stupin, J.H. Early postnatal life as a critical time window for determination of long-term metabolic health. Best. Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 641–653. [Google Scholar] [CrossRef]
- Verduci, E.; Banderali, G.; Barberi, S.; Radaelli, G.; Lops, A.; Betti, F.; Riva, E.; Giovannini, M. Epigenetic effects of human breast milk. Nutrients 2014, 6, 1711–1724. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Xiao, X.; Zhang, Q.; Yu, M. DNA methylation: The pivotal interaction between early-life nutrition and glucose metabolism in later life. Br. J. Nutr. 2014, 112, 1850–1857. [Google Scholar] [CrossRef]
- Arechederra, M.; Daian, F.; Yim, A.; Bazai, S.K.; Richelme, S.; Dono, R.; Saurin, A.J.; Habermann, B.H.; Maina, F. Hypermethylation of gene body CpG islands predicts high dosage of functional oncogenes in liver cancer. Nat. Commun. 2018, 9, 3164, Erratum in Nat. Commun. 2018, 9, 3976. [Google Scholar] [CrossRef]
- Russo, G.; Tramontano, A.; Iodice, I.; Chiariotti, L.; Pezone, A. Epigenome chaos: Stochastic and deterministic DNA methylation events drive cancer evolution. Cancers 2021, 13, 1800. [Google Scholar] [CrossRef]
- Obermann-Borst, S.A.; Eilers, P.H.; Tobi, E.W.; de Jong, F.H.; Slagboom, P.E.; Heijmans, B.T.; Steegers-Theunissen, R.P. Duration of breastfeeding and gender are associated with methylation of the LEPTIN gene in very young children. Pediatr. Res. 2013, 74, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, W.B.; Bion, V.; Lockett, G.A.; Ziyab, A.H.; Soto-Ramírez, N.; Mukherjee, N.; Kurukulaaratchy, R.J.; Ewart, S.; Zhang, H.; Arshad, S.H.; et al. Duration of breastfeeding is associated with leptin (LEP) DNA methylation profiles and BMI in 10-year-old children. Clin. Epigenetics 2019, 11, 128. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, F.P.; Loret de Mola, C.; Davies, N.M.; Victora, C.G.; Relton, C.L. Breastfeeding effects on DNA methylation in the offspring: A systematic literature review. PLoS ONE 2017, 12, e0173070, Erratum in PLoS ONE 2017, 12, e0175604. [Google Scholar]
- Pauwels, S.; Symons, L.; Vanautgaerden, E.L.; Ghosh, M.; Duca, R.C.; Bekaert, B.; Freson, K.; Huybrechts, I.; Langie, S.A.S.; Koppen, G.; et al. The influence of the duration of breastfeeding on the infant’s metabolic epigenome. Nutrients 2019, 11, 1408. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, W.B.; Kothalawala, D.M.; Kadalayil, L.; Ewart, S.; Zhang, H.; Karmaus, W.; Arshad, S.H.; Holloway, J.W.; Rezwan, F.I. Epigenome-wide association study reveals duration of breastfeeding is associated with epigenetic differences in children. Int. J. Environ. Res. Public Health 2020, 17, 3569. [Google Scholar] [CrossRef] [PubMed]
- Briollais, L.; Rustand, D.; Allard, C.; Wu, Y.; Xu, J.; Rajan, S.G.; Hivert, M.F.; Doyon, M.; Bouchard, L.; McGowan, P.O.; et al. DNA methylation mediates the association between breastfeeding and early-life growth trajectories. Clin. Epigenetics 2021, 13, 231. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Schmitz, G. Milk’s role as an epigenetic regulator in health and disease. Diseases 2017, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Trinchese, G.; Feola, A.; Cavaliere, G.; Cimmino, F.; Catapano, A.; Penna, E.; Scala, G.; Greco, L.; Bernardo Porcellini, A.; Crispino, M.; et al. Mitochondrial metabolism and neuroinflammation in the cerebral cortex and cortical synapses of rats: Effect of milk intake through DNA methylation. J. Nutr. Biochem. 2024, 128, 109624. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. Milk: An epigenetic amplifier of FTO-mediated transcription? Implications for Western diseases. J. Transl. Med. 2015, 13, 385. [Google Scholar] [CrossRef]
- Kucher, A.N. The FTO gene and diseases: The role of genetic polymorphism, epigenetic modifications, and environmental factors. Russ. J. Genet. 2020, 56, 1025–1043. [Google Scholar] [CrossRef]
- Popović, A.M.; Huđek Turković, A.; Žuna, K.; Bačun-Družina, V.; Rubelj, I.; Matovinović, M. FTO gene polymorphisms at the crossroads of metabolic pathways of obesity and epigenetic influences. Food Technol. Biotechnol. 2023, 61, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Czogała, W.; Strojny, W.; Schab, M.; Grabowska, A.; Miklusiak, K.; Kowalczyk, W.; Łazarczyk, A.; Tomasik, P.; Skoczeń, S. FTO and PLAG1 genes expression and FTO methylation predict changes in circulating levels of adipokines and gastrointestinal peptides in children. Nutrients 2021, 13, 3585. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.G.; Finer, S.; Lindgren, C.M.; Wilson, G.A.; Rakyan, V.K.; Teschendorff, A.E.; Akan, P.; Stupka, E.; Down, T.A.; Prokopenko, I.; et al. Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS ONE 2010, 5, e14040. [Google Scholar] [CrossRef] [PubMed]
- Czogała, W.; Czogała, M.; Strojny, W.; Wątor, G.; Wołkow, P.; Wójcik, M.; Bik Multanowski, M.; Tomasik, P.; Wędrychowicz, A.; Kowalczyk, W.; et al. Methylation and expression of FTO and PLAG1 genes in childhood obesity: Insight into anthropometric parameters and glucose-lipid metabolism. Nutrients 2021, 13, 1683. [Google Scholar] [CrossRef] [PubMed]
- Yeo, G.S.; O’Rahilly, S. Uncovering the biology of FTO. Mol. Metab. 2012, 1, 32–36. [Google Scholar] [CrossRef]
- Fawcett, K.A.; Barroso, I. The genetics of obesity: FTO leads the way. Trends Genet. 2010, 26, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.L.; Yeo, G.S.H.; O’Rahilly, S.; Coll, A.P. Obesity and FTO: Changing focus at a complex locus. Cell Metab. 2014, 20, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yu, G.L.; Zhu, X.; Peng, T.H.; Lv, Y.C. Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes Dis. 2021, 9, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Velloso, L.A.; Schwartz, M.W. Altered hypothalamic function in diet-induced obesity. Int. J. Obes. 2011, 35, 1455–1465. [Google Scholar] [CrossRef]
- Stratigopoulos, G.; Padilla, S.L.; LeDuc, C.A.; Watson, E.; Hattersley, A.T.; McCarthy, M.I.; Zeltser, L.M.; Chung, W.K.; Leibel, R.L. Regulation of Fto/Ftm gene expression in mice and humans. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R1185–R1196, Erratum in Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R1360–R1363. [Google Scholar] [CrossRef]
- Stratigopoulos, G.; LeDuc, C.A.; Cremona, M.L.; Chung, W.K.; Leibel, R.L. Cut-like homeobox 1 (CUX1) regulates expression of the fat mass and obesity-associated and retinitis pigmentosa GTPase regulator-interacting protein-1-like (RPGRIP1L) genes and coordinates leptin receptor signaling. J. Biol. Chem. 2011, 286, 2155–2170. [Google Scholar] [CrossRef]
- Stratigopoulos, G.; Burnett, L.C.; Rausch, R.; Gill, R.; Penn, D.B.; Skowronski, A.A.; LeDuc, C.A.; Lanzano, A.J.; Zhang, P.; Storm, D.R.; et al. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J. Clin. Investig. 2016, 126, 1897–1910. [Google Scholar] [CrossRef]
- Tung, Y.C.; Gulati, P.; Liu, C.H.; Rimmington, D.; Dennis, R.; Ma, M.; Saudek, V.; O’Rahilly, S.; Coll, A.P.; Yeo, G.S. FTO is necessary for the induction of leptin resistance by high-fat feeding. Mol. Metab. 2015, 4, 287–298. [Google Scholar] [CrossRef]
- Zhu, Z.M.; Huo, F.C.; Pei, D.S. Function and evolution of RNA N6-methyladenosine modification. Int. J. Biol. Sci. 2020, 16, 1929–1940. [Google Scholar] [CrossRef]
- Karthiya, R.; Khandelia, P. m6A RNA methylation: Ramifications for gene expression and human health. Mol. Biotechnol. 2020, 62, 467–484. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Sendinc, E.; Shi, Y. RNA m6A methylation across the transcriptome. Mol. Cell 2023, 83, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Ben-Haim, M.S.; Moshitch-Moshkovitz, S.; Rechavi, G. FTO: Linking m6A demethylation to adipogenesis. Cell Res. 2015, 25, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Azzam, S.K.; Alsafar, H.; Sajini, A.A. FTO m6A demethylase in obesity and cancer: Implications and underlying molecular mechanisms. Int. J. Mol. Sci. 2022, 23, 3800. [Google Scholar] [CrossRef]
- Sun, M.; Zhang, X. Epigenetic regulation of N6-methyladenosine modifications in obesity. J. Diabetes Investig. 2021, 12, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wang, X. Epigenetic regulation of adipose tissue expansion and adipogenesis by N6-methyladenosine. Obes. Rev. 2021, 22, e13124. [Google Scholar] [CrossRef] [PubMed]
- Cheshmeh, S.; Nachvak, S.M.; Rezvani, N.; Saber, A. Effects of breastfeeding and formula feeding on the expression level of FTO, CPT1A and PPAR-α genes in healthy infants. Diabetes Metab. Syndr. Obes. 2020, 13, 2227–2237. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, A.; Liu, X.; Yang, Y.; Zhao, R.; Jia, Y. m6A-Mediated PPARA translational suppression contributes to corticosterone-induced visceral fat deposition in chickens. Int. J. Mol. Sci. 2022, 23, 15761. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, J.; Tang, M.; Wang, X.; Fan, N.; Peng, Y. Fat mass and obesity-associated protein promotes liver steatosis by targeting PPARα. Lipids Health Dis. 2022, 21, 29. [Google Scholar] [CrossRef] [PubMed]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular actions of PPARα in lipid metabolism and inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Sun, B.F.; Zhao, Y.L.; Yang, Y.G. FTO and obesity: Mechanisms of association. Curr. Diab. Rep. 2014, 14, 486. [Google Scholar] [CrossRef]
- Deng, X.; Su, R.; Stanford, S.; Chen, J. Critical enzymatic functions of FTO in obesity and cancer. Front. Endocrinol. 2018, 9, 396. [Google Scholar] [CrossRef]
- Han, Z.; Niu, T.; Chang, J.; Lei, X.; Zhao, M.; Wang, Q.; Cheng, W.; Wang, J.; Feng, Y.; Chai, J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 2010, 464, 1205–1209. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887, Erratum in Nat. Chem. Biol. 2012, 8, 1008. [Google Scholar] [CrossRef]
- Karra, E.; O’Daly, O.G.; Choudhury, A.I.; Yousseif, A.; Millership, S.; Neary, M.T.; Scott, W.R.; Chandarana, K.; Manning, S.; Hess, M.E.; et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J. Clin. Investig. 2013, 123, 3539–3551. [Google Scholar] [CrossRef] [PubMed]
- Merkestein, M.; McTaggart, J.S.; Lee, S.; Kramer, H.B.; McMurray, F.; Lafond, M.; Boutens, L.; Cox, R.; Ashcroft, F.M. Changes in gene expression associated with FTO overexpression in mice. PLoS ONE 2014, 9, e97162. [Google Scholar] [CrossRef] [PubMed]
- Berulava, T.; Horsthemke, B. The obesity-associated SNPs in intron 1 of the FTO gene affect primary transcript levels. Eur. J. Hum. Genet. 2010, 18, 1054–1056. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Lye, S.; Briollais, L. The role of early life growth development, the FTO gene and exclusive breastfeeding on child BMI trajectories. Int. J. Epidemiol. 2017, 46, 1512–1522. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; D’Souza, N.J.; Atkins, L.; Ghobadi, S.; Laws, R.; Szymlek-Gay, E.A.; Grimes, C.; Baker, P.; He, Q.Q.; Campbell, K.J. Breastfeeding and the longitudinal changes of body mass index in childhood and adulthood: A systematic review. Adv. Nutr. 2024, 15, 100152. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef]
- Erson-Bensan, A.E.; Begik, O. m6A Modification and Implications for microRNAs. MicroRNA 2017, 6, 97–101. [Google Scholar] [CrossRef]
- Bartosovic, M.; Molares, H.C.; Gregorova, P.; Hrossova, D.; Kudla, G.; Vanacova, S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017, 45, 11356–11370. [Google Scholar] [CrossRef]
- Huang, J.; Yin, P. Structural insights into N6-methyladenosine (m6A) modification in the transcriptome. Genom. Proteom. Bioinform. 2018, 16, 85–98. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624, Erratum in Nat. Rev. Mol. Cell Biol. 2023, 24, 770. [Google Scholar] [CrossRef]
- He, P.C.; He, C. m6 A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.; Nsengimana, B.; Awan, U.A.; Ji, X.-Y.; Ji, S.; Dong, J. Regulatory roles of N6-methyladenosine (m6A) methylation in RNA processing and non-communicable diseases. Cancer Gene Ther. 2024; epub ahead of print. [Google Scholar] [CrossRef]
- Merkestein, M.; Laber, S.; McMurray, F.; Andrew, D.; Sachse, G.; Sanderson, J.; Li, M.; Usher, S.; Sellayah, D.; Ashcroft, F.M.; et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat. Commun. 2015, 6, 6792. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Y.; Ma, J.; Guo, F.; Cao, Q.; Zhang, Y.; Zhou, B.; Chai, J.; Zhao, W.; Zhao, R. The demethylase activity of FTO (fat mass and obesity associated protein) is required for preadipocyte differentiation. PLoS ONE 2015, 10, e0133788. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Huang, W.; Wang, Z.; Nie, Q. m6A demethylase FTO regulate CTNNB1 to promote adipogenesis of chicken preadipocyte. J. Anim. Sci. Biotechnol. 2022, 13, 147. [Google Scholar] [CrossRef] [PubMed]
- Martin Carli, J.F.; LeDuc, C.A.; Zhang, Y.; Stratigopoulos, G.; Leibel, R.L. FTO mediates cell-autonomous effects on adipogenesis and adipocyte lipid content by regulating gene expression via 6mA DNA modifications. J. Lipid Res. 2018, 59, 1446–1460. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Ai, Y.; Zhu, T.; Li, J.; Ren, Z. FTO promoted adipocyte differentiation by regulating ADRB1 gene through m6A modification in Hycole rabbits. Anim. Biotechnol. 2023, 34, 2565–2570. [Google Scholar] [CrossRef] [PubMed]
- Claussnitzer, M.; Dankel, S.N.; Kim, K.H.; Quon, G.; Meuleman, W.; Haugen, C.; Glunk, V.; Sousa, I.S.; Beaudry, J.L.; Puviindran, V.; et al. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 2015, 373, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Shi, H.; Wang, F.; Wang, Y. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 2020, 16, 1221–1235. [Google Scholar] [CrossRef]
- Song, T.; Yang, Y.; Wei, H.; Xie, X.; Lu, J.; Zeng, Q.; Peng, J.; Zhou, Y.; Jiang, S.; Peng, J. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation. Nucleic Acids Res. 2019, 47, 6130–6144. [Google Scholar] [CrossRef]
- Wang, L.; Song, C.; Wang, N.; Li, S.; Liu, Q.; Sun, Z.; Wang, K.; Yu, S.C.; Yang, Q. NADP modulates RNA m6A methylation and adipogenesis via enhancing FTO activity. Nat. Chem. Biol. 2020, 16, 1394–1402, Erratum in Nat. Chem. Biol. 2023, 19, 1286. [Google Scholar] [CrossRef]
- Luo, G.; Hong, T.; Yu, L.; Ren, Z. FTO regulated intramuscular fat by targeting APMAP gene via an m6A-YTHDF2-dependent manner in Rex rabbits. Cells 2023, 12, 369. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Chen, J.; Li, Z.; Li, L.; Chen, J.; Guo, Y. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J. Exp. Clin. Cancer Res. 2020, 39, 240. [Google Scholar] [CrossRef] [PubMed]
- Deisenroth, C.; Black, M.B.; Pendse, S.; Pluta, L.; Witherspoon, S.M.; McMullen, P.D.; Thomas, R.S. MYC is an early response regulator of human adipogenesis in adipose stem cells. PLoS ONE 2014, 9, e114133. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, L.; Chen, J.; Wang, Y. mRNA m⁶A methylation downregulates adipogenesis in porcine adipocytes. Biochem. Biophys. Res. Commun. 2015, 459, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Liu, Y.; Yao, Y.; Zhao, Y.; Bi, Z.; Jiang, Q.; Liu, Q.; Cai, M.; Wang, F.; Wang, Y.; et al. FTO regulates adipogenesis by controlling cell cycle progression via m6A-YTHDF2 dependent mechanism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Yao, Y.; Jiang, Q.; Cai, M.; Liu, Q.; Wang, Y.; Wang, X. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m6A-YTHDF2-dependent manner. Int. J. Obes. 2018, 42, 1378–1388. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Chen, Y.; Liu, Y.; Zhuang, L.; Chen, W.; Zeng, B.; Liao, X.; Guo, G.; Wang, Y.; Wang, X. m6A methylation promotes white-to-beige fat transition by facilitating Hif1a translation. EMBO Rep. 2021, 22, e52348. [Google Scholar] [CrossRef] [PubMed]
- Ronkainen, J.; Mondini, E.; Cinti, F.; Cinti, S.; Sebért, S.; Savolainen, M.J.; Salonurmi, T. Fto-deficiency affects the gene and microRNA expression involved in brown adipogenesis and browning of white adipose tissue in mice. Int. J. Mol. Sci. 2016, 17, 1851. [Google Scholar] [CrossRef] [PubMed]
- Tews, D.; Fischer-Posovszky, P.; Fromme, T.; Klingenspor, M.; Fischer, J.; Rüther, U.; Marienfeld, R.; Barth, T.F.; Möller, P.; Debatin, K.M.; et al. FTO deficiency induces UCP-1 expression and mitochondrial uncoupling in adipocytes. Endocrinology 2013, 154, 3141–3151. [Google Scholar] [CrossRef]
- Liao, X.; Liu, J.; Chen, Y.; Liu, Y.; Chen, W.; Zeng, B.; Liu, Y.; Luo, Y.; Huang, C.; Guo, G.; et al. Metformin combats obesity by targeting FTO in an m6A-YTHDF2-dependent manner. J. Drug Target. 2022, 30, 983–991. [Google Scholar] [CrossRef]
- Chen, A.; Chen, X.; Cheng, S.; Shu, L.; Yan, M.; Yao, L.; Wang, B.; Huang, S.; Zhou, L.; Yang, Z.; et al. FTO promotes SREBP1c maturation and enhances CIDEC transcription during lipid accumulation in HepG2 cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Zhang, Z.; Yu, L.; Li, Y.; Liang, M.; Zhou, L. FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation. J. Cell. Biochem. 2018, 119, 5676–5685. [Google Scholar] [CrossRef]
- Tang, Z.; Sun, C.; Yan, Y.; Niu, Z.; Li, Y.; Xu, X.; Zhang, J.; Wu, Y.; Li, Y.; Wang, L.; et al. Aberrant elevation of FTO levels promotes liver steatosis by decreasing the m6A methylation and increasing the stability of SREBF1 and ChREBP mRNAs. J. Mol. Cell Biol. 2023, 14, mjac061. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.M. Fat mass and obesity associated (FTO) gene and hepatic glucose and lipid metabolism. Nutrients 2018, 10, 1600. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Feng, Y.; Zhang, L.; Jia, Y.; Cai, D.; Qian, S.B.; Du, M.; Zhao, R. GR-mediated FTO transactivation induces lipid accumulation in hepatocytes via demethylation of m6A on lipogenic mRNAs. RNA Biol. 2020, 17, 930–942. [Google Scholar] [CrossRef]
- Sun, D.; Zhao, T.; Zhang, Q.; Wu, M.; Zhang, Z. Fat mass and obesity-associated protein regulates lipogenesis via m6 A modification in fatty acid synthase mRNA. Cell Biol. Int. 2021, 45, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Xiao, W.; Ju, D.; Sun, B.; Hou, N.; Liu, Q.; Wang, Y.; Zhao, H.; Gao, C.; Zhang, S.; et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 2019, 11, eaau7116. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Feng, J.; Jiang, D.; Zhou, X.; Jiang, Q.; Cai, M.; Wang, X.; Shan, T.; Wang, Y. AMPK regulates lipid accumulation in skeletal muscle cells through FTO-dependent demethylation of N6-methyladenosine. Sci. Rep. 2017, 7, 41606. [Google Scholar] [CrossRef]
- Mo, W.; Chen, Z.; Zhang, X.; Dai, G.; Ma, D.; Pan, J.; Zhang, X.; Wu, G.; Fan, W. N6-methyladenosine demethylase FTO (fat mass and obesity-associated protein) as a novel mediator of statin effects in human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 644–658. [Google Scholar] [CrossRef]
- Banerjee, S.S.; Feinberg, M.W.; Watanabe, M.; Gray, S.; Haspel, R.L.; Denkinger, D.J.; Kawahara, R.; Hauner, H.; Jain, M.K. The Krüppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J. Biol. Chem. 2003, 278, 2581–2584. [Google Scholar] [CrossRef]
- Siddiq, A.; Khan, R.A.; Baig, S.G. Does atorvastatin work more effectively than biguanides in reducing cardiovascular risk factors? J. Pharm. Bioallied Sci. 2011, 3, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Yu, X.; Yang, L.; Liu, X.; Gao, B.; Huang, B.; Dou, X.; Liu, J.; Zou, Z.; Cui, X.L.; et al. FTO mediates LINE1 m6A demethylation and chromatin regulation in mESCs and mouse development. Science 2022, 376, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fu, Q.; Yang, J.; Liu, J.L.; Hou, S.M.; Huang, X.; Cao, J.S.; Liu, T.L.; Wang, K.Z. RNA N6-methyladenosine demethylase FTO promotes osteoporosis through demethylating Runx2 mRNA and inhibiting osteogenic differentiation. Aging 2021, 13, 21134–21141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Li, X.; Qian, S.W.; Guo, L.; Huang, H.Y.; He, Q.; Liu, Y.; Ma, C.G.; Tang, Q.Q. Down-regulation of type I Runx2 mediated by dexamethasone is required for 3T3-L1 adipogenesis. Mol. Endocrinol. 2012, 26, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Cawthorn, W.P.; Li, Y.; Zhao, G.; Macdougald, O.A.; Franceschi, R.T. Reciprocal control of osteogenic and adipogenic differentiation by ERK/MAP kinase phosphorylation of Runx2 and PPARγ transcription factors. J. Cell. Physiol. 2016, 231, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Rahim, F.; Abbasi Pashaki, P.; Jafarisani, M.; Ghorbani, F.; Ebrahimi, A. Runx2 silencing promotes adipogenesis via down-regulation of DLK1 in chondrogenic differentiating MSCs. J. Gene Med. 2020, 22, e3244. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Hata, A. The role of microRNAs in cell fate determination of mesenchymal stem cells: Balancing adipogenesis and osteogenesis. BMB Rep. 2015, 48, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, M.; Wei, F.Y.; Chujo, T.; Oki, S.; Yakita, M.; Kobayashi, D.; Araki, N.; Takahashi, N.; Yoshida, R.; Nakayama, H.; et al. FTO demethylates cyclin D1 mRNA and controls cell-cycle progression. Cell Rep. 2020, 31, 107464. [Google Scholar] [CrossRef] [PubMed]
- Zang, S.; Yin, X.; Li, P. FTO-mediated m6A demethylation regulates GnRH expression in the hypothalamus via the PLCβ3/Ca2+/CAMK signalling pathway. Commun. Biol. 2023, 6, 1297. [Google Scholar] [CrossRef]
- Totzauer, M.; Escribano, J.; Closa-Monasterolo, R.; Luque, V.; Verduci, E.; ReDionigi, A.; Langhendries, J.P.; Martin, F.; Xhonneux, A.; Gruszfeld, D.; et al. Different protein intake in the first year and its effects on adiposity rebound and obesity throughout childhood: 11 years follow-up of a randomized controlled trial. Pediatr. Obes. 2022, 17, e12961. [Google Scholar] [CrossRef] [PubMed]
- Koletzko, B.; von Kries, R.; Closa, R.; Escribano, J.; Scaglioni, S.; Giovannini, M.; Beyer, J.; Demmelmair, H.; Anton, B.; Gruszfeld, D.; et al. Can infant feeding choices modulate later obesity risk? Am. J. Clin. Nutr. 2009, 89, 1502S–1508S, Erratum in Am. J. Clin. Nutr. 2009, 90, 248. [Google Scholar] [CrossRef] [PubMed]
- Koletzko, B.; Demmelmair, H.; Grote, V.; Totzauer, M. Optimized protein intakes in term infants support physiological growth and promote long-term health. Semin. Perinatol. 2019, 43, 151153. [Google Scholar] [CrossRef] [PubMed]
- Koletzko, B. 1.4.2 Early nutrition impact on long-term health. World Rev. Nutr. Diet. 2022, 124, 87–93. [Google Scholar] [PubMed]
- Inostroza, J.; Haschke, F.; Steenhout, P.; Grathwohl, D.; Nelson, S.E.; Ziegler, E.E. Low-protein formula slows weight gain in infants of overweight mothers. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Haschke, F.; Grathwohl, D.; Detzel, P.; Steenhout, P.; Wagemans, N.; Erdmann, P. Postnatal high protein intake can contribute to accelerated weight gain of infants and increased obesity risk. Nestle Nutr. Inst. Workshop Ser. 2016, 8, 101–109. [Google Scholar]
- Doaei, S.; Kalantari, N.; Izadi, P.; Salonurmi, T.; Jarrahi, A.M.; Rafieifar, S.; Azizi Tabesh, G.; Rahimzadeh, G.; Gholamalizadeh, M.; Goodarzi, M.O. Interactions between macro-nutrients’ intake, FTO and IRX3 gene expression, and FTO genotype in obese and overweight male adolescents. Adipocyte 2019, 8, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.K.; Gulati, P.; O’Rahilly, S.; Yeo, G.S. FTO expression is regulated by availability of essential amino acids. Int. J. Obes. 2013, 37, 744–747. [Google Scholar] [CrossRef]
- Gulati, P.; Yeo, G.S. The biology of FTO: From nucleic acid demethylase to amino acid sensor. Diabetologia 2013, 56, 2113–2121. [Google Scholar] [CrossRef]
- Gulati, P.; Cheung, M.K.; Antrobus, R.; Church, C.D.; Harding, H.P.; Tung, Y.C.; Rimmington, D.; Ma, M.; Ron, D.; Lehner, P.J.; et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc. Natl. Acad. Sci. USA 2013, 110, 2557–2562. [Google Scholar] [CrossRef]
- Gulati, P.; Avezov, E.; Ma, M.; Antrobus, R.; Lehner, P.; O’Rahilly, S.; Yeo, G.S. Fat mass and obesity-related (FTO) shuttles between the nucleus and cytoplasm. Biosci. Rep. 2014, 34, e00144. [Google Scholar] [CrossRef] [PubMed]
- Han, J.M.; Jeong, S.J.; Park, M.C.; Kim, G.; Kwon, N.H.; Kim, H.K.; Ha, S.H.; Ryu, S.H.; Kim, S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012, 149, 410–424. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Kim, J.H.; Yoon, I.; Lee, C.; Fallahi Sichani, M.; Kang, J.S.; Kang, J.; Guo, M.; Lee, K.Y.; Han, G.; et al. Coordination of the leucine-sensing Rag GTPase cycle by leucyl-tRNA synthetase in the mTORC1 signaling pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E5279–E5288. [Google Scholar] [CrossRef]
- D’Hulst, G.; Masschelein, E.; De Bock, K. Resistance exercise enhances long-term mTORC1 sensitivity to leucine. Mol. Metab. 2022, 66, 101615. [Google Scholar] [CrossRef]
- Heng, J.; Wu, Z.; Tian, M.; Chen, J.; Song, H.; Chen, F.; Guan, W.; Zhang, S. Excessive BCAA regulates fat metabolism partially through the modification of m6A RNA methylation in weanling piglets. Nutr. Metab. 2020, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ren, Y.; Mao, K.; Hua, F.; Yang, Y.; Wei, N.; Yue, C.; Li, D.; Zhang, H. FTO is involved in Alzheimer’s disease by targeting TSC1-mTOR-Tau signaling. Biochem. Biophys. Res. Commun. 2018, 498, 234–239. [Google Scholar] [CrossRef]
- Huang, J.; Manning, B.D. The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem. J. 2008, 412, 179–190. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 2013, 126, 1713–1719. [Google Scholar] [CrossRef]
- Farmer, S.R. Regulation of PPARgamma activity during adipogenesis. Int. J. Obes. 2005, 29, S13–S16. [Google Scholar] [CrossRef]
- Ayala-Sumuano, J.T.; Velez-Delvalle, C.; Beltrán-Langarica, A.; Marsch-Moreno, M.; Cerbón-Solorzano, J.; Kuri-Harcuch, W. Srebf1a is a key regulator of transcriptional control for adipogenesis. Sci. Rep. 2011, 1, 178. [Google Scholar] [CrossRef]
- Bravard, A.; Lefai, E.; Meugnier, E.; Pesenti, S.; Disse, E.; Vouillarmet, J.; Peretti, N.; Rabasa-Lhoret, R.; Laville, M.; Vidal, H.; et al. FTO is increased in muscle during type 2 diabetes, and its overexpression in myotubes alters insulin signaling, enhances lipogenesis and ROS production, and induces mitochondrial dysfunction. Diabetes 2011, 60, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; He, R.; Han, Z.; Wu, Y.; Wang, Q.; Zhu, X.; Huang, Z.; Ye, J.; Tang, Y.; Huang, H.; et al. Cooperation of ATF4 and CTCF promotes adipogenesis through transcriptional regulation. Cell Biol. Toxicol. 2022, 38, 741–763. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Wang, H.V.; Ruiz, D.; Bixler, B.J.; Linsenbaum, H.; Xiang, J.F.; Forestier, S.; Shafik, A.M.; Jin, P.; Corces, V.G. Recruitment of CTCF to an Fto enhancer is responsible for transgenerational inheritance of BPA-induced obesity. Proc. Natl. Acad. Sci. USA 2022, 119, e2214988119. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Huang, Z.; Du, Y.; Cheng, Y.; Chen, S.; Guo, F. ATF4 regulates lipid metabolism and thermogenesis. Cell Res. 2010, 20, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Yuan, F.; Guo, Y.; Xiao, Y.; Niu, Y.; Deng, Y.; Han, X.; Guan, Y.; Chen, S.; Guo, F. Deletion of ATF4 in AgRP neurons promotes fat loss mainly via increasing energy expenditure. Diabetes 2017, 66, 640–650, Erratum in Diabetes 2017, 66, 3142. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Fortuno, E.S., 3rd; Suh, J.M.; Stenesen, D.; Tang, W.; Parks, E.J.; Adams, C.M.; Townes, T.; Graff, J.M. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes 2009, 58, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Avruch, J.; Long, X.; Ortiz-Vega, S.; Rapley, J.; Papageorgiou, A.; Dai, N. Amino acid regulation of TOR complex 1. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E592–E602. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Inoki, K. Spatial regulation of the mTORC1 system in amino acids sensing pathway. Acta Biochim. Biophys. Sin. 2011, 43, 671–679. [Google Scholar] [CrossRef]
- Jewell, J.L.; Russell, R.C.; Guan, K.L. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 2013, 14, 133–139. [Google Scholar] [CrossRef]
- Park, Y.; Reyna-Neyra, A.; Philippe, L.; Thoreen, C.C. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 2017, 19, 1083–1090. [Google Scholar] [CrossRef]
- Dibble, C.C.; Cantley, L.C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015, 25, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.H.; Huang, J.; Düvel, K.; Boback, B.; Wu, S.; Squillace, R.M.; Wu, C.L.; Manning, B.D. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS ONE 2009, 4, e6189. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, X.; Tang, Q.Q. Transcriptional regulation of adipocyte differentiation: A central role for CCAAT/enhancer-binding protein (C/EBP) β. J. Biol. Chem. 2015, 290, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Zidek, L.M.; Ackermann, T.; Hartleben, G.; Eichwald, S.; Kortman, G.; Kiehntopf, M.; Leutz, A.; Sonenberg, N.; Wang, Z.Q.; von Maltzahn, J.; et al. Deficiency in mTORC1-controlled C/EBPβ-mRNA translation improves metabolic health in mice. EMBO Rep. 2015, 16, 1022–1036. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Saunders, R.A.; Szkudlarek-Mikho, M.; Serna Ide, L.; Chin, K.V. The obesity-associated Fto gene is a transcriptional coactivator. Biochem. Biophys. Res. Commun. 2010, 401, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Ronkainen, J.; Huusko, T.J.; Soininen, R.; Mondini, E.; Cinti, F.; Mäkelä, K.A.; Kovalainen, M.; Herzig, K.H.; Järvelin, M.R.; Sebert, S.; et al. Fat mass- and obesity-associated gene Fto affects the dietary response in mouse white adipose tissue. Sci. Rep. 2015, 5, 9233. [Google Scholar] [CrossRef] [PubMed]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Ricoult, S.J.; Manning, B.D. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 2013, 14, 242–251. [Google Scholar] [CrossRef]
- Magdalon, J.; Festuccia, W.T. Regulation of adiposity by mTORC1. Einstein 2017, 15, 507–511. [Google Scholar] [CrossRef]
- Carnevalli, L.S.; Masuda, K.; Frigerio, F.; Le Bacquer, O.; Um, S.H.; Gandin, V.; Topisirovic, I.; Sonenberg, N.; Thomas, G.; Kozma, S.C. S6K1 plays a critical role in early adipocyte differentiation. Dev. Cell 2010, 18, 763–774. [Google Scholar] [CrossRef]
- Melnik, B.C. Excessive leucine-mTORC1-signalling of cow milk-based infant formula: The missing link to understand early childhood obesity. J. Obes. 2012, 2012, 197653. [Google Scholar] [CrossRef]
- Luque, V.; Closa-Monasterolo, R.; Escribano, J.; Ferré, N. Early programming by protein intake: The effect of protein on adiposity development and the growth and functionality of vital organs. Nutr. Metab. Insights 2016, 8, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.A.; Um, S.H.; Lee, J.; Yoo, J.H.; Bang, S.Y.; Park, E.K.; Lee, M.G.; Nam, K.H.; Jeon, Y.J.; Park, J.W.; et al. S6K1 phosphorylation of H2B mediates EZH2 trimethylation of H3: A determinant of early adipogenesis. Mol. Cell 2016, 62, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.A.; Han, J.; Han, J.W. Epigenetic role of nuclear S6K1 in early adipogenesis. BMB Rep. 2016, 49, 401–402. [Google Scholar] [CrossRef]
- de Winter, T.J.J.; Nusse, R. Running against the Wnt: How Wnt/β-catenin suppresses adipogenesis. Front. Cell. Dev. Biol. 2021, 9, 627429. [Google Scholar] [CrossRef]
- Ross, S.E.; Hemati, N.; Longo, K.A.; Bennett, C.N.; Lucas, P.C.; Erickson, R.L.; MacDougald, O.A. Inhibition of adipogenesis by Wnt signaling. Science 2000, 289, 950–953. [Google Scholar] [CrossRef]
- Bennett, C.N.; Ross, S.E.; Longo, K.A.; Bajnok, L.; Hemati, N.; Johnson, K.W.; Harrison, S.D.; MacDougald, O.A. Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 2002, 277, 30998–31004. [Google Scholar] [CrossRef] [PubMed]
- Cawthorn, W.P.; Bree, A.J.; Yao, Y.; Du, B.; Hemati, N.; Martinez-Santibañez, G.; MacDougald, O.A. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone 2012, 50, 477–489. [Google Scholar] [CrossRef]
- Yi, S.A.; Jeon, Y.J.; Lee, M.G.; Nam, K.H.; Ann, S.; Lee, J.; Han, J.W. S6K1 controls adiponectin expression by inducing a transcriptional switch: BMAL1-to-EZH2. Exp. Mol. Med. 2022, 54, 324–333. [Google Scholar] [CrossRef]
- Bauzá-Thorbrügge, M.; Vujičić, M.; Chanclón, B.; Palsdottir, V.; Pillon, N.J.; Benrick, A.; Wernstedt Asterholm, I. Adiponectin stimulates Sca1 + CD34—adipocyte precursor cells associated with hyperplastic expansion and beiging of brown and white adipose tissue. Metabolism 2024, 151, 155716. [Google Scholar] [CrossRef]
- Lluch, A.; Veiga, S.R.; Latorre, J.; Moreno-Navarrete, J.M.; Bonifaci, N.; Dien Nguyen, V.; Zhou, Y.; Höring, M.; Liebisch, G.; Olkkonen, V.M.; et al. A compound directed against S6K1 hampers fat mass expansion and mitigates diet-induced hepatosteatosis. JCI Insight 2022, 7, e150461. [Google Scholar] [CrossRef]
- Cho, Y.M.; Kim, T.M.; Hun Kim, D.; Hee Kim, D.; Jeong, S.W.; Kwon, O.J. miR-148a is a downstream effector of X-box-binding protein 1 that silences Wnt10b during adipogenesis of 3T3-L1 cells. Exp. Mol. Med. 2016, 48, e226. [Google Scholar] [CrossRef]
- TargetScanHuman Release 8.0, Human WNT10B ENST00000407467.1. Available online: https://www.targetscan.org/cgi-bin/targetscan/vert_80/targetscan.cgi?species=Human&gid=WNT10b&mir_sc=&mir_c=&mir_nc=&mir_vnc=&mirg= (accessed on 21 July 2024).
- Qi, Q.; Downer, M.K.; Kilpeläinen, T.O.; Taal, H.R.; Barton, S.J.; Ntalla, I.; Standl, M.; Boraska, V.; Huikari, V.; Kiefte-de Jong, J.C.; et al. Dietary intake, FTO about:blankgenetic variants, and adiposity: A combined analysis of over 16,000 children and adolescents. Diabetes 2015, 64, 2467–2476. [Google Scholar] [CrossRef]
- Benmoussa, A.; Provost, P. Milk microRNAs in health and disease. Compr. Rev. Food Sci. Food Saf. 2019, 18, 703–722. [Google Scholar] [CrossRef]
- Tingö, L.; Ahlberg, E.; Johansson, L.; Pedersen, S.A.; Chawla, K.; Sætrom, P.; Cione, E.; Simpson, M.R. Non-coding RNAs in human breast milk: A systematic review. Front. Immunol. 2021, 12, 725323. [Google Scholar] [CrossRef]
- Zempleni, J.; Sukreet, S.; Zhou, F.; Wu, D.; Mutai, E. Milk-derived exosomes and metabolic regulation. Annu. Rev. Anim. Biosci. 2019, 7, 245–262. [Google Scholar] [CrossRef]
- Słyk-Gulewska, P.; Kondracka, A.; Kwaśniewska, A. MicroRNA as a new bioactive component in breast milk. Noncoding RNA Res. 2023, 8, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Hatmal, M.M.; Al-Hatamleh, M.A.I.; Olaimat, A.N.; Alshaer, W.; Hasan, H.; Albakri, K.A.; Alkhafaji, E.; Issa, N.N.; Al-Holy, M.A.; Abderrahman, S.M.; et al. Immunomodulatory properties of human breast milk: MicroRNA contents and potential epigenetic effects. Biomedicines 2022, 10, 1219. [Google Scholar] [CrossRef]
- Weil, P.P.; Reincke, S.; Hirsch, C.A.; Giachero, F.; Aydin, M.; Scholz, J.; Jönsson, F.; Hagedorn, C.; Nguyen, D.N.; Thymann, T.; et al. Uncovering the gastrointestinal passage, intestinal epithelial cellular uptake, and AGO2 loading of milk miRNAs in neonates using xenomiRs as tracers. Am. J. Clin. Nutr. 2023, 117, 1195–1210. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.R.; Zhao, J.; Huang, H.Y.; Lin, Y.C.; Lee, T.Y.; Huang, H.D.; Yang, Y.; Wang, Y.F. Recent insights into breast milk microRNA: Their role as functional regulators. Front. Nutr. 2024, 11, 1366435. [Google Scholar] [CrossRef] [PubMed]
- Weiskirchen, R.; Schröder, S.K.; Weiskirchen, S.; Buhl, E.M.; Melnik, B. Isolation of bovine and human milk extracellular vesicles. Biomedicines 2023, 11, 2715. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Ngu, A.; Wang, S.; Wang, H.; Khanam, A.; Zempleni, J. Milk exosomes in nutrition and drug delivery. Am. J. Physiol. Cell Physiol. 2022, 322, C865–C874. [Google Scholar] [CrossRef] [PubMed]
- Leiferman, A.; Shu, J.; Upadhyaya, B.; Cui, J.; Zempleni, J. Storage of extracellular vesicles in human milk, and microRNA profiles in human milk exosomes and infant formulas. J. Pediatr. Gastroenterol. Nutr. 2019, 69, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Vélez-Ixta, J.M.; Benítez-Guerrero, T.; Aguilera-Hernández, A.; Martínez-Corona, H.; Corona-Cervantes, K.; Juárez-Castelán, C.J.; Rangel-Calvillo, M.N.; García-Mena, J. Detection and quantification of immunoregulatory miRNAs in human milk and infant milk formula. BioTech 2022, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.A.; Al-Saigh, N.N.; Saqallah, F.G. Regulation of adipogenesis by exosomal milk miRNA. Rev. Endocr. Metab. Disord. 2023, 24, 297–316. [Google Scholar] [CrossRef] [PubMed]
- Golan-Gerstl, R.; Elbaum Shiff, Y.; Moshayoff, V.; Schecter, D.; Leshkowitz, D.; Reif, S. Characterization and biological function of milk-derived miRNAs. Mol. Nutr. Food Res. 2017, 61, 10. [Google Scholar] [CrossRef]
- Chiba, T.; Takaguri, A.; Kooka, A.; Kowatari, K.; Yoshizawa, M.; Fukushi, Y.; Hongo, F.; Sato, H.; Fujisawa, M.; Wada, S.; et al. Suppression of milk-derived miR-148a caused by stress plays a role in the decrease in intestinal ZO-1 expression in infants. Clin. Nutr. 2022, 41, 2691–2698. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.M.; Zhang, K.; Zhang, J.H. Human breast milk-derived exosomal miR-148a-3p protects against necrotizing enterocolitis by regulating p53 and sirtuin 1. Inflammation 2022, 45, 1254–1268. [Google Scholar] [CrossRef]
- Smyczynska, U.; Bartlomiejczyk, M.A.; Stanczak, M.M.; Sztromwasser, P.; Wesolowska, A.; Barbarska, O.; Pawlikowska, E.; Fendler, W. Impact of processing method on donated human breast milk microRNA content. PLoS ONE 2020, 15, e0236126. [Google Scholar] [CrossRef]
- Chiba, T.; Kooka, A.; Kowatari, K.; Yoshizawa, M.; Chiba, N.; Takaguri, A.; Fukushi, Y.; Hongo, F.; Sato, H.; Wada, S. Expression profiles of hsa-miR-148a-3p and hsa-miR-125b-5p in human breast milk and infant formulae. Int. Breastfeed. J. 2022, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, R.; Shan, W.; Yu, L.; Xue, B.; Shi, H. DNA methylation biphasically regulates 3T3-L1 preadipocyte differentiation. Mol. Endocrinol. 2016, 30, 677–687. [Google Scholar] [CrossRef] [PubMed]
- TargetScanHuman Release 8.0, Human DNMT3A ENST00000380746.4. Available online: https://www.targetscan.org/cgi-bin/targetscan/vert_80/view_gene.cgi?rs=ENST00000380746.4&taxid=9606&members=&showcnc=0&shownc=0&showncf1=&showncf2=&subset=1 (accessed on 21 July 2024).
- Pan, W.; Zhu, S.; Yuan, M.; Cui, H.; Wang, L.; Luo, X.; Li, J.; Zhou, H.; Tang, Y.; Shen, N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 2010, 184, 6773–6781. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Lönnerdal, B. Milk-derived miR-22-3p promotes proliferation of human intestinal epithelial cells (HIECs) by regulating gene expression. Nutrients 2022, 14, 4901. [Google Scholar] [CrossRef] [PubMed]
- Ahlberg, E.; Al-Kaabawi, A.; Thune, R.; Simpson, M.R.; Pedersen, S.A.; Cione, E.; Jenmalm, M.C.; Tingö, L. Breast milk microRNAs: Potential players in oral tolerance development. Front. Immunol. 2023, 14, 1154211. [Google Scholar] [CrossRef]
- Kahn, S.; Liao, Y.; Du, X.; Xu, W.; Li, J.; Lönnerdal, B. Exosomal microRNAs in milk from mothers delivering preterm infants survive in vitro digestion and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2018, 62, e1701050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y.; Zhao, H.; Han, X.; Zhao, T.; Qu, P.; Li, G.; Wang, W. Extracellular vesicle-encapsulated miR-22-3p from bone marrow mesenchymal stem cell promotes osteogenic differentiation via FTO inhibition. Stem Cell Res. Ther. 2020, 11, 227. [Google Scholar] [CrossRef] [PubMed]
- Lou, P.; Bi, X.; Tian, Y.; Li, G.; Kang, Q.; Lv, C.; Song, Y.; Xu, J.; Sheng, X.; Yang, X.; et al. MiR-22 modulates brown adipocyte thermogenesis by synergistically activating the glycolytic and mTORC1 signaling pathways. Theranostics 2021, 11, 3607–3623. [Google Scholar] [CrossRef] [PubMed]
- Hishida, T.; Nishizuka, M.; Osada, S.; Imagawa, M. The role of C/EBPdelta in the early stages of adipogenesis. Biochimie 2009, 91, 654–657. [Google Scholar] [CrossRef]
- Chen, S.; Song, P.; Wang, Y.; Wang, Z.; Xue, J.; Jiang, Y.; Zhou, Y.; Zhao, J.; Tang, L. CircMAPK9 promotes adipogenesis through modulating hsa-miR-1322/FTO axis in obesity. iScience 2023, 26, 107756. [Google Scholar] [CrossRef]
- Zamanillo, R.; Sánchez, J.; Serra, F.; Palou, A. Breast milk supply of microRNA associated with leptin and adiponectin is affected by maternal overweight/obesity and influences infancy BMI. Nutrients 2019, 11, 2589. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Jiang, X.; Li, R.; Chen, M.; Song, W.; Li, X. The levels of human milk microRNAs and their association with maternal weight characteristics. Eur. J. Clin. Nutr. 2016, 70, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.B.; Chernausek, S.D.; Garman, L.D.; Pezant, N.P.; Plows, J.F.; Kharoud, H.K.; Demerath, E.W.; Fields, D.A. Human milk exosomal microRNA: Associations with maternal overweight/obesity and infant body composition at 1 month of life. Nutrients 2021, 13, 1091. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.B.; Fields, D.A.; Pezant, N.P.; Kharoud, H.K.; Gulati, S.; Jacobs, K.; Gale, C.A.; Kharbanda, E.O.; Nagel, E.M.; Demerath, E.W.; et al. Gestational diabetes mellitus is associated with altered abundance of exosomal microRNAs in human milk. Clin. Ther. 2022, 44, 172–185, Erratum in Clin. Ther. 2022, 44, 1034. [Google Scholar] [CrossRef] [PubMed]
- TargetScanHuman Release 8.0, FTO Human ENST00000471389.1. Available online: https://www.targetscan.org/cgi-bin/targetscan/vert_80/targetscan.cgi?species=Human&gid=DNMT3a&mir_sc=&mir_c=&mir_nc=&mir_vnc=&mirg= (accessed on 21 July 2024).
- Sun, L.; Gao, M.; Qian, Q.; Guo, Z.; Zhu, P.; Wang, X.; Wang, H. Triclosan-induced abnormal expression of miR-30b regulates fto-mediated m6A methylation level to cause lipid metabolism disorder in zebrafish. Sci. Total Environ. 2021, 770, 145285. [Google Scholar] [CrossRef] [PubMed]
- Caruso, V.; Chen, H.; Morris, M.J. Early hypothalamic FTO overexpression in response to maternal obesity--potential contribution to postweaning hyperphagia. PLoS ONE 2011, 6, e25261. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Wang, M.; Xiao, T.; Yin, B.; He, L.; Meng, W.; Dong, M.; Liu, F. miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes 2015, 64, 2056–2068. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Kiskinis, E.; Debevec, D.; Leonardsson, G.; White, R.; Parker, M.G. RIP140-targeted repression of gene expression in adipocytes. Mol. Cell. Biol. 2005, 25, 9383–9391. [Google Scholar] [CrossRef] [PubMed]
- Leonardsson, G.; Steel, J.H.; Christian, M.; Pocock, V.; Milligan, S.; Bell, J.; So, P.W.; Medina-Gomez, G.; Vidal-Puig, A.; White, R.; et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. USA 2004, 101, 8437–8442. [Google Scholar] [CrossRef]
- Kim, J.; Okla, M.; Erickson, A.; Carr, T.; Natarajan, S.K.; Chung, S. Eicosapentaenoic acid potentiates brown thermogenesis through FFAR4-dependent up-regulation of miR-30b and miR-378. J. Biol. Chem. 2016, 291, 20551–20562. [Google Scholar] [CrossRef]
- Ortega-Molina, A.; Efeyan, A.; Lopez-Guadamillas, E.; Muñoz-Martin, M.; Gómez-López, G.; Cañamero, M.; Mulero, F.; Pastor, J.; Martinez, S.; Romanos, E.; et al. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metab. 2012, 15, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Nakae, J.; Cao, Y.; Oki, M.; Orba, Y.; Sawa, H.; Kiyonari, H.; Iskandar, K.; Suga, K.; Lombes, M.; Hayashi, Y. Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes 2008, 57, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Kodani, N.; Nakae, J. Tissue-specific metabolic regulation of FOXO-binding protein: FOXO does not act alone. Cells 2020, 9, 702. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.; Ayuso, E.; Shan, X.; Bosch, F.; O’Rahilly, S.; Coll, A.P.; Yeo, G.S. Hypothalamic-specific manipulation of Fto, the ortholog of the human obesity gene FTO, affects food intake in rats. PLoS ONE 2010, 5, e8771. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, P.K.; Fredriksson, R.; Olszewska, A.M.; Stephansson, O.; Alsiö, J.; Radomska, K.J.; Levine, A.S.; Schiöth, H.B. Hypothalamic FTO is associated with the regulation of energy intake not feeding reward. BMC Neurosci. 2009, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, P.K.; Radomska, K.J.; Ghimire, K.; Klockars, A.; Ingman, C.; Olszewska, A.M.; Fredriksson, R.; Levine, A.S.; Schiöth, H.B. Fto immunoreactivity is widespread in the rodent brain and abundant in feeding-related sites, but the number of Fto-positive cells is not affected by changes in energy balance. Physiol. Behav. 2011, 103, 248–253. [Google Scholar] [CrossRef]
- Heinig, M.J.; Nommsen, L.A.; Peerson, J.M.; Lonnerdal, B.; Dewey, K.G. Energy and protein intakes of breast-fed and formula-fed infants during the first year of life and their association with growth velocity: The DARLING Study. Am. J. Clin. Nutr. 1993, 58, 152–161. [Google Scholar] [CrossRef]
- Johansson, A. Leucine Intake Affects Brain Activity and Central Expression of Genes Associated with Food Intake, Energy Homeostasis and Reward; Institutionen för Neurovetenskap, Uppsala Universitet: Uppsala, Sweden, 2011; Available online: https://mdh.diva-portal.org/smash/get/diva2:447251/FULLTEXT01.pdf (accessed on 25 June 2024).
- Church, C.; Moir, L.; McMurray, F.; Girard, C.; Banks, G.T.; Teboul, L.; Wells, S.; Brüning, J.C.; Nolan, P.M.; Ashcroft, F.M.; et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 2010, 42, 1086–1092. [Google Scholar] [CrossRef]
- Yeo, G.S. FTO and obesity: A problem for a billion people. J. Neuroendocrinol. 2012, 24, 393–394. [Google Scholar] [CrossRef]
- Mehrdad, M.; Doaei, S.; Gholamalizadeh, M.; Eftekhari, M.H. The association between FTO genotype with macronutrients and calorie intake in overweight adults. Lipids Health Dis. 2020, 19, 197. [Google Scholar] [CrossRef]
- Melhorn, S.J.; Askren, M.K.; Chung, W.K.; Kratz, M.; Bosch, T.A.; Tyagi, V.; Webb, M.F.; De Leon, M.R.B.; Grabowski, T.J.; Leibel, R.L.; et al. FTO genotype impacts food intake and corticolimbic activation. Am. J. Clin. Nutr. 2018, 107, 145–154. [Google Scholar] [CrossRef]
- Tanofsky-Kraff, M.; Han, J.C.; Anandalingam, K.; Shomaker, L.B.; Columbo, K.M.; Wolkoff, L.E.; Kozlosky, M.; Elliott, C.; Ranzenhofer, L.M.; Roza, C.A.; et al. The FTO gene rs9939609 obesity-risk allele and loss of control over eating. Am. J. Clin. Nutr. 2009, 90, 1483–1488. [Google Scholar] [CrossRef] [PubMed]
- Obregón Rivas, A.M.; Santos, J.L.; Valladares, M.A.; Cameron, J.; Goldfield, G. Association of the FTO fat mass and obesity-associated gene rs9939609 polymorphism with rewarding value of food and eating behavior in Chilean children. Nutrition 2018, 54, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Ranzenhofer, L.M.; Mayer, L.E.S.; Davis, H.A.; Mielke-Maday, H.K.; McInerney, H.; Korn, R.; Gupta, N.; Brown, A.J.; Schebendach, J.; Tanofsky-Kraff, M.; et al. The FTO gene and measured food intake in 5- to 10-year-old children without obesity. Obesity 2019, 27, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lu, P.; Tang, H.L.; Yan, H.J.; Jiang, W.; Shi, H.; Chen, S.Y.; Gao, M.M.; Zeng, X.D.; Long, Y.S. Valproate-induced epigenetic upregulation of hypothalamic Fto expression potentially linked with weight gain. Cell. Mol. Neurobiol. 2021, 41, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Tang, H.L.; He, Q.; Lu, P.; Fu, T.; Xu, X.L.; Su, T.; Gao, M.M.; Duan, S.; Luo, Y.; et al. FTO is a transcriptional repressor to auto-regulate its own gene and potentially associated with homeostasis of body weight. J. Mol. Cell Biol. 2019, 11, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Song, S.; Wang, S.; Cai, T.; Qin, L.; Wang, X.; Zhu, G.; Wang, H.; Yang, W.; Fang, C.; et al. Hypothalamic FTO promotes high-fat diet-induced leptin resistance in mice through increasing CX3CL1 expression. J. Nutr. Biochem. 2024, 123, 109512. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, X.; Shen, Z.; Chen, Z.; Wang, H.; Zhang, X. GnRH receptor mediates lipid storage in female adipocytes via AMPK pathway. Int. J. Med. Sci. 2022, 19, 1442–1450. [Google Scholar] [CrossRef]
- Busby, E.R.; Sherwood, N.M. Gonadotropin-releasing hormone receptor (Gnrhr) gene knock out: Normal growth and development of sensory, motor and spatial orientation behavior but altered metabolism in neonatal and prepubertal mice. PLoS ONE 2017, 12, e0174452. [Google Scholar] [CrossRef]
- Benedict, C.; Axelsson, T.; Söderberg, S.; Larsson, A.; Ingelsson, E.; Lind, L.; Schiöth, H.B. Fat mass and obesity-associated gene (FTO) is linked to higher plasma levels of the hunger hormone ghrelin and lower serum levels of the satiety hormone leptin in older adults. Diabetes 2014, 63, 3955–3959. [Google Scholar] [CrossRef]
- Savino, F.; Fissore, M.F.; Grassino, E.C.; Nanni, G.E.; Oggero, R.; Silvestro, L. Ghrelin, leptin and IGF-I levels in breast-fed and formula-fed infants in the first years of life. Acta Paediatr. 2005, 94, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Wiedmer, P.; Nogueiras, R.; Broglio, F.; D’Alessio, D.; Tschöp, M.H. Ghrelin, obesity and diabetes. Nat. Clin. Pract. Endocrinol. Metab. 2007, 3, 705–712. [Google Scholar] [CrossRef] [PubMed]
- De Vriese, C.; Delporte, C. Influence of ghrelin on food intake and energy homeostasis. Curr. Opin. Clin. Nutr. Metab. Care 2007, 10, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Pulkkinen, L.; Ukkola, O.; Kolehmainen, M.; Uusitupa, M. Ghrelin in diabetes and metabolic syndrome. Int. J. Pept. 2010, 2010, 248948. [Google Scholar] [CrossRef] [PubMed]
- De Vriese, C.; Perret, J.; Delporte, C. Focus on the short- and long-term effects of ghrelin on energy homeostasis. Nutrition 2010, 26, 579–584. [Google Scholar] [CrossRef]
- Davis, J.F.; Perello, M.; Choi, D.L.; Magrisso, I.J.; Kirchner, H.; Pfluger, P.T.; Tschoep, M.; Zigman, J.M.; Benoit, S.C. GOAT induced ghrelin acylation regulates hedonic feeding. Horm. Behav. 2012, 62, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Yang, Q.; Yin, W.; Su, W.; Lin, H.; Feng, M.; Pan, K.; Yin, Y.; Zhang, W. ETV5 regulates GOAT/ghrelin system in an mTORC1-dependent manner. Mol. Cell. Endocrinol. 2019, 485, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Micioni Di Bonaventura, E.; Botticelli, L.; Del Bello, F.; Giorgioni, G.; Piergentili, A.; Quaglia, W.; Cifani, C.; Micioni Di Bonaventura, M.V. Assessing the role of ghrelin and the enzyme ghrelin O-acyltransferase (GOAT) system in food reward, food motivation, and binge eating behavior. Pharmacol. Res. 2021, 172, 105847. [Google Scholar] [CrossRef] [PubMed]
- Wellman, M.; Abizaid, A. Knockdown of central ghrelin O-acyltransferase by vivo-morpholino reduces body mass of rats fed a high-fat diet. Peptides 2015, 70, 17–22. [Google Scholar] [CrossRef]
- TargetScanHuman Release 8.0; Human ETV5 ENST00000306376.5. Available online: https://www.targetscan.org/cgibin/tagetscan/vert_80/view_gene.cgi?rs=ENST00000306376.5&taxid=9606&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1 (accessed on 21 July 2024).
- Ofori, J.K.; Karagiannopoulos, A.; Nagao, M.; Westholm, E.; Ramadan, S.; Wendt, A.; Esguerra, J.L.S.; Eliasson, L. Human islet microRNA-200c is elevated in type 2 diabetes and targets the transcription factor ETV5 to reduce insulin secretion. Diabetes 2022, 71, 275–284. [Google Scholar] [CrossRef]
- van Herwijnen, M.J.C.; Driedonks, T.A.P.; Snoek, B.L.; Kroon, A.M.T.; Kleinjan, M.; Jorritsma, R.; Pieterse, C.M.J.; Hoen, E.N.M.N.; Wauben, M.H.M. Abundantly present miRNAs in milk-derived extracellular vesicles are conserved between mammals. Front. Nutr. 2018, 5, 81. [Google Scholar] [CrossRef]
- Cho, Y.E.; Vorn, R.; Chimenti, M.; Crouch, K.; Shaoshuai, C.; Narayanaswamy, J.; Harken, A.; Schmidt, R.; Gill, J.; Lee, H. Extracellular vesicle miRNAs in breast milk of obese mothers. Front. Nutr. 2022, 9, 976886. [Google Scholar] [CrossRef] [PubMed]
- Cavadas, C.; Aveleira, C.A.; Souza, G.F.; Velloso, L.A. The pathophysiology of defective proteostasis in the hypothalamus—From obesity to ageing. Nat. Rev. Endocrinol. 2016, 12, 723–733. [Google Scholar] [CrossRef] [PubMed]
- de Araujo, T.M.; Razolli, D.S.; Correa-da-Silva, F.; de Lima-Junior, J.C.; Gaspar, R.S.; Sidarta-Oliveira, D.; Victorio, S.C.; Donato, J., Jr.; Kim, Y.B.; Velloso, L.A. The partial inhibition of hypothalamic IRX3 exacerbates obesity. EBioMedicine 2019, 39, 448–460. [Google Scholar] [CrossRef]
- de Araújo, T.M.; Velloso, L.A. Hypothalamic IRX3: A new player in the development of obesity. Trends Endocrinol. Metab. 2020, 31, 368–377. [Google Scholar] [CrossRef]
- Sievers, E.; Oldigs, H.D.; Santer, R.; Schaub, J. Feeding patterns in breast-fed and formula-fed infants. Ann. Nutr. Metab. 2002, 46, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Olejniczak, I.; Pilorz, V.; Oster, H. Circle(s) of life: The circadian clock from birth to death. Biology 2023, 12, 383. [Google Scholar] [CrossRef]
- Wong, S.D.; Wright, K.P., Jr.; Spencer, R.L.; Vetter, C.; Hicks, L.M.; Jenni, O.G.; LeBourgeois, M.K. Development of the circadian system in early life: Maternal and environmental factors. J. Physiol. Anthropol. 2022, 41, 22. [Google Scholar] [CrossRef]
- Sánchez, C.L.; Cubero, J.; Sánchez, J.; Franco, L.; Rodríguez, A.B.; Rivero, M.; Barriga, C. Evolution of the circadian profile of human milk amino acids during breastfeeding. J. Appl. Biomed. 2013, 11, 59–70. [Google Scholar] [CrossRef]
- Cubero, J.; Valero, V.; Sánchez, J.; Rivero, M.; Parvez, H.; Rodríguez, A.B.; Barriga, C. The circadian rhythm of tryptophan in breast milk affects the rhythms of 6-sulfatoxymelatonin and sleep in newborn. Neuro. Endocrinol. Lett. 2005, 26, 657–661. [Google Scholar]
- Lodemore, M.R.; Petersen, S.A.; Wailoo, M.P. Factors affecting the development of night time temperature rhythms. Arch. Dis. Child. 1992, 67, 1259–1261. [Google Scholar] [CrossRef] [PubMed]
- Caba-Flores, M.D.; Ramos-Ligonio, A.; Camacho-Morales, A.; Martínez-Valenzuela, C.; Viveros-Contreras, R.; Caba, M. Breast milk and the importance of chrononutrition. Front. Nutr. 2022, 9, 867507. [Google Scholar] [CrossRef] [PubMed]
- Fustin, J.M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Hastings, M.H. m(6)A mRNA methylation: A new circadian pacesetter. Cell 2013, 155, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Yu, J.; Frazier, K.; Weng, X.; Li, Y.; Cham, C.M.; Dolan, K.; Zhu, X.; Hubert, N.; Tao, Y.; et al. Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation. Cell Rep. 2018, 25, 1816–1828.e4. [Google Scholar] [CrossRef] [PubMed]
- Lokody, I. Gene regulation: RNA methylation regulates the circadian clock. Nat. Rev. Genet. 2014, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Shie, S.S.; Hsieh, I.C.; Tsai, M.L.; Wen, M.S. FTO modulates circadian rhythms and inhibits the CLOCK-BMAL1-induced transcription. Biochem. Biophys. Res. Commun. 2015, 464, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Reinke, H.; Asher, G. Circadian clock control of liver metabolic functions. Gastroenterology 2016, 150, 574–580. [Google Scholar] [CrossRef]
- Yang, Y.; Feng, Y.; Hu, Y.; Liu, J.; Shi, H.; Zhao, R. Exposure to constant light impairs cognition with FTO inhibition and m6A-dependent TrκB repression in mouse hippocampus. Environ. Pollut. 2021, 283, 117037. [Google Scholar] [CrossRef]
- Chong, Y.S.; Lek, N.; Chan, J.K.Y.; Goh, D.Y.T.; Chong, M.F.; Yap, F. Circadian feeding patterns of 12-month-old infants. Br. J. Nutr. 2017, 117, 1702–1710. [Google Scholar]
- Li, Y.; Ma, J.; Yao, K.; Su, W.; Tan, B.; Wu, X.; Huang, X.; Li, T.; Yin, Y.; Tosini, G.; et al. Circadian rhythms and obesity: Timekeeping governs lipid metabolism. J. Pineal Res. 2020, 69, e12682. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Kannan, N.N. Only time will tell: The interplay between circadian clock and metabolism. Chronobiol. Int. 2021, 38, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Mota, S.; Zhang, B. Circadian clock regulation on lipid metabolism and metabolic diseases. Adv. Exp. Med. Biol. 2020, 1276, 53–66. [Google Scholar] [PubMed]
- Albrecht, U. The circadian clock, metabolism and obesity. Obes. Rev. 2017, 18, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Sebti, Y.; Hebras, A.; Pourcet, B.; Staels, B.; Duez, H. The circadian clock and obesity. Handb. Exp. Pharmacol. 2022, 274, 29–56. [Google Scholar] [PubMed]
- Aparicio, S.; Garau, C.; Esteban, S.; Nicolau, M.C.; Rivero, M.; Rial, R.V. Chrononutrition: Use of dissociated day/night infant milk formulas to improve the development of the wake-sleep rhythms. Effects of tryptophan. Nutr. Neurosci. 2007, 10, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Richard, D.M.; Dawes, M.A.; Mathias, C.W.; Acheson, A.; Hill-Kapturczak, N.; Dougherty, D.M. L-Tryptophan: Basic metabolic functions, behavioral research and therapeutic Indications. Int. J. Tryptophan Res. 2009, 2, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. Milk proteins as a source of tryptophan-containing bioactive peptides. Food Funct. 2015, 6, 2115–2127. [Google Scholar] [CrossRef] [PubMed]
- Castro-Portuguez, R.; Sutphin, G.L. Kynurenine pathway, NAD+ synthesis, and mitochondrial function: Targeting tryptophan metabolism to promote longevity and healthspan. Exp. Gerontol. 2020, 132, 110841. [Google Scholar] [CrossRef]
- Zhang, Z.; Adelman, A.S.; Rai, D.; Boettcher, J.; Lőnnerdal, B. Amino acid profiles in term and preterm human milk through lactation: A systematic review. Nutrients 2013, 5, 4800–4821. [Google Scholar] [CrossRef]
- Chailurkit, L.O.; Chanprasertyothin, S.; Thongmung, N.; Sritara, P.; Ongphiphadhanakul, B. Targeted metabolomics suggests a probable role of the FTO gene in the kynurenine pathway in prediabetes. PeerJ 2022, 10, e13612. [Google Scholar] [CrossRef] [PubMed]
- Favennec, M.; Hennart, B.; Caiazzo, R.; Leloire, A.; Yengo, L.; Verbanck, M.; Arredouani, A.; Marre, M.; Pigeyre, M.; Bessede, A.; et al. The kynurenine pathway is activated in human obesity and shifted toward kynurenine monooxygenase activation. Obesity 2015, 23, 2066–2074, Erratum in Obesity 2016, 24, 1821. [Google Scholar] [CrossRef] [PubMed]
- Marszalek-Grabska, M.; Stachniuk, A.; Iwaniak, P.; Gawel, K.; Sumara, A.; Kocki, T.; Fornal, E.; Milart, P.; Paluszkiewicz, P.; Turski, W. Unexpected content of kynurenine in mother’s milk and infant formulas. Sci. Rep. 2022, 12, 6464. [Google Scholar] [CrossRef]
- Badawy, A.A. Kynurenine pathway of tryptophan metabolism: Regulatory and functional aspects. Int. J. Tryptophan Res. 2017, 10, 1178646917691938. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Luo, Y.; Zeng, B.; Chen, Y.; Liu, Y.; Chen, W.; Liao, X.; Liu, Y.; Wang, Y.; Wang, X. Branched-chain amino acids prevent obesity by inhibiting the cell cycle in an NADPH-FTO-m6A coordinated manner. J. Nutr. Biochem. 2023, 122, 109437. [Google Scholar] [CrossRef]
- Rongvaux, A.; Shea, R.J.; Mulks, M.H.; Gigot, D.; Urbain, J.; Leo, O.; Andris, F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 2002, 32, 3225–3234. [Google Scholar] [CrossRef]
- Revollo, J.R.; Grimm, A.A.; Imai, S. The regulation of nicotinamide adenine dinucleotide biosynthesis by Nampt/PBEF/visfatin in mammals. Curr. Opin. Gastroenterol. 2007, 23, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Garten, A.; Schuster, S.; Penke, M.; Gorski, T.; de Giorgis, T.; Kiess, W. Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat. Rev. Endocrinol. 2015, 11, 535–546. [Google Scholar] [CrossRef]
- Carbone, F.; Liberale, L.; Bonaventura, A.; Vecchiè, A.; Casula, M.; Cea, M.; Monacelli, F.; Caffa, I.; Bruzzone, S.; Montecucco, F.; et al. Regulation and function of extracellular nicotinamide phosphoribosyltransferase/visfatin. Compr. Physiol. 2017, 7, 603–621. [Google Scholar]
- López-Bermejo, A.; Petry, C.J.; Díaz, M.; Sebastiani, G.; de Zegher, F.; Dunger, D.B.; Ibáñez, L. The association between the FTO gene and fat mass in humans develops by the postnatal age of two weeks. J. Clin. Endocrinol. Metab. 2008, 93, 1501–1505. [Google Scholar] [CrossRef]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, K.M.; Yoshino, J.; Brace, C.S.; Abrassart, D.; Kobayashi, Y.; Marcheva, B.; Hong, H.K.; Chong, J.L.; Buhr, E.D.; Lee, C.; et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009, 324, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.Y.; Park, Y.J. Function of NAD metabolism in white adipose tissue: Lessons from mouse models. Adipocyte 2024, 13, 2313297. [Google Scholar] [CrossRef] [PubMed]
- Basse, A.L.; Nielsen, K.N.; Karavaeva, I.; Ingerslev, L.R.; Ma, T.; Havelund, J.F.; Nielsen, T.S.; Frost, M.; Peics, J.; Dalbram, E.; et al. NAMPT-dependent NAD+ biosynthesis controls circadian metabolism in a tissue-specific manner. Proc. Natl. Acad. Sci. USA 2023, 120, e2220102120. [Google Scholar] [CrossRef] [PubMed]
- Appleton, J.; Russell, C.G.; Laws, R.; Fowler, C.; Campbell, K.; Denney-Wilson, E. Infant formula feeding practices associated with rapid weight gain: A systematic review. Matern. Child Nutr. 2018, 14, e12602. [Google Scholar] [CrossRef] [PubMed]
- Mehta, N.; Cheng, H.Y. Micro-managing the circadian clock: The role of microRNAs in biological timekeeping. J. Mol. Biol. 2013, 425, 3609–3624. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tian, G.; Li, Z.; Zheng, L. The crosstalk between miRNA and mammalian circadian clock. Curr. Med. Chem. 2015, 22, 1582–1588. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhang, Y. Emerging roles for microRNA in the regulation of Drosophila circadian clock. BMC Neurosci. 2018, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Anna, G.; Kannan, N.N. Post-transcriptional modulators and mediators of the circadian clock. Chronobiol. Int. 2021, 38, 1244–1261. [Google Scholar] [CrossRef]
- Jacovetti, C.; Rodriguez-Trejo, A.; Guay, C.; Sobel, J.; Gattesco, S.; Petrenko, V.; Saini, C.; Dibner, C.; Regazzi, R. MicroRNAs modulate core-clock gene expression in pancreatic islets during early postnatal life in rats. Diabetologia 2017, 60, 2011–2020. [Google Scholar] [CrossRef]
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. MicroRNAs in breastmilk and the lactating breast: Potential immunoprotectors and developmental regulators for the infant and the mother. Int. J. Environ. Res. Public Health 2015, 12, 13981–14020. [Google Scholar] [CrossRef] [PubMed]
- Chutipongtanate, S.; Morrow, A.L.; Newburg, D.S. Human milk extracellular vesicles: A biological system with clinical implications. Cells 2022, 11, 2345. [Google Scholar] [CrossRef]
- Gialeli, G.; Panagopoulou, O.; Liosis, G.; Siahanidou, T. Potential epigenetic effects of human milk on infants’ neurodevelopment. Nutrients 2023, 15, 3614. [Google Scholar] [CrossRef] [PubMed]
- Freiría-Martínez, L.; Iglesias-Martínez-Almeida, M.; Rodríguez-Jamardo, C.; Rivera-Baltanás, T.; Comís-Tuche, M.; Rodrígues-Amorím, D.; Fernández-Palleiro, P.; Blanco-Formoso, M.; Diz-Chaves, Y.; González-Freiria, N.; et al. Human breast milk microRNAs, potential players in the regulation of nervous system. Nutrients 2023, 15, 3284. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, J.; Sun, S.; Cao, D.; Shi, H.; Loor, J.J. miR-148a and miR-17-5p synergistically regulate milk TAG synthesis via PPARGC1A and PPARA in goat mammary epithelial cells. RNA Biol. 2017, 14, 326–338. [Google Scholar] [CrossRef]
- Tan, L.; Chen, Z.; Ruan, Y.; Xu, H. Differential regulatory roles of microRNAs during intramuscular adipogenesis in Chinese Guizhou Congjiang Xiang pigs. Epigenetics 2022, 17, 1800–1819. [Google Scholar] [CrossRef] [PubMed]
- TargetScanHuman Release 8.0: Human PPARGC1A ENST00000264867.2. Available online: https://www.targetscan.org/cgi-bin/targetscan/vert_80/view_gene.cgi?rs=ENST00000264867.2&taxid=9606&members=&showcnc=0&shownc=0&showncf1=&showncf2=&subset=1#miR-148-3p/152-3p (accessed on 21 July 2024).
- Liu, C.; Li, S.; Liu, T.; Borjigin, J.; Lin, J.D. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature 2007, 447, 477–481. [Google Scholar] [CrossRef]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef]
- Gao, Q.; Zhou, L.; Yang, S.Y.; Cao, J.M. A novel role of microRNA 17-5p in the modulation of circadian rhythm. Sci. Rep. 2016, 6, 30070. [Google Scholar] [CrossRef]
- Yang, W.; Xie, L.; Wang, P.; Zhuang, C. MiR-155 regulates m6A level and cell progression by targeting FTO in clear cell renal cell carcinoma. Cell. Signal. 2022, 91, 110217. [Google Scholar] [CrossRef]
- Kondracka, A.; Gil-Kulik, P.; Kondracki, B.; Frąszczak, K.; Oniszczuk, A.; Rybak-Krzyszkowska, M.; Staniczek, J.; Kwaśniewska, A.; Kocki, J. Occurrence, role, and challenges of microRNA in human breast milk: A scoping review. Biomedicines 2023, 11, 248. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, C.; Kathale, N.D.; Liu, D.; Lee, C.; Freeman, D.A.; Hogenesch, J.B.; Cao, R.; Liu, A.C. mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet. 2018, 14, e1007369. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.; Zhong, Z.X.; Peng, Y.D.; Jiang, S.W. The emerging role of Zfp217 in adipogenesis. Int. J. Mol. Sci. 2017, 18, 1367. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.F.; Walsh, M.J.; Aguiló, F. ZNF217/ZFP217 meets chromatin and RNA. Trends Biochem. Sci. 2016, 41, 986–988. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhao, Y.; Wu, R.; Jiang, Q.; Cai, M.; Bi, Z.; Liu, Y.; Yao, Y.; Feng, J.; Wang, Y.; et al. ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3-m6A dependent manner. RNA Biol. 2019, 16, 1785–1793. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Wang, N.; Zhang, Y.; Yang, Y.; Li, S.; Zheng, R.; Chai, J.; Qiao, T.; Jiang, S. Partial deficiency of Zfp217 resists high-fat diet-induced obesity by increasing energy metabolism in mice. Int. J. Mol. Sci. 2021, 22, 5390. [Google Scholar] [CrossRef] [PubMed]
- TargetScanHuman Release 8.0: Human ZNF217 ENST00000371471.2. Available online: https://www.targetscan.org/cgi-bin/targetscan/vert_80/view_gene.cgi?rs=ENST00000371471.2&taxid=9606&showcnc=0&shownc=0&shownc_nc=&showncf1=&showncf2=&subset=1 (accessed on 21 July 2024).
- Liu, X.; Zhang, Z.; Song, Y.; Xie, H.; Dong, M. An update on brown adipose tissue and obesity intervention: Function, regulation and therapeutic implications. Front. Endocrinol. 2023, 13, 1065263. [Google Scholar] [CrossRef] [PubMed]
- Bienboire-Frosini, C.; Wang, D.; Marcet-Rius, M.; Villanueva-García, D.; Gazzano, A.; Domínguez-Oliva, A.; Olmos-Hernández, A.; Hernández-Ávalos, I.; Lezama-García, K.; Verduzco-Mendoza, A.; et al. The role of brown adipose tissue and energy metabolism in mammalian thermoregulation during the perinatal period. Animals 2023, 13, 2173. [Google Scholar] [CrossRef]
- Lidell, M.E. Brown adipose tissue in human infants. Handb. Exp. Pharmacol. 2019, 251, 107–123. [Google Scholar]
- Yulita, E.; Mappaware, N.A.; Nontji, W.; Usman, A.N. The effect of breastfeeding on body temperature, body weight, and jaundice of 0-72h old infants. Gac. Sanit. 2021, 35, S254–S257. [Google Scholar] [CrossRef]
- Su, Y.; Xu, R.; Zhang, R.; Qu, Y.; Zuo, W.; Ji, Z.; Geng, H.; Pan, M.; Ma, G. N6-methyladenosine methyltransferase plays a role in hypoxic preconditioning partially through the interaction with lncRNA H19. Acta Biochim. Biophys. Sin. 2020, 52, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.; Dhaouadi, I.; Gaziano, I.; Oliverio, M.; Klemm, P.; Awazawa, M.; Mitterer, G.; Fernandez-Rebollo, E.; Pradas-Juni, M.; Wagner, W.; et al. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat. Commun. 2018, 9, 3622. [Google Scholar] [CrossRef] [PubMed]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Wang, J.H.; Fan, W.J.; Meng, Y.T.; Li, M.M.; Li, T.T.; Cui, B.; Wang, H.F.; Zhao, Y.; An, F.; et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 2018, 37, 1062–1074, Erratum in Oncogene 2018, 37, 1119. [Google Scholar] [CrossRef] [PubMed]
- Kouwenhoven, S.M.P.; Muts, J.; Finken, M.J.J.; Goudoever, J.B.V. Low-protein infant formula and obesity risk. Nutrients 2022, 14, 2728. [Google Scholar] [CrossRef] [PubMed]
- Savitz, J. The kynurenine pathway: A finger in every pie. Mol. Psychiatry 2020, 25, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Ramprasath, T.; Wang, H.; Zou, M.H. Abnormal kynurenine pathway of tryptophan catabolism in cardiovascular diseases. Cell. Mol. Life Sci. 2017, 74, 2899–2916. [Google Scholar] [CrossRef] [PubMed]
- Mangge, H.; Summers, K.L.; Meinitzer, A.; Zelzer, S.; Almer, G.; Prassl, R.; Schnedl, W.J.; Reininghaus, E.; Paulmichl, K.; Weghuber, D.; et al. Obesity-related dysregulation of the tryptophan-kynurenine metabolism: Role of age and parameters of the metabolic syndrome. Obesity 2014, 22, 195–201. [Google Scholar] [CrossRef]
- Boulet, M.M.; Chevrier, G.; Grenier-Larouche, T.; Pelletier, M.; Nadeau, M.; Scarpa, J.; Prehn, C.; Marette, A.; Adamski, J.; Tchernof, A. Alterations of plasma metabolite profiles related to adipose tissue distribution and cardiometabolic risk. Am. J. Physiol. Endocrinol. Metab. 2015, 309, E736–E746. [Google Scholar] [CrossRef]
- Huang, T.; Song, J.; Gao, J.; Cheng, J.; Xie, H.; Zhang, L.; Wang, Y.H.; Gao, Z.; Wang, Y.; Wang, X.; et al. Adipocyte-derived kynurenine promotes obesity and insulin resistance by activating the AhR/STAT3/IL-6 signaling. Nat. Commun. 2022, 13, 3489. [Google Scholar] [CrossRef]
- Lacerda, J.Z.; Ferreira, L.C.; Lopes, B.C.; Aristizábal-Pachón, A.F.; Bajgelman, M.C.; Borin, T.F.; Zuccari, D.A.P.C. Therapeutic potential of melatonin in the regulation of miR-148a-3p and angiogenic factors in breast cancer. Microrna 2019, 8, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Hannan, F.M.; Elajnaf, T.; Vandenberg, L.N.; Kennedy, S.H.; Thakker, R.V. Hormonal regulation of mammary gland development and lactation. Nat. Rev. Endocrinol. 2023, 19, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Gutman-Ido, E.; Reif, S.; Musseri, M.; Schabes, T.; Golan-Gerstl, R. Oxytocin regulates the expression of selected colostrum-derived microRNAs. J. Pediatr. Gastroenterol. Nutr. 2022, 74, e8–e15. [Google Scholar] [CrossRef]
- Bryder, L. From breast to bottle: A history of modern infant feeding. Endeavour 2009, 33, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenberg, S.J.; Georgieff, M.K.; Committee on Nutrition. Advocacy for improving nutrition in the first 1000 days to support childhood development and adult health. Pediatrics 2018, 141, e20173716. [Google Scholar] [CrossRef] [PubMed]
- Mameli, C.; Mazzantini, S.; Zuccotti, G.V. Nutrition in the first 1000 days: The origin of childhood obesity. Int. J. Environ. Res. Public Health 2016, 13, 838. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Escamilla, R.; Tomori, C.; Hernández-Cordero, S.; Baker, P.; Barros, A.J.D.; Bégin, F.; Chapman, D.J.; Grummer-Strawn, L.M.; McCoy, D.; Menon, P.; et al. Breastfeeding: Crucially important, but increasingly challenged in a market-driven world. Lancet 2023, 401, 472–485. [Google Scholar] [CrossRef]
- Rollins, N.; Piwoz, E.; Baker, P.; Kingston, G.; Mabaso, K.M.; McCoy, D.; Ribeiro Neves, P.A.; Pérez-Escamilla, R.; Richter, L.; Russ, K.; et al. Marketing of commercial milk formula: A system to capture parents, communities, science, and policy. Lancet 2023, 401, 486–502. [Google Scholar] [CrossRef]
- Bassols, J.; Prats-Puig, A.; Vázquez-Ruíz, M.; García-González, M.M.; Martínez-Pascual, M.; Avellí, P.; Martínez-Martínez, R.; Fàbrega, R.; Colomer-Virosta, C.; Soriano-Rodríguez, P.; et al. Placental FTO expression relates to fetal growth. Int. J. Obes. 2010, 34, 1365–1370. [Google Scholar] [CrossRef]
- Capuco, A.V.; Akers, R.M. The origin and evolution of lactation. J. Biol. 2009, 8, 37. [Google Scholar] [CrossRef]
- Fewtrell, M.S.; Mohd Shukri, N.H.; Wells, J.C.K. ‘Optimising’ breastfeeding: What can we learn from evolutionary, comparative and anthropological aspects of lactation? BMC Med. 2020, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ma, L.; Zhang, H.; Cao, Y.; Wang, C.; Hou, N.; Huang, N.; von Deneen, K.M.; Zhao, C.; Shi, Y.; et al. Fto deficiency reduces anxiety- and depression-like behaviors in mice via alterations in gut microbiota. Theranostics 2019, 9, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wang, F.; Guo, H.; Cheng, J.; Du, J.; Kan, J. Gut microbiota modulates differential lipid metabolism outcomes associated with FTO gene polymorphisms in response to personalized nutrition intervention. Front. Nutr. 2022, 9, 985723. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Paz, H.A.; Sadri, M.; Cui, J.; Kachman, S.D.; Fernando, S.C.; Zempleni, J. Dietary bovine milk exosomes elicit changes in bacterial communities in C57BL/6 mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G618–G624. [Google Scholar] [CrossRef] [PubMed]
- Pammi, M.; Cope, J.; Tarr, P.I.; Warner, B.B.; Morrow, A.L.; Mai, V.; Gregory, K.E.; Kroll, J.S.; McMurtry, V.; Ferris, M.J.; et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: A systematic review and meta-analysis. Microbiome 2017, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, X.; Yan, X.; Yu, Z.; Zhang, J.; Han, S. The emerging role of exosomes in the pathogenesis, prognosis and treatment of necrotizing enterocolitis. Am. J. Transl. Res. 2020, 12, 7020–7033. [Google Scholar] [PubMed]
- Yan, X.; Liu, L.; Yao, S.; Chen, Y.; Yu, Q.; Jiang, C.; Chen, W.; Chen, X.; Han, S. LncRNA and mRNA profiles of human milk-derived exosomes and their possible roles in protecting against necrotizing enterocolitis. Food Funct. 2022, 13, 12953–12965. [Google Scholar] [CrossRef]
- Stremmel, W.; Weiskirchen, R.; Melnik, B.C. Milk exosomes prevent intestinal inflammation in a genetic mouse model of ulcerative colitis: A pilot experiment. Inflamm. Intest. Dis. 2020, 5, 117–123. [Google Scholar] [CrossRef]

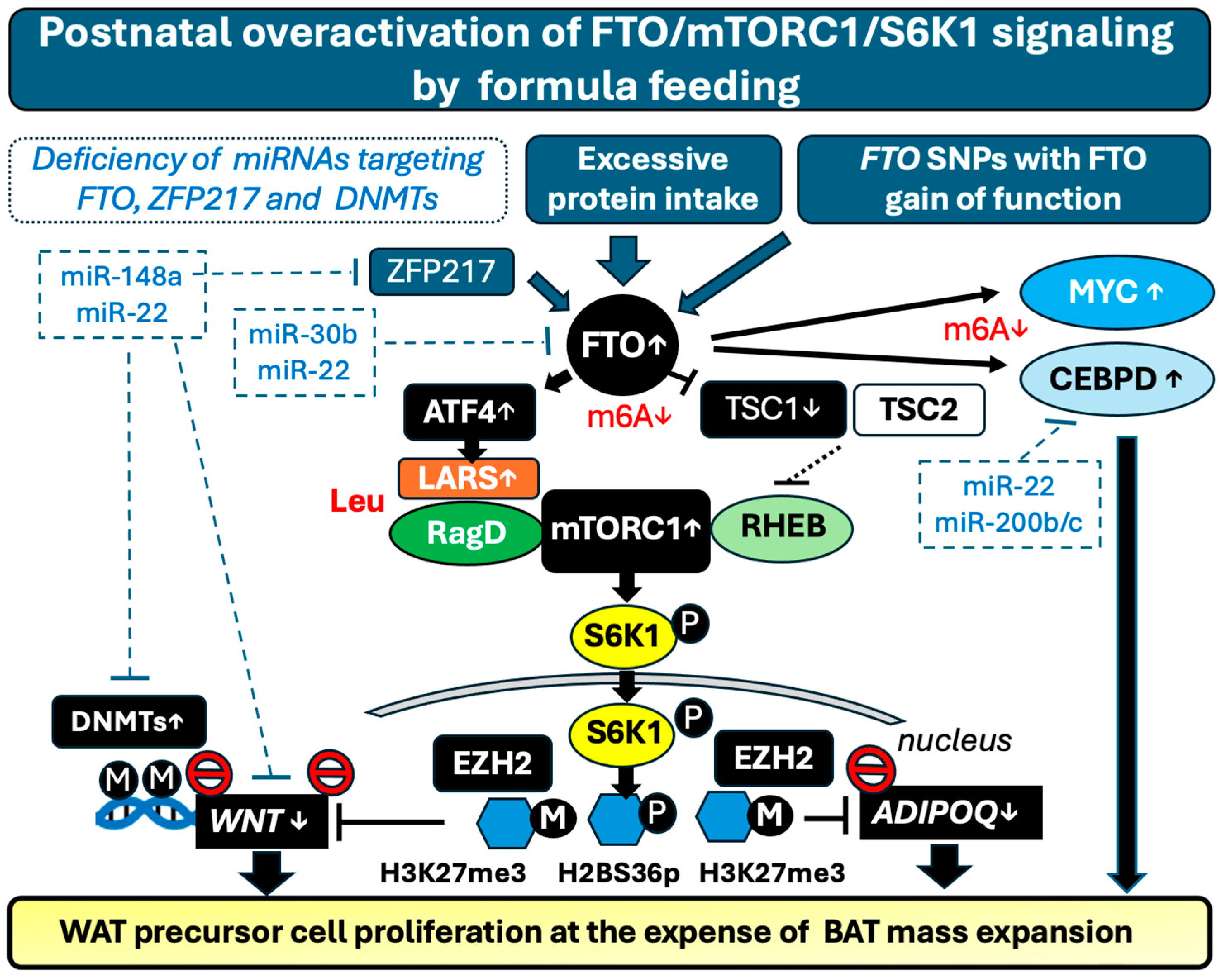

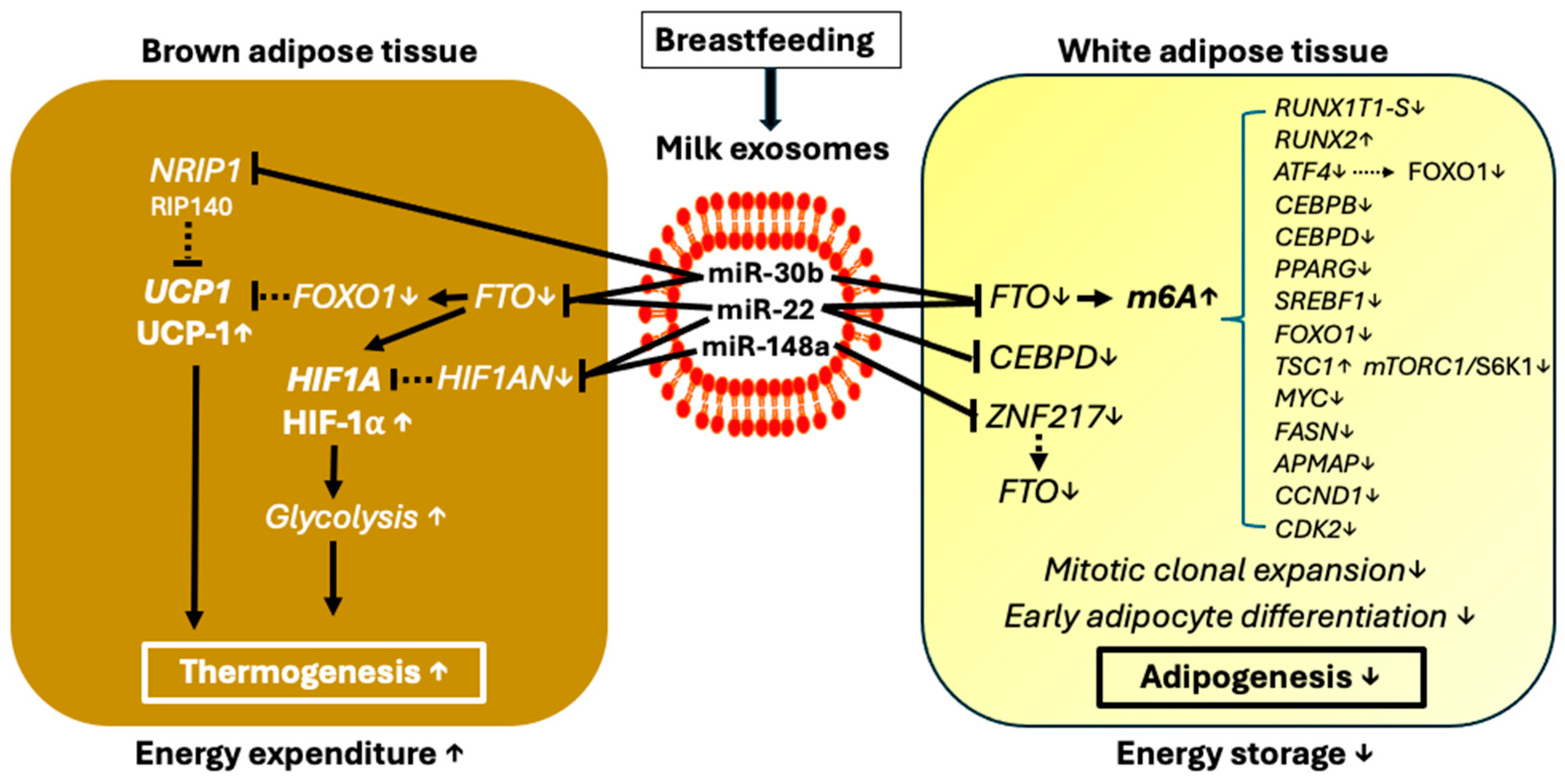
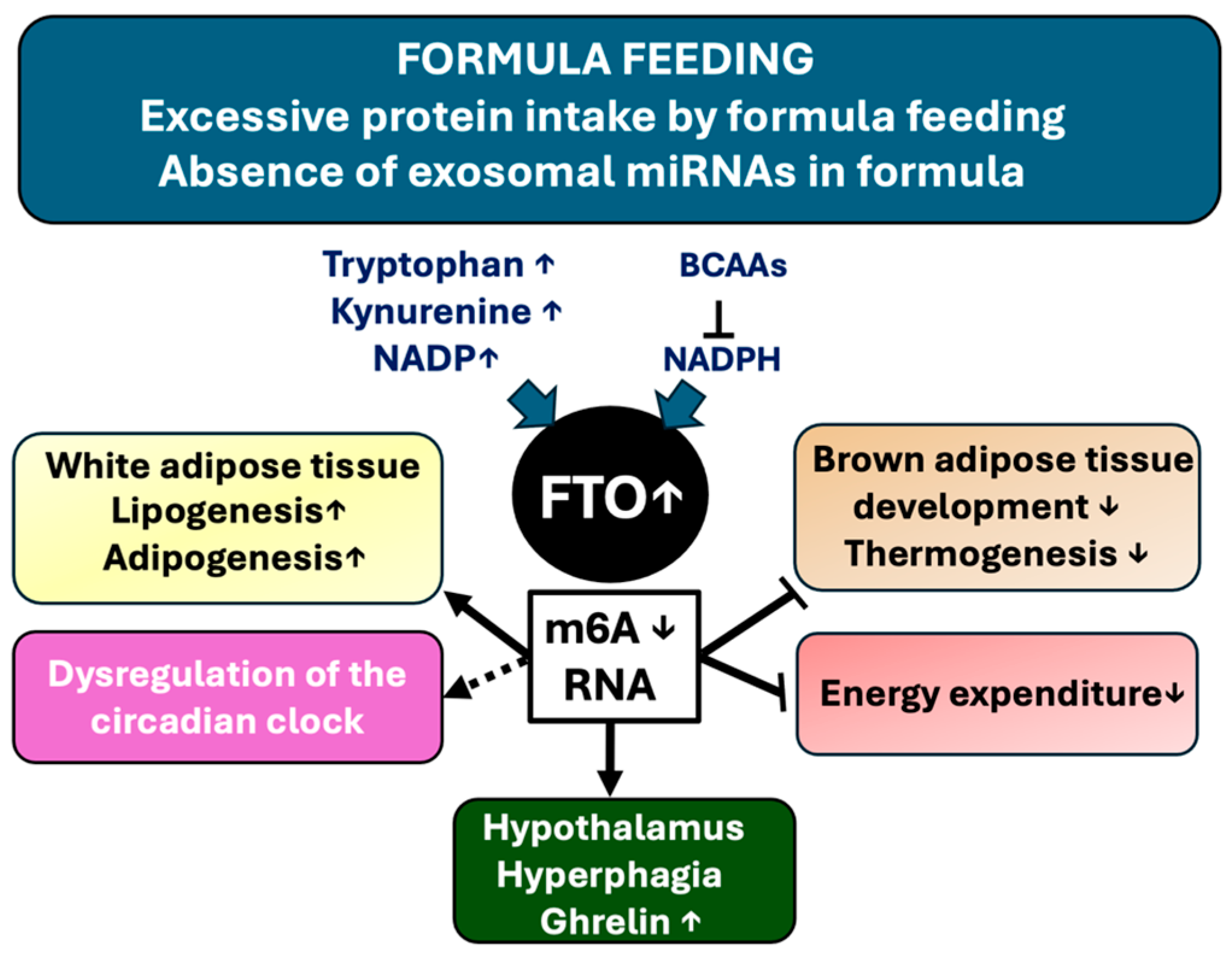
| Cell Type | Finding | Reference |
|---|---|---|
| Adipocytes | Inverse correlation during adipogenesis between m6A RNA levels and FTO gene expression. The adipogenic role of FTO is due to the splicing of RUNX1T1 to the short splice variant RUNX1T1-S | [61] |
| The adipogenic effect of FTO is transmitted by mitotic clonal expansion (MCE), an early stage in adipogenesis | [85] | |
| The demethylase activity of FTO is required for preadipocyte differentiation | [86] | |
| FTO enhances catenin (CTNNB) expression, promoting lipogenesis in a m6A demethylation-dependent manner | [87] | |
| FTO promotes adipogenesis and maintenance of lipid content in mature adipocytes by enabling CEBPB-driven transcription and expression of CEBPD | [88] | |
| FTO promotes adipocyte differentiation via -adrenergic receptor (ADRB1) gene regulation through m6A modification in Hycole rabbits | [89] | |
| FTO SNP rs1421085 increases the number of white adipocytes and lipid storage but reduces mitochondrial thermogenesis | [90] | |
| FTO regulates autophagy and adipogenesis | [91] | |
| Zinc finger protein 217 (ZFP217) stimulates adipogenesis via activation of FTO transcription and by posttranscriptional interaction with YTHDF2 | [92] | |
| NADP binds to the FTO protein, increasing its activity and thereby promoting m6A demethylation and adipogenesis | [93] | |
| FTO promotes the expression of the adipocyte plasma membrane-associated protein (APMAP) gene by YTHDF2 recognition, stimulating adipocyte differentiation | [94] | |
| Via m6A modification, FTO upregulates MYC, a critical transcription factor promoting multipotent adipocyte stem cells to the adipogenic lineage | [95,96] | |
| FTO knockdown shows that increased mRNA m6A methylation downregulates porcine adipogenesis | [97] | |
| Epigallocatechin gallate-mediated FTO inhibition suppresses adipogenesis by inhibiting MCE in an m6A-YTHDF2-dependent manner | [98,99] | |
| FTO deficiency promotes thermogenesis and the transition of white-to-beige adipocytes via YTHDC2-mediated translation and increased expression of hypoxia-inducible factor-1 (HIF1A) | [100] | |
| FTO deficiency in mice modifies gene and miRNA expression involved in brown adipogenesis and browning of WAT | [101] | |
| FTO deficiency upregulates uncoupling protein 1 (UCP1) and subsequently enhances mitochondrial uncoupling and energy expenditure, resulting in the induction of the brown adipocyte phenotype | [102] | |
| Metformin inhibits FTO and counteracts high-fat diet (HFD)-induced obesity in mice in an m6A-YTHDF2-dependent manner | [103] | |
| Hepatocytes | FTO enhances lipid accumulation in hepatocytes by upregulation of the nuclear translocation and maturation of SREBP1c, thereby enhancing the transcriptional activity of the lipid droplet-associated protein CIDEC | [104] |
| FTO-mediated RNA demethylation reduces mitochondria numbers and stimulates hepatic fat accumulation | [105] | |
| Elevation of FTO stimulates liver steatosis by m6A demethylation and increases the stability of sterol regulatory element-binding transcription factor 1 (SREBF1) and carbohydrate response element-binding protein (CHREBP; MLXIPL) mRNAs | [106] | |
| FTO promotes hepatic gluconeogenesis and lipid accumulation by increasing the expression of activating transcription factor 4 (ATF4), which works in conjunction with forkhead box O1 (FoxO1) | [107] | |
| Glucocorticoid-mediated FTO transactivation enhances lipid accumulation in hepatocytes by m6A demethylation of lipogenic mRNAs | [108] | |
| FTO induces hepatic lipogenesis through an m6A demethylation-dependent increase of fatty acid synthase (FASN) mRNA | [109] | |
| Entacapone, a chemical FTO inhibitor, mediates metabolic regulation through FOXO1 in HepG2 cells. FTO via m6A demethylation upregulates FOXO1 expression | [110] | |
| Skeletal muscle cells | AMP-activated protein kinase (AMPK) stimulates lipid accumulation in skeletal muscle cells through FTO-dependent m6A demethylation | [111] |
| Human endothelial cells | Atorvastatin reduces FTO protein expression via a geranylgeranyl pyrophosphate–dependent and proteasome-dependent mechanism. After FTO silencing, KrüKrüüppel-like factor 2 (KLF2) transcripts with higher levels of m6A modification in their 3′ untranslated regions are captured by YTHDF3 stabilizing KLF2 mRNA, increasing KLF2 expression. KLF2 is a neg-ative regulator of PPAR- and adipogenesis, a potential explanation for atorvastatin-induced weight loss | [112,113,114] |
| Blood mononuclear cells | FTO reduces ghrelin m6A levels and subsequently enhances ghrelin (GHRL) mRNA abundance in FTO SNP rs9939609 (risk allele A) carriers | [73] |
| Embryonic stem cells | FTO demethylates the m6A level of long-interspersed element-1 (LINE1) RNA in mouse embryonic stem cells, enhancing LINE1 RNA abundance and modifying the local chromatin state, which changes the transcription of LINE1-containing genes | [115] |
| Bone marrow mesenchymal stem cells | FTO overexpression decreases the m6A RNA and total level of runt-related transcription factor 2 (RUNX2) mRNA, suppressing osteogenic differentiation. Decreased expression of RUNX2 promotes adipogenic differentiation. In contrast, intake of cow’s milk including bovine milk miRNAs increases RUNX2 expression in peripheral blood mononuclear cells of healthy adult human volunteers | [116,117,118,119,120,121] |
| Genes upregulated by FTO-mediated m6A mRNA demethylation: |
|
| Genes upregulated by suppression of FTO-mediated m6A mRNA demethylation: |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melnik, B.C.; Weiskirchen, R.; Stremmel, W.; John, S.M.; Schmitz, G. Risk of Fat Mass- and Obesity-Associated Gene-Dependent Obesogenic Programming by Formula Feeding Compared to Breastfeeding. Nutrients 2024, 16, 2451. https://doi.org/10.3390/nu16152451
Melnik BC, Weiskirchen R, Stremmel W, John SM, Schmitz G. Risk of Fat Mass- and Obesity-Associated Gene-Dependent Obesogenic Programming by Formula Feeding Compared to Breastfeeding. Nutrients. 2024; 16(15):2451. https://doi.org/10.3390/nu16152451
Chicago/Turabian StyleMelnik, Bodo C., Ralf Weiskirchen, Wolfgang Stremmel, Swen Malte John, and Gerd Schmitz. 2024. "Risk of Fat Mass- and Obesity-Associated Gene-Dependent Obesogenic Programming by Formula Feeding Compared to Breastfeeding" Nutrients 16, no. 15: 2451. https://doi.org/10.3390/nu16152451








