Multi-Omics Profiles of Small Intestine Organoids in Reaction to Breast Milk and Different Infant Formula Preparations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Breast Milk Donors
2.2. Breast Milk Collection
2.3. Infant Formulas
2.4. Induced Differentiation of Small Intestine Organoids
2.5. In Vitro Digestion Simulation of Milk
2.6. Establishment of the Absorption Metabolism Model
2.7. RNA Sequencing
2.8. Untargeted Metabolite Sequencing
2.9. Data Analysis of Transcriptome
2.10. Data Analysis of Untargeted Metabolomics
2.11. Data Processing and Statistical Analysis
2.12. RT-qPCR
3. Results
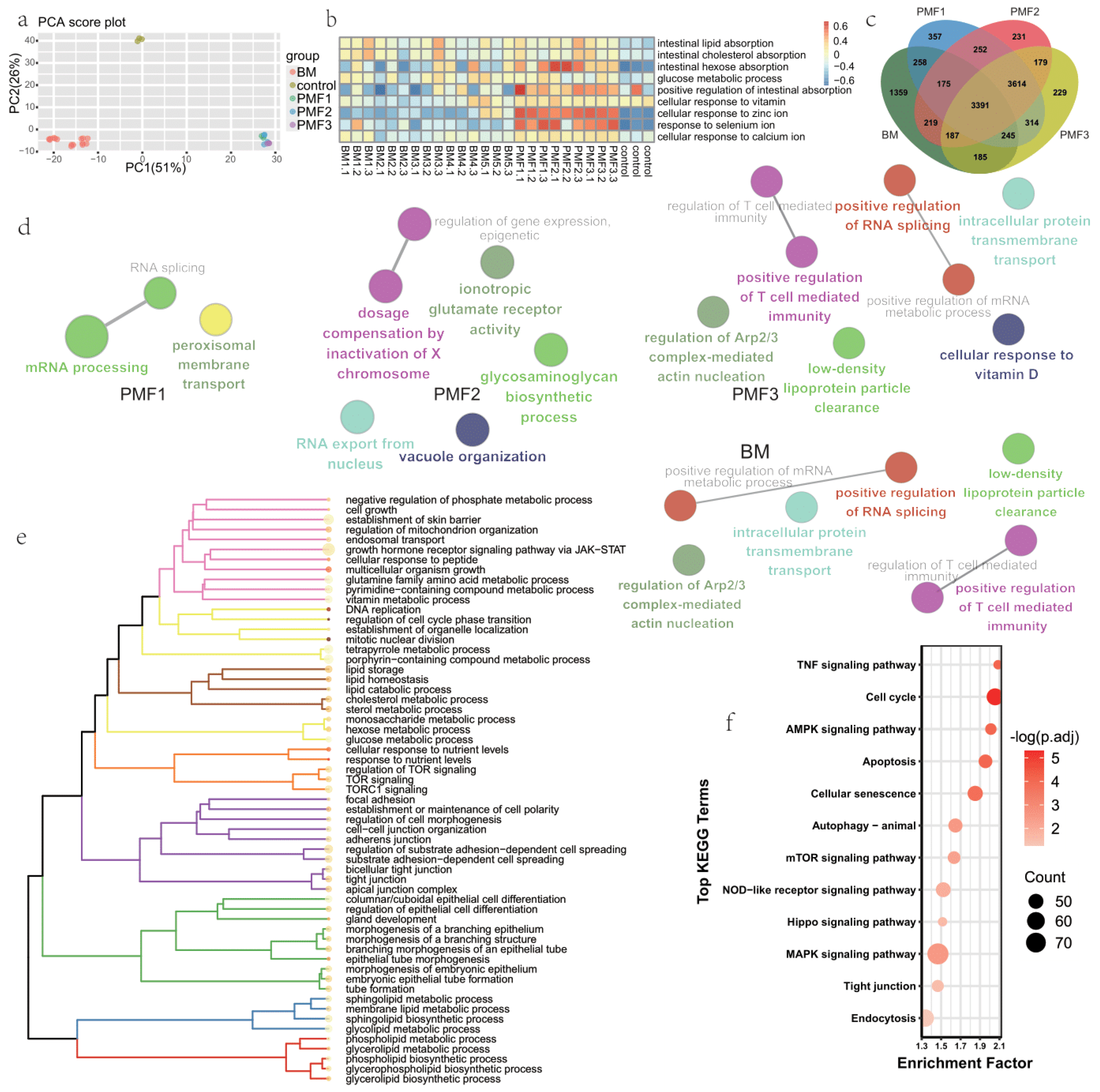
3.1. Transcriptome Profiles of Organoids Feeding by Breast Milk and Different Infant Formulas
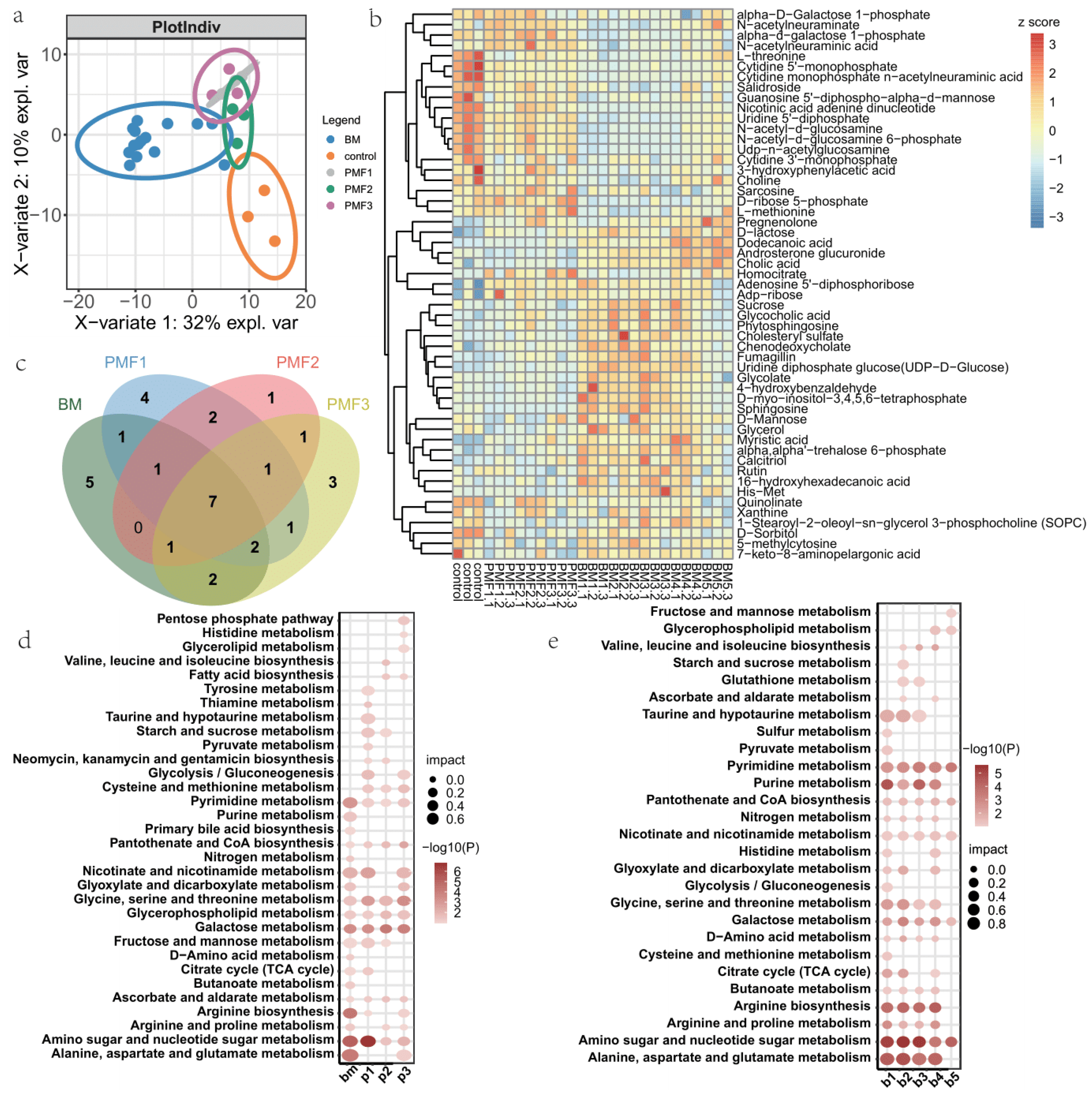
3.2. Metabolite Profiles of Breast Milk and Different Infant Formulas Treated Cells
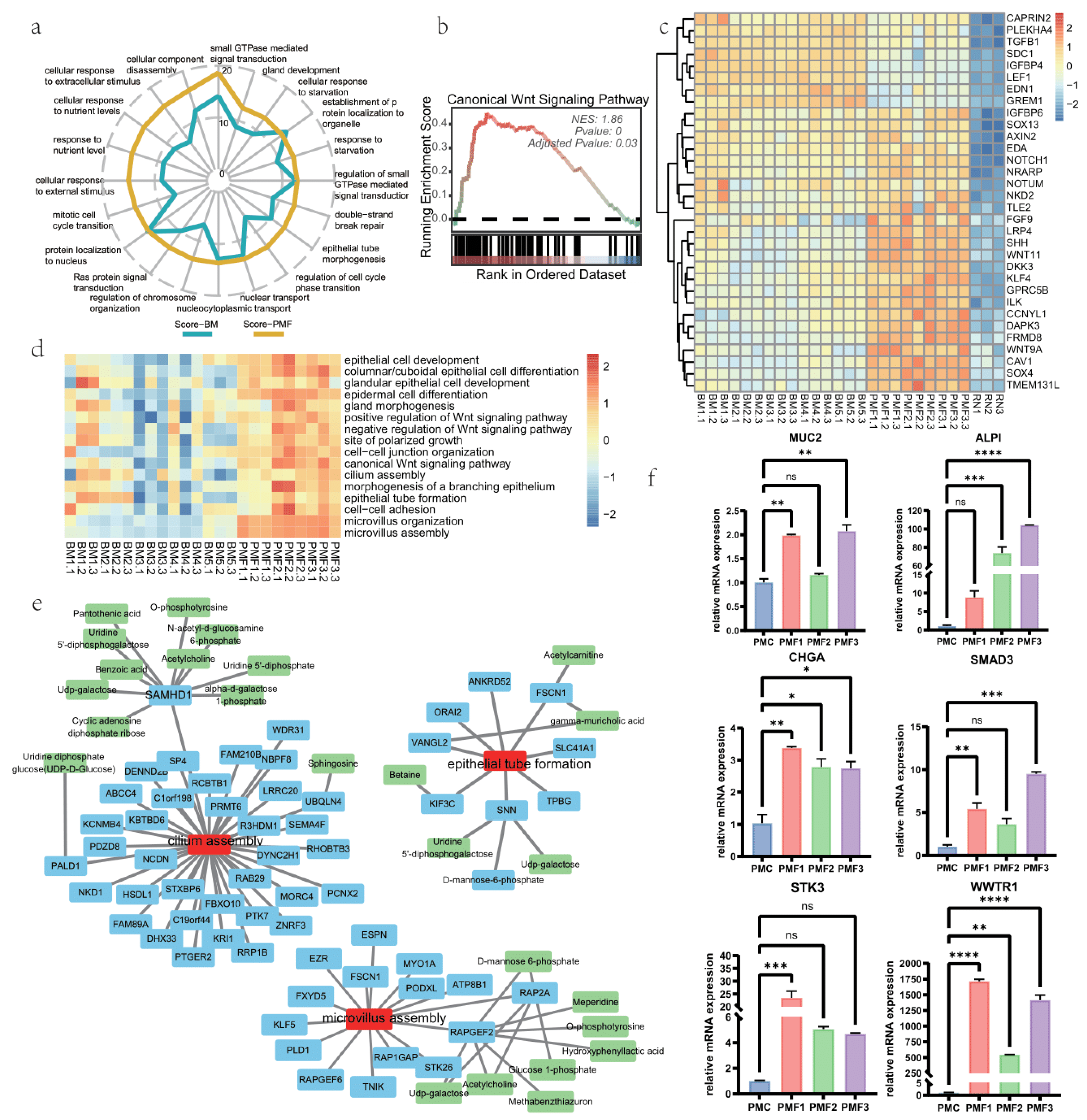
3.3. Pro-Development Effects of Breast Milk and Different Infant Formulae on Intestine Organoid
3.4. Effects on Cell Junction of Different Infant Formulae and Breast Milk
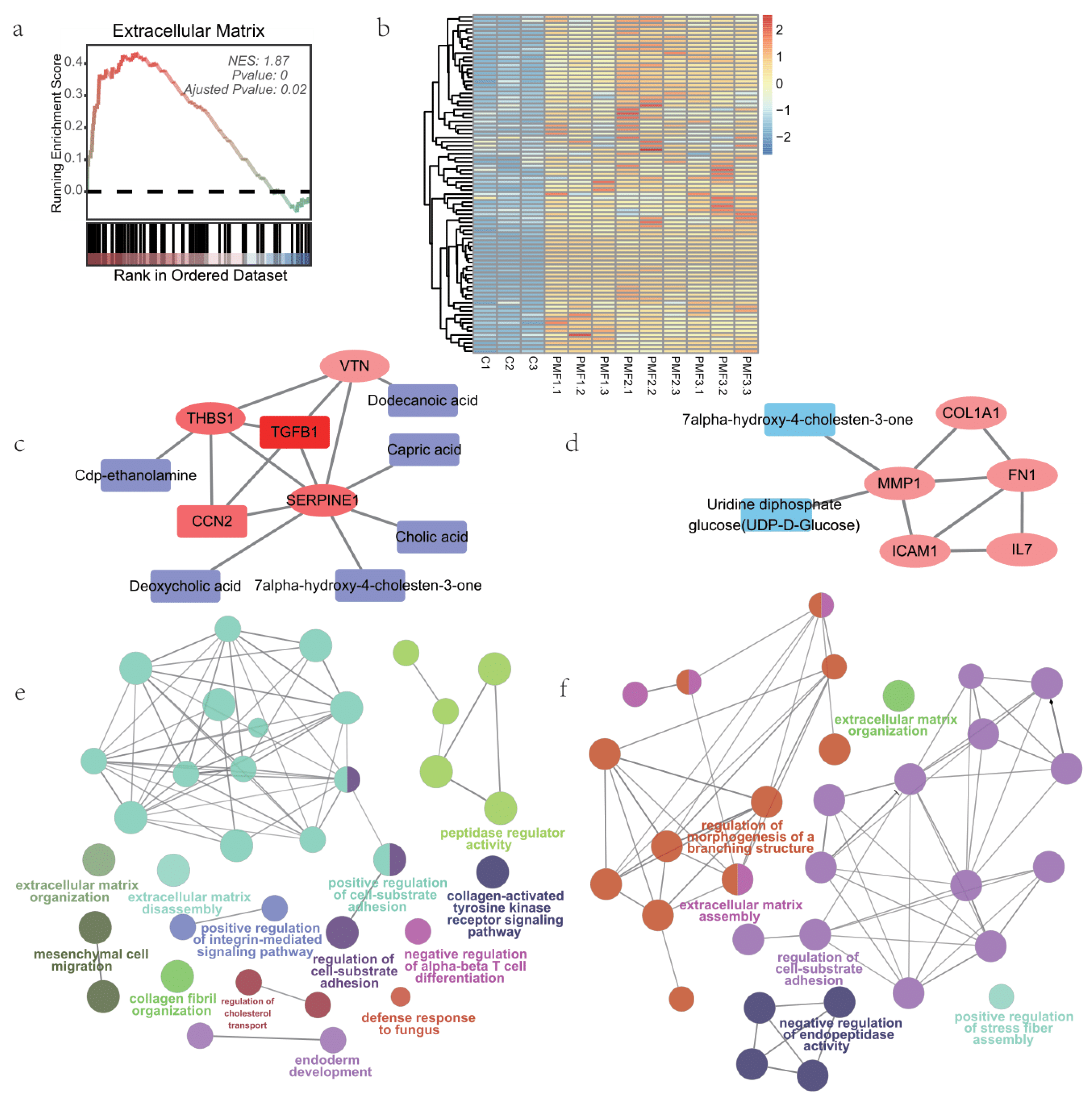
3.5. Extracellular Matrix Processes of Small Intestine Organoids Feeding by Different Infant Formulae and Breast Milk
4. Discussion
4.1. Application of Induced Pluripotent Stem Cell-Derived Organoid Models
4.2. Nutritional Absorption Effects Revealed by Transcriptomics and Metabolomics of Formula Feeding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ESC | embryo stem cell |
| FA% | % of fatty acids |
| DHA | Docosahexaenoic acid |
| ARA | arachidonic acid |
| CACO-2 | a widely used human colorectal adenocarcinoma cell line, simulating intestine epithelial cells |
| BM | breast milk |
| PMF1, PMF2, PMF3 (p1, p2, p3) | experimental groups in our study, representing infant formulas groups |
| BM1~5 (b1~5) | experimental groups in our study, representing breast milk groups |
| DEG | differential expressed genes |
| GSVA | Gene Set Variation Analysis |
| GSEA | Gene Set Enrichment analysis |
| KEGG | kyoto encyclopedia of genes and genomes |
| GO | gene ontology |
| IPA | Ingenuity Pathway Analysis, a software application for analyzing omics data |
| STRING | a database of functional protein association networks |
| OPO | 1,3-Dioleoyl-2-palmitoyl glycerol |
| ECM | extracellular matrix |
| HMO | Human Milk Oligosaccharides |
| SEM | standard error of the mean |
| NES | normalized enrichment score |
References
- Lanigan, J.; Singhal, A. Early nutrition and long-term health: A practical approach: Symposium on ‘Early nutrition and later disease: Current concepts, research and implications’. Proc. Nutr. Soc. 2009, 68, 422–429. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Nutrition Targets 2025: Breastfeeding Policy Brief (WHO/NMH/NHD14. 7); World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Hennet, T.; Borsig, L. Breastfed at Tiffany’s. Trends Biochem. Sci. 2016, 41, 508–518. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on the essential composition of infant and follow-on formulae. EFSA J. 2014, 12, 3760. [Google Scholar] [CrossRef]
- Lordan, C.; Roche, A.K.; Delsing, D.; Nauta, A.; Groeneveld, A.; MacSharry, J.; Cotter, P.D.; van Sinderen, D. Linking human milk oligosaccharide metabolism and early life gut microbiota: Bifidobacteria and beyond. Microbiol. Mol. Biol. Rev. 2024, 88, e0009423. [Google Scholar] [CrossRef] [PubMed]
- Tenenbaum Weiss, Y.; Ovnat Tamir, S.; Globus, O.; Marom, T. Protective Characteristics of Human Breast Milk on Early Childhood Otitis Media: A Narrative Review. Breastfeed Med. 2024, 19, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Nucci, A.M.; Virtanen, S.M.; Becker, D.J. Infant feeding and timing of complementary foods in the development of type 1 diabetes. Curr. Diabetes Rep. 2015, 15, 62. [Google Scholar] [CrossRef]
- Gabrielli, O.; Zampini, L.; Galeazzi, T.; Padella, L.; Santoro, L.; Peila, C.; Giuliani, F.; Bertino, E.; Fabris, C.; Coppa, G.V. Preterm milk oligosaccharides during the first month of lactation. Pediatrics 2011, 128, e1520–e1531. [Google Scholar] [CrossRef]
- Belderbos, M.; Houben, M.; Van Bleek, G.; Schuijff, L.; Van Uden, N.; Bloemen-Carlier, E.; Kimpen, J.; Eijkemans, M.; Rovers, M.; Bont, L. Breastfeeding modulates neonatal innate immune responses: A prospective birth cohort study. Pediatr. Allergy Immunol. 2012, 23, 65–74. [Google Scholar] [CrossRef]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef]
- Green Corkins, K.; Shurley, T. What’s in the bottle? A review of infant formulas. Nutr. Clin. Pract. 2016, 31, 723–729. [Google Scholar] [CrossRef]
- World Health Organization. Implementation Guidance: Protecting, Promoting, and Supporting Breastfeeding in Facilities Providing Maternity and Newborn Services: The Revised Baby-Friendly Hospital Initiative 2018; World Health Organization & United Nations Children’s Fund (UNICEF): Geneve, Switzerland, 2018. [Google Scholar]
- Barreiro, R.; Regal, P.; Díaz-Bao, M.; Fente, C.A.; Cepeda, A. Analysis of Naturally Occurring Steroid Hormones in Infant Formulas by HPLC-MS/MS and Contribution to Dietary Intake. Foods 2015, 4, 605–621. [Google Scholar] [CrossRef] [PubMed]
- Lönnerdal, B. Infant formula and infant nutrition: Bioactive proteins of human milk and implications for composition of infant formulas. Am. J. Clin. Nutr. 2014, 99, 712S–717S. [Google Scholar] [CrossRef] [PubMed]
- Lönnerdal, B.; Hernell, O. An opinion on “staging” of infant formula: A developmental perspective on infant feeding. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.R.; Ling, P.R.; Blackburn, G.L. Review of Infant Feeding: Key Features of Breast Milk and Infant Formula. Nutrients 2016, 8, 279. [Google Scholar] [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Underwood, M.A.; Dallas, D.C. Differences and Similarities in the Peptide Profile of Preterm and Term Mother’s Milk, and Preterm and Term Infant Gastric Samples. Nutrients 2020, 12, 2825. [Google Scholar] [CrossRef]
- Bourlieu, C.; Ménard, O.; Bouzerzour, K.; Mandalari, G.; Macierzanka, A.; Mackie, A.R.; Dupont, D. Specificity of infant digestive conditions: Some clues for developing relevant in vitro models. Crit. Rev. Food Sci. Nutr. 2014, 54, 1427–1457. [Google Scholar] [CrossRef]
- Ahern, G.J.; Hennessy, A.A.; Ryan, C.A.; Ross, R.P.; Stanton, C. Advances in Infant Formula Science. Annu. Rev. Food Sci. Technol. 2019, 10, 75–102. [Google Scholar] [CrossRef]
- Hernell, O. Human milk vs. cow’s milk and the evolution of infant formulas. Nestle Nutr. Workshop Ser. Pediatr. Program 2011, 67, 17–28. [Google Scholar] [CrossRef]
- Akkerman, R.; Faas, M.M.; de Vos, P. Non-digestible carbohydrates in infant formula as substitution for human milk oligosaccharide functions: Effects on microbiota and gut maturation. Crit. Rev. Food Sci. Nutr. 2019, 59, 1486–1497. [Google Scholar] [CrossRef]
- Farquharson, J.; Jamieson, E.; Logan, R.; Cockburn, F.; Patrick, W.A. Infant cerebral cortex phospholipid fatty-acid composition and diet. Lancet 1992, 340, 810–813. [Google Scholar] [CrossRef]
- Lauritzen, L.; Brambilla, P.; Mazzocchi, A.; Harsløf, L.B.S.; Ciappolino, V.; Agostoni, C. DHA Effects in Brain Development and Function. Nutrients 2016, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Hadley, K.B.; Ryan, A.S.; Forsyth, S.; Gautier, S.; Salem, N. The Essentiality of Arachidonic Acid in Infant Development. Nutrients 2016, 8, 216. [Google Scholar] [CrossRef]
- Rumbo, M.; Schiffrin, E.J. Intestinal epithelial barrier and mucosal immunity: Ontogeny of intestinal epithelium immune functions: Developmental and environmental regulation. Cell. Mol. Life Sci. 2005, 62, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.; Ahluwalia, A. Advances and current challenges in intestinal in vitro model engineering: A digest. Front. Bioeng. Biotechnol. 2019, 7, 144. [Google Scholar] [CrossRef] [PubMed]
- Pageot, L.P.; Perreault, N.; Basora, N.; Francoeur, C.; Magny, P.; Beaulieu, J.F. Human cell models to study small intestinal functions: Recapitulation of the crypt-villus axis. Microsc. Res. Tech. 2000, 49, 394–406. [Google Scholar] [CrossRef]
- Hernandez-Gordillo, V.; Koppes, A.N.; Griffith, L.G.; Breault, D.T.; Carrier, R.L. Engineering the Niche for Intestinal Regeneration. In Biology and Engineering of Stem Cell Niches; Elsevier: Amsterdam, The Netherlands, 2017; pp. 601–615. [Google Scholar]
- Perreault, N.; Beaulieu, J.-F. Use of the dissociating enzyme thermolysin to generate viable human normal intestinal epithelial cell cultures. Exp. Cell Res. 1996, 224, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Fleisher, D. Transport processes in pharmaceutical systems. Inf. Healthc. 1999, 147–184. [Google Scholar] [CrossRef]
- Woodcock, J.; Woosley, R. The FDA critical path initiative and its influence on new drug development. Annu. Rev. Med. 2008, 59, 1–12. [Google Scholar] [CrossRef]
- Zachos, N.C.; Kovbasnjuk, O.; Foulke-Abel, J.; In, J.; Blutt, S.E.; de Jonge, H.R.; Estes, M.K.; Donowitz, M. Human Enteroids/Colonoids and Intestinal Organoids Functionally Recapitulate Normal Intestinal Physiology and Pathophysiology. J. Biol. Chem. 2016, 291, 3759–3766. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Hrvatin, S.; O’Donnell, C.W.; Deng, F.; Millman, J.R.; Pagliuca, F.W.; DiIorio, P.; Rezania, A.; Gifford, D.K.; Melton, D.A. Differentiated human stem cells resemble fetal, not adult, β cells. Proc. Natl. Acad. Sci. USA 2014, 111, 3038–3043. [Google Scholar] [CrossRef]
- Camp, J.G.; Badsha, F.; Florio, M.; Kanton, S.; Gerber, T.; Wilsch-Bräuninger, M.; Lewitus, E.; Sykes, A.; Hevers, W.; Lancaster, M.; et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 2015, 112, 15672–15677. [Google Scholar] [CrossRef] [PubMed]
- Dye, B.R.; Hill, D.R.; Ferguson, M.A.; Tsai, Y.H.; Nagy, M.S.; Dyal, R.; Wells, J.M.; Mayhew, C.N.; Nattiv, R.; Klein, O.D.; et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 2015, 4, 05098. [Google Scholar] [CrossRef] [PubMed]
- Gerli, M.F.M.; Calà, G.; Beesley, M.A.; Sina, B.; Tullie, L.; Sun, K.Y.; Panariello, F.; Michielin, F.; Davidson, J.R.; Russo, F.M.; et al. Single-cell guided prenatal derivation of primary fetal epithelial organoids from human amniotic and tracheal fluids. Nat. Med. 2024, 30, 875–887. [Google Scholar] [CrossRef]
- Spitzer, J.; Buettner, A. Characterization of aroma changes in human milk during storage at −19 °C. Food Chem. 2010, 120, 240–246. [Google Scholar] [CrossRef]
- Yu, M.; Xie, Q.; Song, H.; Wang, L.; Sun, H.; Jiang, S.; Zhang, Y.; Zheng, C. Characterization of the odor compounds in human milk by DHS/GC × GC-O-MS: A feasible and efficient method. Food Res. Int. 2023, 174, 113597. [Google Scholar] [CrossRef]
- D’Amour, K.A.; Agulnick, A.D.; Eliazer, S.; Kelly, O.G.; Kroon, E.; Baetge, E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005, 23, 1534–1541. [Google Scholar] [CrossRef]
- Zhang, X.; McGrath, P.S.; Salomone, J.; Rahal, M.; McCauley, H.A.; Schweitzer, J.; Kovall, R.; Gebelein, B.; Wells, J.M. A Comprehensive Structure-Function Study of Neurogenin3 Disease-Causing Alleles during Human Pancreas and Intestinal Organoid Development. Dev. Cell 2019, 50, 367–380.e367. [Google Scholar] [CrossRef]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Marini, F.; Ludt, A.; Linke, J.; Strauch, K. GeneTonic: An R/Bioconductor package for streamlining the interpretation of RNA-seq data. BMC Bioinform. 2021, 22, 610. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; KA, L.C. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef] [PubMed]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-L.; Narvaez-Ortiz, H.Y.; Miner, M.; Kiemel, J.; Oberhelman, N.; Watt, A.; Wagner, A.R.; Luan, Q.; Helgeson, L.A.; Nolen, B.J. Analysis of functional surfaces on the actin nucleation promoting factor Dip1 required for Arp2/3 complex activation and endocytic actin network assembly. J. Biol. Chem. 2022, 298, 102019. [Google Scholar] [CrossRef]
- Lee, J.S.; Wang, R.X.; Alexeev, E.E.; Lanis, J.M.; Battista, K.D.; Glover, L.E.; Colgan, S.P. Hypoxanthine is a checkpoint stress metabolite in colonic epithelial energy modulation and barrier function. J. Biol. Chem. 2018, 293, 6039–6051. [Google Scholar] [CrossRef]
- Onozato, D.; Ogawa, I.; Kida, Y.; Mizuno, S.; Hashita, T.; Iwao, T.; Matsunaga, T. Generation of budding-like intestinal organoids from human induced pluripotent stem cells. J. Pharm. Sci. 2021, 110, 2637–2650. [Google Scholar] [CrossRef] [PubMed]
- Clinton, J.; McWilliams-Koeppen, P. Initiation, expansion, and cryopreservation of human primary tissue-derived normal and diseased organoids in embedded three-dimensional culture. Curr. Protoc. Cell Biol. 2019, 82, e66. [Google Scholar] [CrossRef]
- Li, V.S.W. Modelling intestinal inflammation and infection using ‘mini-gut’ organoids. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 89–90. [Google Scholar] [CrossRef]
- Dou, Y.; Pizarro, T.; Zhou, L. Organoids as a Model System for Studying Notch Signaling in Intestinal Epithelial Homeostasis and Intestinal Cancer. Am. J. Pathol. 2022, 192, 1347–1357. [Google Scholar] [CrossRef]
- Merenda, A.; Fenderico, N.; Maurice, M.M. Wnt Signaling in 3D: Recent Advances in the Applications of Intestinal Organoids. Trends Cell Biol. 2020, 30, 60–73. [Google Scholar] [CrossRef]
- Zietek, T.; Rath, E.; Haller, D.; Daniel, H. Intestinal organoids for assessing nutrient transport, sensing and incretin secretion. Sci. Rep. 2015, 5, 16831. [Google Scholar] [CrossRef]
- Angus, H.C.K.; Butt, A.G.; Schultz, M.; Kemp, R.A. Intestinal Organoids as a Tool for Inflammatory Bowel Disease Research. Front. Med. 2019, 6, 334. [Google Scholar] [CrossRef] [PubMed]
- Yung, C.; Zhang, Y.; Kuhn, M.; Armstrong, R.J.; Olyaei, A.; Aloia, M.; Scottoline, B.; Andres, S.F. Neonatal enteroids absorb extracellular vesicles from human milk-fed infant digestive fluid. J. Extracell. Vesicles 2024, 13, e12422. [Google Scholar] [CrossRef]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Martinez-Silgado, A.; Akkerman, N.; Saftien, A.; Boot, C.; de Waal, A.; Beumer, J.; Dutta, D.; Heo, I.; et al. Intestinal organoid cocultures with microbes. Nat. Protoc. 2021, 16, 4633–4649. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Zhu, Y.; Ni, D.; Chen, J.; Zhang, W.; Mu, W. Infant formulae—Key components, nutritional value, and new perspectives. Food Chem. 2023, 424, 136393. [Google Scholar] [CrossRef]
- Ghide, M.K.; Yan, Y. 1,3-Dioleoyl-2-palmitoyl glycerol (OPO)-Enzymatic synthesis and use as an important supplement in infant formulas. J. Food Biochem. 2021, 45, e13799. [Google Scholar] [CrossRef]
- Aumeistere, L.; Ciproviča, I.; Zavadska, D.; Bavrins, K.; Borisova, A. Zinc Content in Breast Milk and Its Association with Maternal Diet. Nutrients 2018, 10, 1438. [Google Scholar] [CrossRef]
- Bae, Y.J.; Kratzsch, J. Vitamin D and calcium in the human breast milk. Best Pract. Res. Clin. Endocrinol. Metab. 2018, 32, 39–45. [Google Scholar] [CrossRef]
- Comparing Infant Formulas with Human Milk. In Infant Formula: Evaluating the Safety of New Ingredients; Institute of Medicine (US) Committee on the Evaluation of the Addition of Ingredients New to Infant Formula, National Academies Press (US): Washington, DC, USA, 2004. Available online: https://www.ncbi.nlm.nih.gov/books/NBK215837/ (accessed on 15 June 2024).
- Allgrove, J. Physiology of calcium, phosphate, magnesium and vitamin D. Calcium Bone Disord. Child. Adolesc. 2015, 28, 7–32. [Google Scholar]
- Nighot, P.; Ma, T. Endocytosis of Intestinal Tight Junction Proteins: In Time and Space. Inflamm. Bowel Dis. 2021, 27, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Rowart, P.; Jouret, F.; Gassaway, B.M.; Rajendran, V.; Rinehart, J.; Caplan, M.J. Mechanisms involved in AMPK-mediated deposition of tight junction components to the plasma membrane. Am. J. Physiol.-Cell Physiol. 2020, 318, C486–C501. [Google Scholar] [CrossRef]
- Sheppe, A.E.F.; Edelmann, M.J. Roles of Eicosanoids in Regulating Inflammation and Neutrophil Migration as an Innate Host Response to Bacterial Infections. Infect Immun. 2021, 89, e0009521. [Google Scholar] [CrossRef] [PubMed]
- Peters, E.; Mehta, R.L.; Murray, P.T.; Hummel, J.; Joannidis, M.; Kellum, J.A.; Arend, J.; Pickkers, P. Study protocol for a multicentre randomised controlled trial: Safety, Tolerability, efficacy and quality of life Of a human recombinant alkaline Phosphatase in patients with sepsis-associated Acute Kidney Injury (STOP-AKI). BMJ Open 2016, 6, e012371. [Google Scholar] [CrossRef] [PubMed]
- Lukas, M.; Drastich, P.; Konecny, M.; Gionchetti, P.; Urban, O.; Cantoni, F.; Bortlik, M.; Duricova, D.; Bulitta, M. Exogenous alkaline phosphatase for the treatment of patients with moderate to severe ulcerative colitis. Inflamm. Bowel Dis. 2010, 16, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Hamarneh, S.R.; Mohamed, M.M.R.; Economopoulos, K.P.; Morrison, S.A.; Phupitakphol, T.; Tantillo, T.J.; Gul, S.S.; Gharedaghi, M.H.; Tao, Q.; Kaliannan, K.; et al. A novel approach to maintain gut mucosal integrity using an oral enzym e supplement. Ann. Surg. 2014, 260, 706–714, discussion 714–715. [Google Scholar] [CrossRef]
- Cario, E. Bacterial interactions with cells of the intestinal mucosa: Toll-like receptors and NOD2. Gut 2005, 54, 1182–1193. [Google Scholar] [CrossRef]
- Gribble, F.M.; Reimann, F. Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat. Rev. Endocrinol. 2019, 15, 226–237. [Google Scholar] [CrossRef]
- Engelstoft, M.S.; Lund, M.L.; Grunddal, K.V.; Egerod, K.L.; Osborne-Lawrence, S.; Poulsen, S.S.; Zigman, J.M.; Schwartz, T.W. Research Resource: A Chromogranin A Reporter for Serotonin and Histamine Secreting Enteroendocrine Cells. Mol. Endocrinol. 2015, 29, 1658–1671. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.R.; Barnett, D.; Gentles, E.; Cairns, L.; Simpson, J.H. Macronutrient content of donor human breast milk. Arch. Dis. Child.-Fetal Neonatal Ed. 2013, 98, F539–F541. [Google Scholar] [CrossRef]
- Marx, C.; Bridge, R.; Wolf, A.K.; Rich, W.; Kim, J.H.; Bode, L. Human Milk Oligosaccharide Composition Differs between Donor Milk and Mother’s Own Milk in the NICU. J. Hum. Lact. 2014, 30, 54–61. [Google Scholar] [CrossRef]
- Martín-Carrasco, I.; Carbonero-Aguilar, P.; Dahiri, B.; Moreno, I.M.; Hinojosa, M. Comparison between pollutants found in breast milk and infant formula in the last decade: A review. Sci. Total Environ. 2023, 875, 162461. [Google Scholar] [CrossRef]
- Hair, A.B.; Ferguson, J.; Grogan, C.; Kim, J.H.; Taylor, S.N. Human milk fortification: The clinician and parent perspectives. Pediatr. Res. 2020, 88, 25–29. [Google Scholar] [CrossRef]
- Gu, X.; Shi, X.; Zhang, L.; Zhou, Y.; Cai, Y.; Jiang, W.; Zhou, Q. Evidence summary of human milk fortifier in preterm infants. Transl. Pediatr. 2021, 10, 3058–3067. [Google Scholar] [CrossRef]
- Berbari, N.F.; O’Connor, A.K.; Haycraft, C.J.; Yoder, B.K. The Primary Cilium as a Complex Signaling Center. Curr. Biol. 2009, 19, R526–R535. [Google Scholar] [CrossRef]
- Hoey, D.A.; Downs, M.E.; Jacobs, C.R. The mechanics of the primary cilium: An intricate structure with complex function. J. Biomech. 2012, 45, 17–26. [Google Scholar] [CrossRef]
- Wood, C.R.; Rosenbaum, J.L. Ciliary ectosomes: Transmissions from the cell’s antenna. Trends Cell Biol. 2015, 25, 276–285. [Google Scholar] [CrossRef]
- Gaeta, I.M.; Meenderink, L.M.; Postema, M.M.; Cencer, C.S.; Tyska, M.J. Direct visualization of epithelial microvilli biogenesis. Curr. Biol. 2021, 31, 2561–2575.e2566. [Google Scholar] [CrossRef]
- Sauvanet, C.; Wayt, J.; Pelaseyed, T.; Bretscher, A. Structure, Regulation, and Functional Diversity of Microvilli on the Apical Domain of Epithelial Cells. Annu. Rev. Cell Dev. Biol. 2015, 31, 593–621. [Google Scholar] [CrossRef] [PubMed]
- Lubarsky, B.; Krasnow, M.A. Tube Morphogenesis: Making and Shaping Biological Tubes. Cell 2003, 112, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Moing, A.; Ebbels, T.M.D.; Maucourt, M.; Tomos, A.D.; Rolin, D.; Hooks, M.A. Correlation Network Analysis reveals a sequential reorganization of metabolic and transcriptional states during germination and gene-metabolite relationships in developing seedlings of Arabidopsis. BMC Syst. Biol. 2010, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Coggins, S.A.; Mahboubi, B.; Schinazi, R.F.; Kim, B. SAMHD1 Functions and Human Diseases. Viruses 2020, 12, 382. [Google Scholar] [CrossRef]
- Coquel, F.; Silva, M.-J.; Técher, H.; Zadorozhny, K.; Sharma, S.; Nieminuszczy, J.; Mettling, C.; Dardillac, E.; Barthe, A.; Schmitz, A.-L.; et al. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 2018, 557, 57–61. [Google Scholar] [CrossRef]
- Meng, Z.; Qiu, Y.; Lin, K.C.; Kumar, A.; Placone, J.K.; Fang, C.; Wang, K.-C.; Lu, S.; Pan, M.; Hong, A.W.; et al. RAP2 mediates mechanoresponses of the Hippo pathway. Nature 2018, 560, 655–660. [Google Scholar] [CrossRef]
- Farag, M.I.; Yoshikawa, Y.; Maeta, K.; Kataoka, T. Rapgef2, a guanine nucleotide exchange factor for Rap1 small GTPases, plays a crucial role in adherence junction (AJ) formation in radial glial cells through ERK-mediated upregulation of the AJ-constituent protein expression. Biochem. Biophys. Res. Commun. 2017, 493, 139–145. [Google Scholar] [CrossRef]
- Ye, T.; Ip, J.P.K.; Fu, A.K.Y.; Ip, N.Y. Cdk5-mediated phosphorylation of RapGEF2 controls neuronal migration in the developing cerebral cortex. Nat. Commun. 2014, 5, 4826. [Google Scholar] [CrossRef]
- Li, N.; Xie, Q.; Zhao, L.; Shi, J.; Evivie, S.E.; Lv, X.; Huo, G.; Li, B. Human milk and infant formula modulate the intestinal microbiota and immune systems of human microbiota-associated mice. Food Funct. 2021, 12, 2784–2798. [Google Scholar] [CrossRef]
- Rowart, P.; Wu, J.; Caplan, M.J.; Jouret, F. Implications of AMPK in the Formation of Epithelial Tight Junctions. Int. J. Mol. Sci. 2018, 19, 2040. [Google Scholar] [CrossRef]
- Walsh, C.; Lane, J.A.; van Sinderen, D.; Hickey, R.M. Human milk oligosaccharides: Shaping the infant gut microbiota and supporting health. J. Funct. Foods 2020, 72, 104074. [Google Scholar] [CrossRef]
- Akbari, P.; Fink-Gremmels, J.; Willems, R.; Difilippo, E.; Schols, H.A.; Schoterman, M.H.C.; Garssen, J.; Braber, S. Characterizing microbiota-independent effects of oligosaccharides on intestinal epithelial cells: Insight into the role of structure and size: Structure-activity relationships of non-digestible oligosaccharides. Eur. J. Nutr. 2017, 56, 1919–1930. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, C.; Zhou, J.; Liang, Q.; He, F.; Li, F.; Li, Y.; Chen, J.; Zhang, F.; Han, C.; et al. Fibronectin 1 activates WNT/β-catenin signaling to induce osteogenic differentiation via integrin β1 interaction. Lab. Investig. 2020, 100, 1494–1502. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Wu, H.Y.; Lin, Y.L.; Tzou, S.C.; Chuang, C.H.; Jian, T.Y.; Chen, P.R.; Chang, Y.C.; Lin, C.H.; Huang, T.H.; et al. Blockade of ITGA2 Induces Apoptosis and Inhibits Cell Migration in Gastric Cancer. Biol. Proced. Online 2018, 20, 10. [Google Scholar] [CrossRef]
- Díaz, M.I.; Díaz, P.; Bennett, J.C.; Urra, H.; Ortiz, R.; Orellana, P.C.; Hetz, C.; Quest, A.F.G. Caveolin-1 suppresses tumor formation through the inhibition of the unfolded protein response. Cell Death Dis. 2020, 11, 648. [Google Scholar] [CrossRef]
- Arumugam, M.K.; Paal, M.C.; Donohue, T.M., Jr.; Ganesan, M.; Osna, N.A.; Kharbanda, K.K. Beneficial Effects of Betaine: A Comprehensive Review. Biology 2021, 10, 456. [Google Scholar] [CrossRef]
- Luz, M.; Knust, E. Fluorescently tagged Lin7c is a dynamic marker for polarity maturation in the zebrafish retinal epithelium. Biol. Open 2013, 2, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Cha, H.J.; Jeong, H.; Lee, S.-N.; Lee, C.-W.; Kim, M.; Yoo, J.; Woo, J.-S. Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM. Nat. Commun. 2023, 14, 931. [Google Scholar] [CrossRef]
- Bhatia, S.; Prabhu, P.N.; Benefiel, A.C.; Miller, M.J.; Chow, J.; Davis, S.R.; Gaskins, H.R. Galacto-oligosaccharides may directly enhance intestinal barrier function through the modulation of goblet cells. Mol. Nutr. Food Res. 2015, 59, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Liang, J.; Noble, P.W. Hyaluronan as an immune regulator in human diseases. Physiol. Rev. 2011, 91, 221–264. [Google Scholar] [CrossRef]
- Hatano, S.; Watanabe, H. Regulation of Macrophage and Dendritic Cell Function by Chondroitin Sulfate in Innate to Antigen-Specific Adaptive Immunity. Front. Immunol. 2020, 11, 232. [Google Scholar] [CrossRef]
- Collins, L.E.; Troeberg, L. Heparan sulfate as a regulator of inflammation and immunity. J. Leukoc. Biol. 2019, 105, 81–92. [Google Scholar] [CrossRef]
- Farrugia, B.L.; Lord, M.S.; Melrose, J.; Whitelock, J.M. The Role of Heparan Sulfate in Inflammation, and the Development of Biomimetics as Anti-Inflammatory Strategies. J. Histochem. Cytochem. 2018, 66, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Gelse, K.; Pöschl, E.; Aigner, T. Collagens—Structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef] [PubMed]
- Howell, S.J.; Doane, K.J. Type VI Collagen Increases Cell Survival and Prevents Anti-β1Integrin-Mediated Apoptosis. Exp. Cell Res. 1998, 241, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Rühl, M.; Sahin, E.; Johannsen, M.; Somasundaram, R.; Manski, D.; Riecken, E.O.; Schuppan, D. Soluble Collagen VI Drives Serum-starved Fibroblasts through S Phase and Prevents Apoptosis via Down-regulation of Bax. J. Biol. Chem. 1999, 274, 34361–34368. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Yang, S.; Zheng, C.; Huang, C.; Yao, H.; Guo, Z.; Wu, Y.; Wang, Z.; Wu, Z.; Ge, R.; et al. Multi-Omics Profiles of Small Intestine Organoids in Reaction to Breast Milk and Different Infant Formula Preparations. Nutrients 2024, 16, 2951. https://doi.org/10.3390/nu16172951
Wang X, Yang S, Zheng C, Huang C, Yao H, Guo Z, Wu Y, Wang Z, Wu Z, Ge R, et al. Multi-Omics Profiles of Small Intestine Organoids in Reaction to Breast Milk and Different Infant Formula Preparations. Nutrients. 2024; 16(17):2951. https://doi.org/10.3390/nu16172951
Chicago/Turabian StyleWang, Xianli, Shangzhi Yang, Chengdong Zheng, Chenxuan Huang, Haiyang Yao, Zimo Guo, Yilun Wu, Zening Wang, Zhenyang Wu, Ruihong Ge, and et al. 2024. "Multi-Omics Profiles of Small Intestine Organoids in Reaction to Breast Milk and Different Infant Formula Preparations" Nutrients 16, no. 17: 2951. https://doi.org/10.3390/nu16172951
APA StyleWang, X., Yang, S., Zheng, C., Huang, C., Yao, H., Guo, Z., Wu, Y., Wang, Z., Wu, Z., Ge, R., Cheng, W., Yan, Y., Jiang, S., Sun, J., Li, X., Xie, Q., & Wang, H. (2024). Multi-Omics Profiles of Small Intestine Organoids in Reaction to Breast Milk and Different Infant Formula Preparations. Nutrients, 16(17), 2951. https://doi.org/10.3390/nu16172951








