Analysis of Gut Bacterial and Fungal Microbiota in Children with Autism Spectrum Disorder and Their Non-Autistic Siblings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohorts
2.2. Stool Samples for Microbiome Testing
2.3. DNA Extraction
2.4. Targeted Amplification
2.5. Library Preparation and Sequencing
2.6. Statistical Analysis
2.7. Statistical Modeling
2.8. Potential Probiotic Screening
2.9. Microbial Isolates
- Media:
- 2.
- Test Fibers:
- 3.
- Bacterial Growth
- 4.
- Evaluation of the ability of strains to break down commercially available common fibers
- 5.
- Delftia biofilm growth in the presence and absence of probiotic filtrates
- 6.
- Experimental Animal Model
3. Results
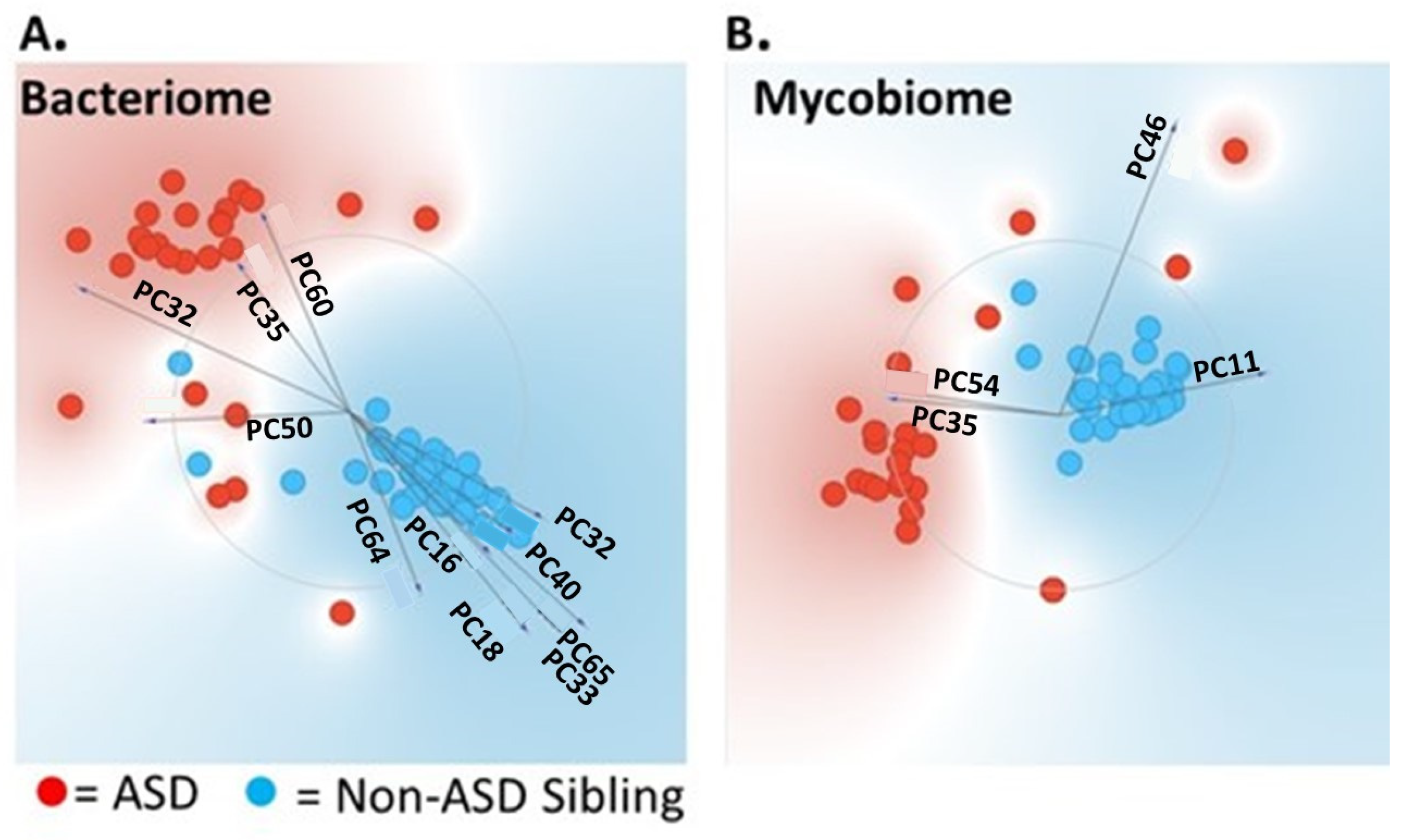
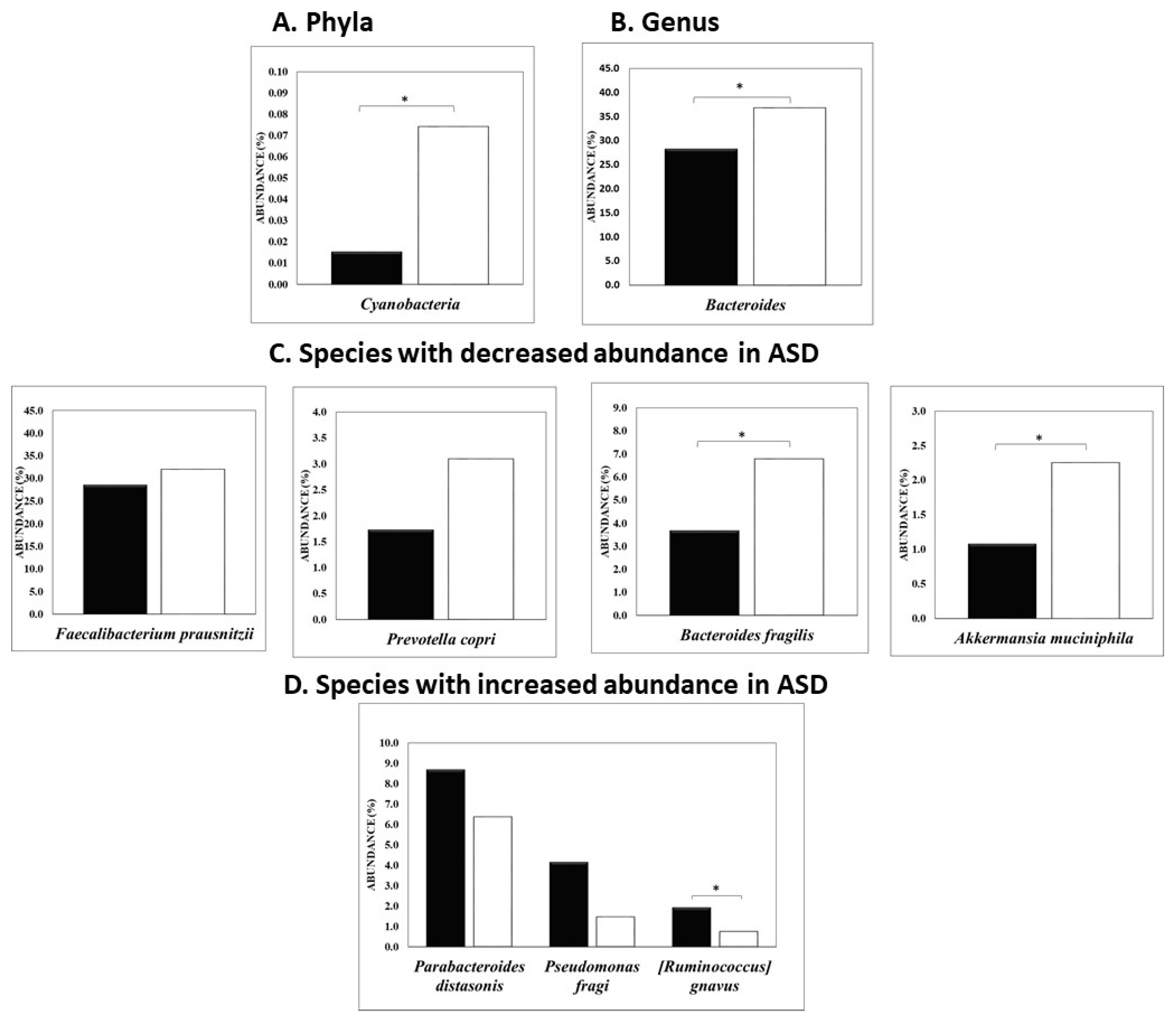
3.1. Univariate and Multivariate Statistical Analysis Used to Differentiate ASD from Non-ASD Sibling Cohort
3.2. Biofilm Formation
3.3. Delftia Influences Mouse Behavioral Changes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Psychiatric Association; Force, D.S.M.T. Diagnostic and Statistical Manual of Mental Disorders: DSM-5; American Psychiatric Association: Arlington, VA, USA, 2017. [Google Scholar]
- Maenner, M.J.; Shaw, K.A.; Baio, J.; Washington, A.; Patrick, M.; DiRienzo, M.; Christensen, D.L.; Wiggins, L.D.; Pettygrove, S.; Andrews, J.G.; et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef] [PubMed]
- National Center on Birth Defects and Developmental Disabilities, Centers for Disease Control and Prevention (CDC). Data & Statistics on Autism Spectrum Disorder. Available online: https://www.cdc.gov/ncbddd/autism/data.html (accessed on 14 June 2023).
- Maenner, M.J.; Warren, Z.; Williams, A.R.; Amoakohene, E.; Bakian, A.V.; Bilder, D.A.; Durkin, M.S.; Fitzgerald, R.T.; Furnier, S.M.; Hughes, M.M. Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network. MMWR Surveill. Summ. 2023, 72, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Newschaffer, C.J.; Falb, M.D.; Gurney, J.G. National autism prevalence trends from United States special education data. Pediatrics 2005, 115, e277–e282. [Google Scholar] [CrossRef]
- Parner, E.T.; Schendel, D.E.; Thorsen, P. Autism prevalence trends over time in Denmark: Changes in prevalence and age at diagnosis. Arch. Pediatr. Adolesc. Med. 2008, 162, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- Kogan, M.D.; Blumberg, S.J.; Schieve, L.A.; Boyle, C.A.; Perrin, J.M.; Ghandour, R.M.; Singh, G.K.; Strickland, B.B.; Trevathan, E.; van Dyck, P.C. Prevalence of parent-reported diagnosis of autism spectrum disorder among children in the US, 2007. Pediatrics 2009, 124, 1395–1403. [Google Scholar] [CrossRef]
- Lundstrom, S.; Reichenberg, A.; Anckarsater, H.; Lichtenstein, P.; Gillberg, C. Autism phenotype versus registered diagnosis in Swedish children: Prevalence trends over 10 years in general population samples. BMJ 2015, 350, h1961. [Google Scholar] [CrossRef] [PubMed]
- Rice, C.E.; Rosanoff, M.; Dawson, G.; Durkin, M.S.; Croen, L.A.; Singer, A.; Yeargin-Allsopp, M. Evaluating Changes in the Prevalence of the Autism Spectrum Disorders (ASDs). Public Health Rev. 2012, 34, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Damiani, S.; Leali, P.; Nosari, G.; Caviglia, M.; Puci, M.V.; Monti, M.C.; Brondino, N.; Politi, P. Association of Autism Onset, Epilepsy, and Behavior in a Community of Adults with Autism and Severe Intellectual Disability. Brain Sci. 2020, 10, 486. [Google Scholar] [CrossRef]
- Buie, T.; Campbell, D.B.; Fuchs, G.J., 3rd; Furuta, G.T.; Levy, J.; Vandewater, J.; Whitaker, A.H.; Atkins, D.; Bauman, M.L.; Beaudet, A.L.; et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: A consensus report. Pediatrics 2010, 125 (Suppl. S1), S1–S18. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, B.S.; Geschwind, D.H. Advances in autism genetics: On the threshold of a new neurobiology. Nat. Rev. Genet. 2008, 9, 341–355. [Google Scholar] [CrossRef]
- Bolton, P.F. Medical conditions in autism spectrum disorders. J. Neurodev. Disord. 2009, 1, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Zafeiriou, D.I.; Ververi, A.; Vargiami, E. Childhood autism and associated comorbidities. Brain Dev. 2007, 29, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Madra, M.; Ringel, R.; Margolis, K.G. Gastrointestinal Issues and Autism Spectrum Disorder. Psychiatr. Clin. N. Am. 2021, 44, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Gjevik, E.; Eldevik, S.; Fjaeran-Granum, T.; Sponheim, E. Kiddie-SADS reveals high rates of DSM-IV disorders in children and adolescents with autism spectrum disorders. J. Autism. Dev. Disord. 2011, 41, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Leyfer, O.T.; Folstein, S.E.; Bacalman, S.; Davis, N.O.; Dinh, E.; Morgan, J.; Tager-Flusberg, H.; Lainhart, J.E. Comorbid psychiatric disorders in children with autism: Interview development and rates of disorders. J. Autism. Dev. Disord. 2006, 36, 849–861. [Google Scholar] [CrossRef]
- Matson, J.L.; Nebel-Schwalm, M.S. Comorbid psychopathology with autism spectrum disorder in children: An overview. Res. Dev. Disabil. 2007, 28, 341–352. [Google Scholar] [CrossRef]
- Buie, T.; Fuchs, G.J., 3rd; Furuta, G.T.; Kooros, K.; Levy, J.; Lewis, J.D.; Wershil, B.K.; Winter, H. Recommendations for evaluation and treatment of common gastrointestinal problems in children with ASDs. Pediatrics 2010, 125 (Suppl. S1), S19–S29. [Google Scholar] [CrossRef]
- D’Eufemia, P.; Celli, M.; Finocchiaro, R.; Pacifico, L.; Viozzi, L.; Zaccagnini, M.; Cardi, E.; Giardini, O. Abnormal intestinal permeability in children with autism. Acta Paediatr. 1996, 85, 1076–1079. [Google Scholar] [CrossRef]
- Coury, D.L.; Ashwood, P.; Fasano, A.; Fuchs, G.; Geraghty, M.; Kaul, A.; Mawe, G.; Patterson, P.; Jones, N.E. Gastrointestinal conditions in children with autism spectrum disorder: Developing a research agenda. Pediatrics 2012, 130 (Suppl. S2), S160–S168. [Google Scholar] [CrossRef]
- Ahearn, W.H.; Castine, T.; Nault, K.; Green, G. An assessment of food acceptance in children with autism or pervasive developmental disorder-not otherwise specified. J. Autism. Dev. Disord. 2001, 31, 505–511. [Google Scholar] [CrossRef]
- Field, D.; Garland, M.; Williams, K. Correlates of specific childhood feeding problems. J. Paediatr. Child Health 2003, 39, 299–304. [Google Scholar] [CrossRef]
- de Magistris, L.; Familiari, V.; Pascotto, A.; Sapone, A.; Frolli, A.; Iardino, P.; Carteni, M.; De Rosa, M.; Francavilla, R.; Riegler, G.; et al. Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 418–424. [Google Scholar] [CrossRef]
- Belizario, J.E.; Faintuch, J. Microbiome and Gut Dysbiosis. Exp. Suppl. 2018, 109, 459–476. [Google Scholar] [CrossRef]
- Garcia-Gutierrez, E.; Narbad, A.; Rodriguez, J.M. Autism Spectrum Disorder Associated with Gut Microbiota at Immune, Metabolomic, and Neuroactive Level. Front. Neurosci. 2020, 14, 578666. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska-Pietruszka, Z.; Figlerowicz, M.; Mazur-Melewska, K. Microbiota in Autism Spectrum Disorder: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 16660. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.T.; Jin, D.M.; Mills, R.H.; Shao, Y.; Rahman, G.; McDonald, D.; Zhu, Q.; Balaban, M.; Jiang, Y.; Cantrell, K.; et al. Multi-level analysis of the gut-brain axis shows autism spectrum disorder-associated molecular and microbial profiles. Nat. Neurosci. 2023, 26, 1208–1217. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Marzal, L.N.; Rojas-Velazquez, D.; Rigters, D.; Prince, N.; Garssen, J.; Kraneveld, A.D.; Perez-Pardo, P.; Lopez-Rincon, A. A robust microbiome signature for autism spectrum disorder across different studies using machine learning. Sci. Rep. 2024, 14, 814. [Google Scholar] [CrossRef]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar]
- Rutsch, A.; Kantsjo, J.B.; Ronchi, F. The Gut-Brain Axis: How Microbiota and Host Inflammasome Influence Brain Physiology and Pathology. Front. Immunol. 2020, 11, 604179. [Google Scholar] [CrossRef]
- Ho, L.K.H.; Tong, V.J.W.; Syn, N.; Nagarajan, N.; Tham, E.H.; Tay, S.K.; Shorey, S.; Tambyah, P.A.; Law, E.C.N. Gut microbiota changes in children with autism spectrum disorder: A systematic review. Gut Pathog. 2020, 12, 6. [Google Scholar] [CrossRef]
- Malkova, N.V.; Yu, C.Z.; Hsiao, E.Y.; Moore, M.J.; Patterson, P.H. Maternal immune activation yields offspring displaying mouse versions of the three core symptoms of autism. Brain Behav. Immun. 2012, 26, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Fowlie, G.; Cohen, N.; Ming, X. The Perturbance of Microbiome and Gut-Brain Axis in Autism Spectrum Disorders. Int. J. Mol. Sci. 2018, 19, 2251. [Google Scholar] [CrossRef]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Cruz, N.J.; Kang, D.W.; Gandal, M.J.; Wang, B.; Kim, Y.M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 2019, 177, 1600–1618.e17. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, S.Y.; El Gendy, Y.G.; Mehanna, N.S.; El-Senousy, W.M.; El-Feki, H.S.A.; Saad, K.; El-Asheer, O.M. The role of probiotics in children with autism spectrum disorder: A prospective, open-label study. Nutr. Neurosci. 2018, 21, 676–681. [Google Scholar] [CrossRef]
- Grossi, E.; Melli, S.; Dunca, D.; Terruzzi, V. Unexpected improvement in core autism spectrum disorder symptoms after long-term treatment with probiotics. SAGE Open Med Case Rep. 2016, 4, 2050313X16666231. [Google Scholar] [CrossRef]
- Kaluzna-Czaplinska, J.; Blaszczyk, S. The level of arabinitol in autistic children after probiotic therapy. Nutrition 2012, 28, 124–126. [Google Scholar] [CrossRef]
- Parracho, H.; Gibson, G.R.; Knott, F.; Bosscher, D.; Kleerebezem, M.; McCartney, A. A double-blind, placebo-controlled, crossover-designed probiotic feeding study in children diagnosed with autistic spectrum disorders. Int. J. Probiotics Prebiotics 2010, 5, 69–74. [Google Scholar]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio 2016, 7, e01250-16. [Google Scholar] [CrossRef]
- Rema, T.; Lawrence, J.R.; Dynes, J.J.; Hitchcock, A.P.; Korber, D.R. Microscopic and spectroscopic analyses of chlorhexidine tolerance in Delftia acidovorans biofilms. Antimicrob. Agents Chemother. 2014, 58, 5673–5686. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Bjorck, I.; Backhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Krajmalnik-Brown, R.; Porazinska, D.L.; Weiss, S.J.; Knight, R. Toward effective probiotics for autism and other neurodevelopmental disorders. Cell 2013, 155, 1446–1448. [Google Scholar] [CrossRef] [PubMed]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [PubMed]
- Salyers, A.A. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 1984, 38, 293–313. [Google Scholar] [CrossRef]
- Sternisa, M.; Klancnik, A.; Smole Mozina, S. Spoilage Pseudomonas biofilm with Escherichia coli protection in fish meat at 5 degrees C. J. Sci. Food Agric. 2019, 99, 4635–4641. [Google Scholar] [CrossRef]
- Grygier, A.; Myszka, K.; Rudzinska, M. Galactomyces geotrichum—Moulds from dairy products with high biotechnological potential. Acta Sci. Pol. Technol. Aliment. 2017, 16, 5–16. [Google Scholar] [CrossRef]
- Lima, R.N.; Porto, A.L. Recent Advances in Marine Enzymes for Biotechnological Processes. Adv. Food Nutr. Res. 2016, 78, 153–192. [Google Scholar] [CrossRef] [PubMed]
- Buie, T. Potential Etiologic Factors of Microbiome Disruption in Autism. Clin. Ther. 2015, 37, 976–983. [Google Scholar] [CrossRef]
- de Theije, C.G.; Wu, J.; da Silva, S.L.; Kamphuis, P.J.; Garssen, J.; Korte, S.M.; Kraneveld, A.D. Pathways underlying the gut-to-brain connection in autism spectrum disorders as future targets for disease management. Eur. J. Pharmacol. 2011, 668 (Suppl. S1), S70–S80. [Google Scholar] [CrossRef]
- Estes, M.L.; McAllister, A.K. Brain, Immunity, Gut: “BIG” Links between Pregnancy and Autism. Immunity 2017, 47, 816–819. [Google Scholar] [CrossRef]
- Hicks, S.D.; Uhlig, R.; Afshari, P.; Williams, J.; Chroneos, M.; Tierney-Aves, C.; Wagner, K.; Middleton, F.A. Oral microbiome activity in children with autism spectrum disorder. Autism. Res. 2018, 11, 1286–1299. [Google Scholar] [CrossRef] [PubMed]
- Israelyan, N.; Margolis, K.G. Serotonin as a link between the gut-brain-microbiome axis in autism spectrum disorders. Pharmacol. Res. 2018, 132, 1–6. [Google Scholar] [CrossRef]
- Kraneveld, A.D.; Szklany, K.; de Theije, C.G.; Garssen, J. Gut-to-Brain Axis in Autism Spectrum Disorders: Central Role for the Microbiome. Int. Rev. Neurobiol. 2016, 131, 263–287. [Google Scholar] [CrossRef]
- Luna, R.A.; Oezguen, N.; Balderas, M.; Venkatachalam, A.; Runge, J.K.; Versalovic, J.; Veenstra-VanderWeele, J.; Anderson, G.M.; Savidge, T.; Williams, K.C. Distinct Microbiome-Neuroimmune Signatures Correlate with Functional Abdominal Pain in Children with Autism Spectrum Disorder. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 218–230. [Google Scholar] [CrossRef]
- Mayer, E.A.; Padua, D.; Tillisch, K. Altered brain-gut axis in autism: Comorbidity or causative mechanisms? Bioessays 2014, 36, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Reichelt, K.L.; Knivsberg, A.M. The possibility and probability of a gut-to-brain connection in autism. Ann. Clin. Psychiatry 2009, 21, 205–211. [Google Scholar] [PubMed]
- Vasquez, A. Biological plausibility of the gut-brain axis in autism. Ann. N. Y. Acad. Sci. 2017, 1408, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.E.; Hsiao, E.Y. Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Biol. Psychiatry 2017, 81, 411–423. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; Labaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xu, X.; Li, J.; Li, F. Association Between Gut Microbiota and Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Front. Psychiatry 2019, 10, 473. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- De Angelis, M.; Francavilla, R.; Piccolo, M.; De Giacomo, A.; Gobbetti, M. Autism spectrum disorders and intestinal microbiota. Gut Microbes 2015, 6, 207–213. [Google Scholar] [CrossRef]
- Su, Q.; Wong, O.W.H.; Lu, W.; Wan, Y.; Zhang, L.; Xu, W.; Li, M.K.T.; Liu, C.; Cheung, C.P.; Ching, J.Y.L.; et al. Multikingdom and functional gut microbiota markers for autism spectrum disorder. Nat. Microbiol. 2024. [Google Scholar] [CrossRef]
- Liu, L.Y.; Seo, J.; McCanna, D.J.; Subbaraman, L.N.; Jones, L.W. Assessment of biofilm formation of E. meningoseptica, D. acidovorans, and S. maltophilia in lens cases and their growth on recovery media. Cont. Lens. Anterior. Eye 2016, 39, 117–123. [Google Scholar] [CrossRef]
- Mahmood, S.; Taylor, K.E.; Overman, T.L.; McCormick, M.I. Acute infective endocarditis caused by Delftia acidovorans, a rare pathogen complicating intravenous drug use. J. Clin. Microbiol. 2012, 50, 3799–3800. [Google Scholar] [CrossRef]
- Deb, A.K.; Chavhan, P.; Chowdhury, S.S.; Sistla, S.; Sugumaran, R.; Panicker, G. Endophthalmitis due to Delftia acidovorans: An unusual ocular pathogen. Indian J. Ophthalmol. 2020, 68, 2591–2594. [Google Scholar] [CrossRef]
- Kam, S.-K.; Lee, W.-S.; Ou, T.-Y.; Teng, S.-O.; Chen, F.-L. Delftia acidovorans Bacteremia Associated with Ascending Urinary Tract Infections Proved by Molecular Method. J. Exp. Clin. Med. 2012, 4, 180–182. [Google Scholar] [CrossRef]
- Khan, S.; Sistla, S.; Dhodapkar, R.; Parija, S.C. Fatal Delftia acidovorans infection in an immunocompetent patient with empyema. Asian Pac. J. Trop. Biomed. 2012, 2, 923–924. [Google Scholar] [CrossRef]
- Bilgin, H.; Sarmis, A.; Tigen, E.; Soyletir, G.; Mulazimoglu, L. Delftia acidovorans: A rare pathogen in immunocompetent and immunocompromised patients. Can. J. Infect. Dis. Med Microbiol. 2015, 26, 277–279. [Google Scholar] [CrossRef]
- Yildiz, H.; Sunnetcioglu, A.; Ekin, S.; Baran, A.I.; Ozgokce, M.; Asker, S.; Uney, I.; Turgut, E.; Akyuz, S. Delftia acidovorans pneumonia with lung cavities formation. Colomb. Med. 2019, 50, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Safak, B.; Altunan, B.; Topcu, B.; Eren Topkaya, A. The gut microbiome in epilepsy. Microb. Pathog. 2020, 139, 103853. [Google Scholar] [CrossRef]
- Jones-Hall, Y.L.; Kozik, A.; Nakatsu, C. Ablation of tumor necrosis factor is associated with decreased inflammation and alterations of the microbiota in a mouse model of inflammatory bowel disease. PLoS ONE 2015, 10, e0119441. [Google Scholar] [CrossRef]
- McNeill, R.M.; DeFoor, W.M.; Goller, C.C.; Ott, L.E. Delftia spp elicit a pro-inflammatory response in monocytes. J. Young Investig. 2015, 29, 41–48. [Google Scholar]
- West, K.A.; Yin, X.; Rutherford, E.M.; Wee, B.; Choi, J.; Chrisman, B.S.; Dunlap, K.L.; Hannibal, R.L.; Hartono, W.; Lin, M.; et al. Multi-angle meta-analysis of the gut microbiome in Autism Spectrum Disorder: A step toward understanding patient subgroups. Sci. Rep. 2022, 12, 17034. [Google Scholar] [CrossRef] [PubMed]
- Demay, J.; Halary, S.; Knittel-Obrecht, A.; Villa, P.; Duval, C.; Hamlaoui, S.; Roussel, T.; Yepremian, C.; Reinhardt, A.; Bernard, C.; et al. Anti-Inflammatory, Antioxidant, and Wound-Healing Properties of Cyanobacteria from Thermal Mud of Balaruc-Les-Bains, France: A Multi-Approach Study. Biomolecules 2020, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Ku, C.S.; Yang, Y.; Park, Y.; Lee, J. Health benefits of blue-green algae: Prevention of cardiovascular disease and nonalcoholic fatty liver disease. J. Med. Food. 2013, 16, 103–111. [Google Scholar] [CrossRef]
- Rojas, V.; Rivas, L.; Cardenas, C.; Guzman, F. Cyanobacteria and Eukaryotic Microalgae as Emerging Sources of Antibacterial Peptides. Molecules 2020, 25, 5804. [Google Scholar] [CrossRef]
- Hu, C.; Rzymski, P. Non-Photosynthetic Melainabacteria (Cyanobacteria) in Human Gut: Characteristics and Association with Health. Life 2022, 12, 476. [Google Scholar] [CrossRef] [PubMed]
- Castro, K.; Faccioli, L.S.; Baronio, D.; Gottfried, C.; Perry, I.S.; Riesgo, R. Feeding behavior and dietary intake of male children and adolescents with autism spectrum disorder: A case-control study. Int. J. Dev. Neurosci. 2016, 53, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Motta, J.P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M. Leaky gut-concept or clinical entity? Curr. Opin. Gastroenterol. 2016, 32, 74–79. [Google Scholar] [CrossRef]
- Mu, Q.; Kirby, J.; Reilly, C.M.; Luo, X.M. Leaky Gut as a Danger Signal for Autoimmune Diseases. Front. Immunol. 2017, 8, 598. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Chen, X.; Chen, Y.; Jiang, W.; Chen, H. The effect of five probiotic lactobacilli strains on the growth and biofilm formation of Streptococcus mutans. Oral Dis. 2015, 21, e128–e134. [Google Scholar] [CrossRef] [PubMed]
- Soderling, E.M.; Marttinen, A.M.; Haukioja, A.L. Probiotic lactobacilli interfere with Streptococcus mutans biofilm formation in vitro. Curr. Microbiol. 2011, 62, 618–622. [Google Scholar] [CrossRef]
- Munoz, J.A.; Chenoll, E.; Casinos, B.; Bataller, E.; Ramon, D.; Genoves, S.; Montava, R.; Ribes, J.M.; Buesa, J.; Fabrega, J.; et al. Novel probiotic Bifidobacterium longum subsp. infantis CECT 7210 strain active against rotavirus infections. Appl. Environ. Microbiol. 2011, 77, 8775–8783. [Google Scholar] [CrossRef]
- Chichlowski, M.; Shah, N.; Wampler, J.L.; Wu, S.S.; Vanderhoof, J.A. Bifidobacterium longum Subspecies infantis (B. infantis) in Pediatric Nutrition: Current State of Knowledge. Nutrients 2020, 12, 1581. [Google Scholar] [CrossRef]
- Bozzi Cionci, N.; Baffoni, L.; Gaggia, F.; Di Gioia, D. Therapeutic Microbiology: The Role of Bifidobacterium breve as Food Supplement for the Prevention/Treatment of Paediatric Diseases. Nutrients 2018, 10, 1723. [Google Scholar] [CrossRef]
- Simone, M.; Gozzoli, C.; Quartieri, A.; Mazzola, G.; Di Gioia, D.; Amaretti, A.; Raimondi, S.; Rossi, M. The probiotic Bifidobacterium breve B632 inhibited the growth of Enterobacteriaceae within colicky infant microbiota cultures. Biomed. Res. Int. 2014, 2014, 301053. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, A.G.; Esaam, A.; Hazaa, M.M. Cell free preparations of probiotics exerted antibacterial and antibiofilm activities against multidrug resistant E. coli. Saudi Pharm. J. 2018, 26, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Barzegari, A.; Kheyrolahzadeh, K.; Hosseiniyan Khatibi, S.M.; Sharifi, S.; Memar, M.Y.; Zununi Vahed, S. The Battle of Probiotics and Their Derivatives Against Biofilms. Infect. Drug Resist. 2020, 13, 659–672. [Google Scholar] [CrossRef]
- O’Mahony, L.; McCarthy, J.; Kelly, P.; Hurley, G.; Luo, F.; Chen, K.; O’Sullivan, G.C.; Kiely, B.; Collins, J.K.; Shanahan, F.; et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: Symptom responses and relationship to cytokine profiles. Gastroenterology 2005, 128, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Whorwell, P.J.; Altringer, L.; Morel, J.; Bond, Y.; Charbonneau, D.; O’Mahony, L.; Kiely, B.; Shanahan, F.; Quigley, E.M.M. Efficacy of an Encapsulated Probiotic Bifidobacterium infantis 35624 in Women with Irritable Bowel Syndrome. Off. J. Am. Coll. Gastroenterol. ACG 2006, 101, 1581–1590. [Google Scholar] [CrossRef]
- Rao, A.V.; Bested, A.C.; Beaulne, T.M.; Katzman, M.A.; Iorio, C.; Berardi, J.M.; Logan, A.C. A randomized, double-blind, placebo-controlled pilot study of a probiotic in emotional symptoms of chronic fatigue syndrome. Gut Pathog. 2009, 1, 6. [Google Scholar] [CrossRef]
- Yang, J.; Yang, H. Antibacterial Activity of Bifidobacterium breve against Clostridioides difficile. Front. Cell. Infect. Microbiol. 2019, 9, 288. [Google Scholar] [CrossRef]
- Groeger, D.; O’Mahony, L.; Murphy, E.F.; Bourke, J.F.; Dinan, T.G.; Kiely, B.; Shanahan, F.; Quigley, E.M. Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes 2013, 4, 325–339. [Google Scholar] [CrossRef]

| Factors | Multivariable Model | |
|---|---|---|
| Odds Ratio (95% CI) | p Value | |
| p__Cyanobacteria (per 0.01 percentage of abundance increase) | 0.76 (0.62, 0.94) | 0.013 |
| s__Prevotella nigrescens (per 0.01 percentage of abundance increase) | 0.84 (0.64, 1.1) | 0.202 |
| g__Anaerostipes (per percentage of abundance increase) | 0.17 (0.01, 2.53) | 0.197 |
| g__Bacteroides (per percentage of abundance increase) | 0.98 (0.93, 1.03) | 0.355 |
| g__Leptothrix (per 0.01 percentage of abundance increase) | 1.26 (0.8, 1.98) | 0.321 |
| g__Delftia (per 0.01 percentage of abundance increase) | 9.99 (1.33, 75) | 0.025 |
| g__Shewanella (per 0.1 percentage of abundance increase) | 0.37 (0.03, 4.6) | 0.443 |
| g__Azospirillum (per 0.01 percentage of abundance increase) | 0.19 (0.05, 0.82) | 0.026 |
| g__Metarhizium (per percentage of abundance increase) | 0.21 (0.01, 7.25) | 0.386 |
| Fish (per serving increase) | 0.29 (0.1, 0.82) | 0.02 |
| Seizure (Yes vs. no) | 1.33 (0.13, 13.78) | 0.809 |
| Sex (Male vs. Female) | 36.79 (2.89, 467.93) | 0.006 |
| upper gastrointestinal disturbances | 2.89 (0.25, 32.87) | 0.392 |
| Probiotic Candidates | Probiotic Fiber Breakdown Score (PFBS) | Inulin | Agave Inulin | FOS | Growth Rate |
|---|---|---|---|---|---|
| Lactobacillus casei | 9 | 3 | 3 | 3 | Good |
| Bifidobacterium longum subps. Infantis | 9 | 3 | 3 | 3 | Good |
| Lactobacillus paracasei | 8 | 2 | 3 | 3 | Good |
| Lactobacillus salivarius | 8 | 3 | 3 | 2 | Good |
| Lactobacillus delbrueckii subsp. bulgaricus | 8 | 2 | 3 | 3 | Poor |
| Bifidobacterium breve | 6 | 0 | 3 | 3 | Good |
| Bifidobacterium bifidum | 5 | 0 | 2 | 3 | Good |
| Pediococcus pentosaceus | 5 | 0 | 3 | 2 | Good |
| Streptococcus thermophilus | 4 | 0 | 3 | 1 | Good |
| Pediococcus acidilactici | 4 | 0 | 3 | 1 | Good |
| Lactobacillus plantarum | 4 | 0 | 2 | 2 | Good |
| Bifidobacterium longum | 3 | 0 | 2 | 1 | Good |
| Lactobacillus plantarum | 3 | 0 | 2 | 1 | Good |
| Lactobacillus rhamnosus | 3 | 0 | 2 | 1 | Good |
| Lactobacillus rhamnosus | 3 | 0 | 2 | 1 | Good |
| Bifidobacterium breve | 3 | 0 | 2 | 1 | Good |
| Saccharomyces boulardii | 3 | 0 | 0 | 3 | Good |
| Lactobacillus gasseri | 3 | 0 | 2 | 1 | Good |
| Lactococcus lactis | 1 | 0 | 1 | 0 | Good |
| Lactobacillus reuteri | 1 | 0 | 1 | 0 | Good |
| Lactobacillus acidophilus | 1 | 0 | 0 | 1 | Good |
| Bifidobacterium longum | 1 | 0 | 1 | 0 | Good |
| Bifidobacterium pseudocatenulatum | 0 | 0 | 0 | 0 | Good |
| Bifidobacterium catenulatum subsp. kashiwanohense | 0 | 0 | 0 | 0 | Good |
| Komagataella pastoris | 0 | 0 | 0 | 0 | Good |
| Delftia Biofilm Inhibition by Various Organisms | Log CFUs of Delftia Biofilm (±SD) | p-Value Compared to the Growth Control |
|---|---|---|
| Untreated Delftia Control | 6.28 ± 0.02 | |
| Lactobacillus casei | 5.17 ± 0.12 | 0.002 |
| Bifidobacterium longum subps. infantis | 5.09 ± 0.34 | 0.025 |
| Lactobacillus paracasei | 5.79 ± 0.13 | 0.017 |
| Lactobacillus salivarius | 6.21 ± 0.04 | 0.057 |
| Lactobacillus delbrueckii subsp. bulgaricus | 6.27 ± 0.03 | 0.608 |
| Bifidobacterium breve | 4.75 ± 0.02 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Retuerto, M.; Al-Shakhshir, H.; Herrada, J.; McCormick, T.S.; Ghannoum, M.A. Analysis of Gut Bacterial and Fungal Microbiota in Children with Autism Spectrum Disorder and Their Non-Autistic Siblings. Nutrients 2024, 16, 3004. https://doi.org/10.3390/nu16173004
Retuerto M, Al-Shakhshir H, Herrada J, McCormick TS, Ghannoum MA. Analysis of Gut Bacterial and Fungal Microbiota in Children with Autism Spectrum Disorder and Their Non-Autistic Siblings. Nutrients. 2024; 16(17):3004. https://doi.org/10.3390/nu16173004
Chicago/Turabian StyleRetuerto, Mauricio, Hilmi Al-Shakhshir, Janet Herrada, Thomas S. McCormick, and Mahmoud A. Ghannoum. 2024. "Analysis of Gut Bacterial and Fungal Microbiota in Children with Autism Spectrum Disorder and Their Non-Autistic Siblings" Nutrients 16, no. 17: 3004. https://doi.org/10.3390/nu16173004




