Exploring Lectin Bioactivity and Total Phenolic Compounds in Kiwifruit (Actinidia deliciosa var. Hayward)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials, Solvents and Reagents
2.2. Methods
2.2.1. Extraction of Phenolic Compounds
2.2.2. Characterization of Kiwifruit Phenolic Extract
Total Phenolic and Flavonoid Content
2.2.3. Identification of Phenolic Compounds by HPLC-DAD-MS/MS
2.2.4. Determination of Antioxidant Activity of the Kiwifruit Phenolic Extract
2.2.5. Protein Extraction
2.2.6. Polypeptide and Lectin Characterization
2.2.7. Hemagglutination Assays
Erythrocyte Cells
Hemagglutination Activity
Hemagglutination Inhibition
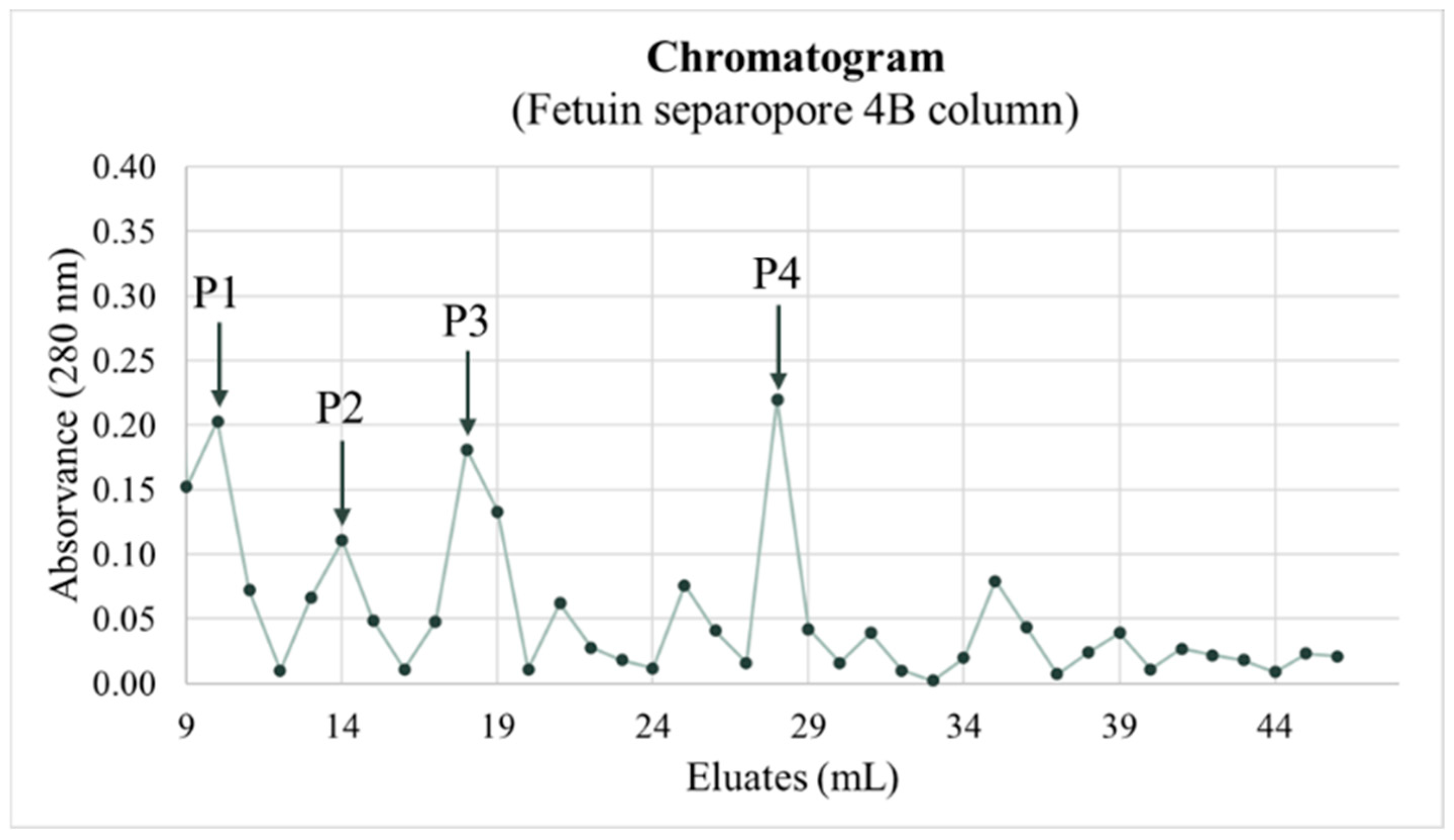
2.2.8. Purification of Sialic Acid Lectins
2.2.9. Lectin Binding to HT-29 and MIA PaCa-2 Cells
Isolation of HT-29 and MIA PaCa-2 Cell Membranes
Affinity Binding of Polypeptides vs. Lectins from Total Protein Extract to HT-29 and MIA PaCa-2 Cells Membranes
2.2.10. Statistical Analysis
3. Results
3.1. Characterization of Phenolic and Flavonoid Compounds
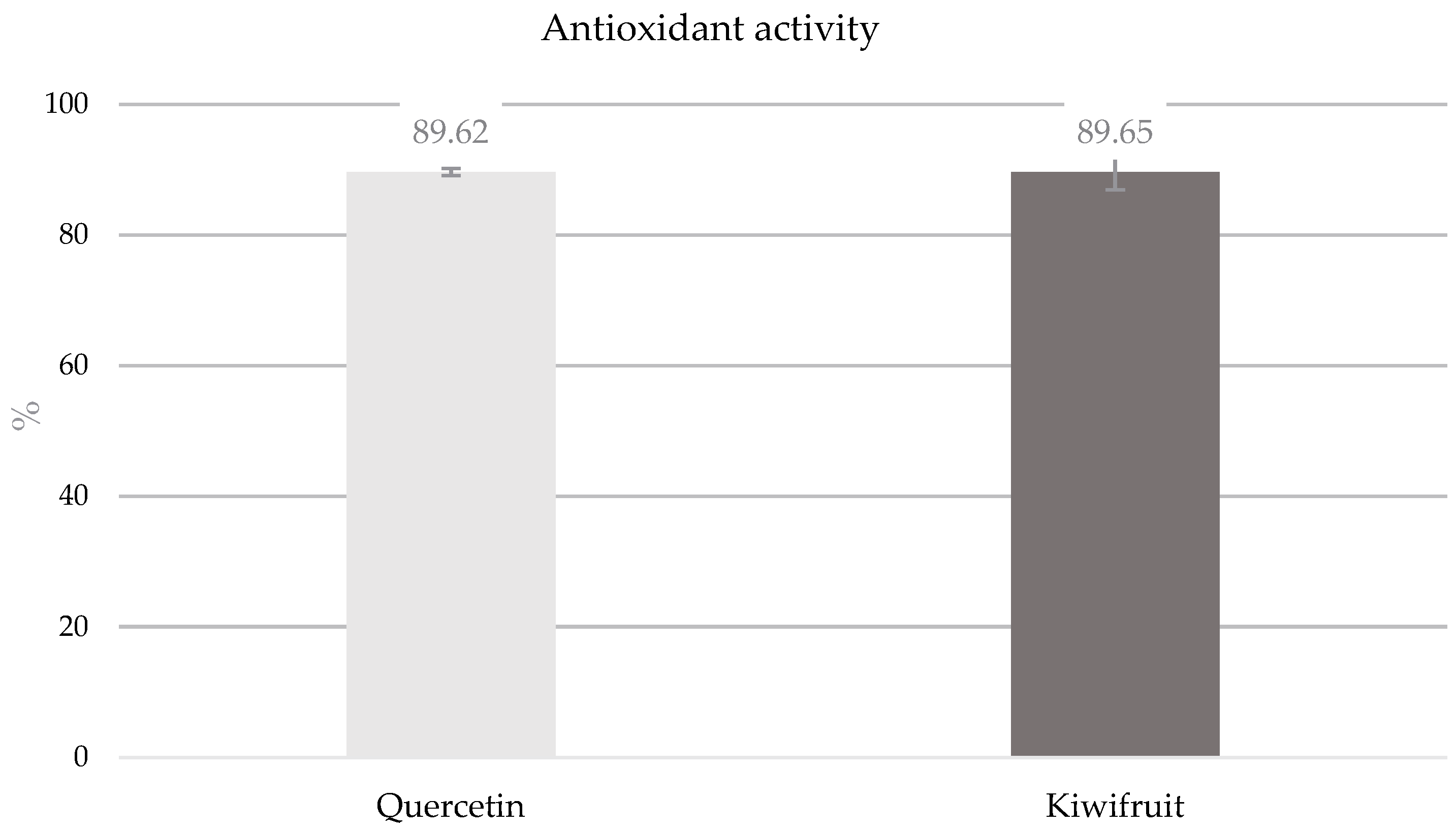
3.2. Determination of Antioxidant Activity (DDPH) of Kiwifruit Phenolic Extract
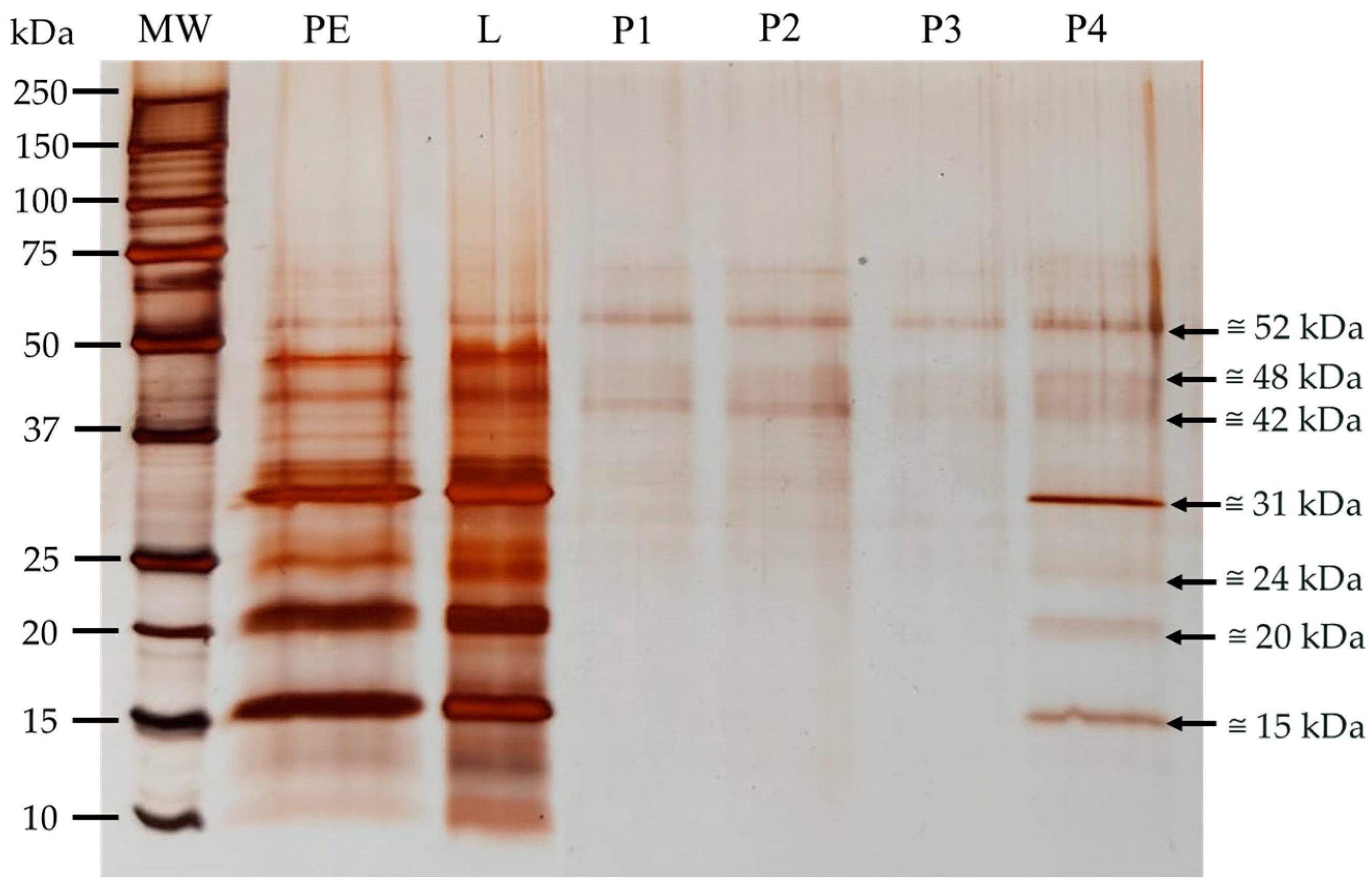
3.3. Polypeptide Characterization
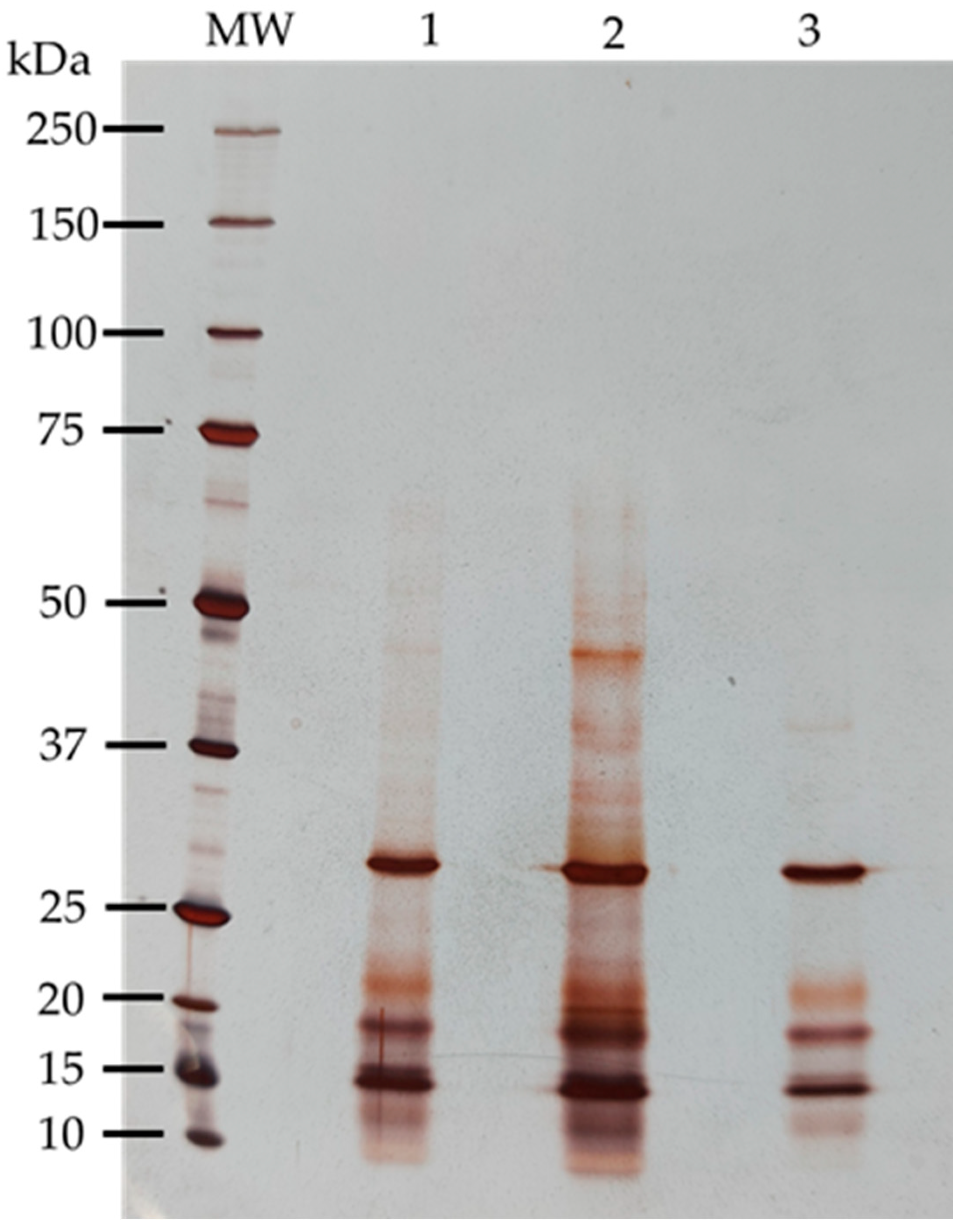
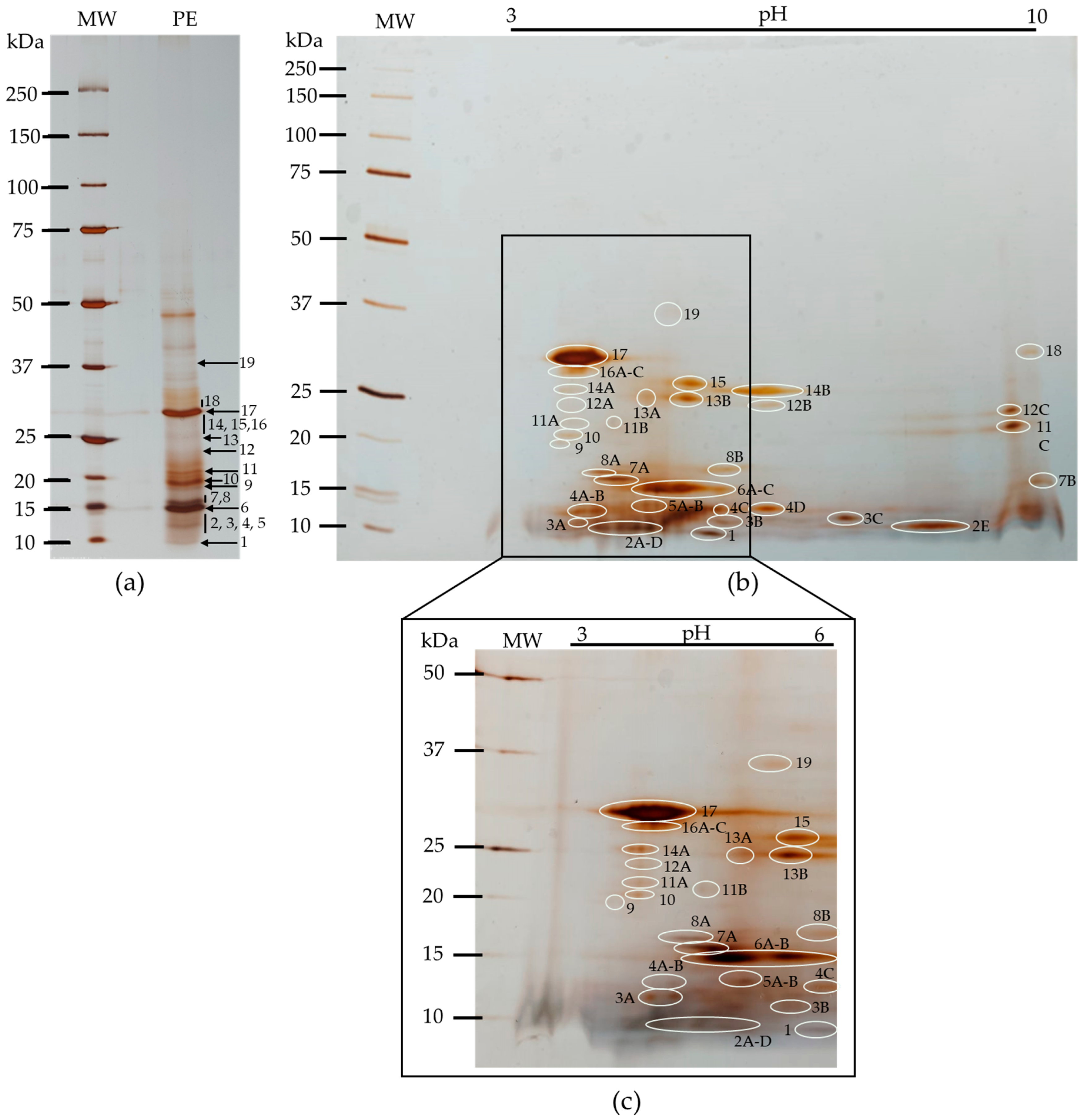
3.3.1. Polypeptide Profile
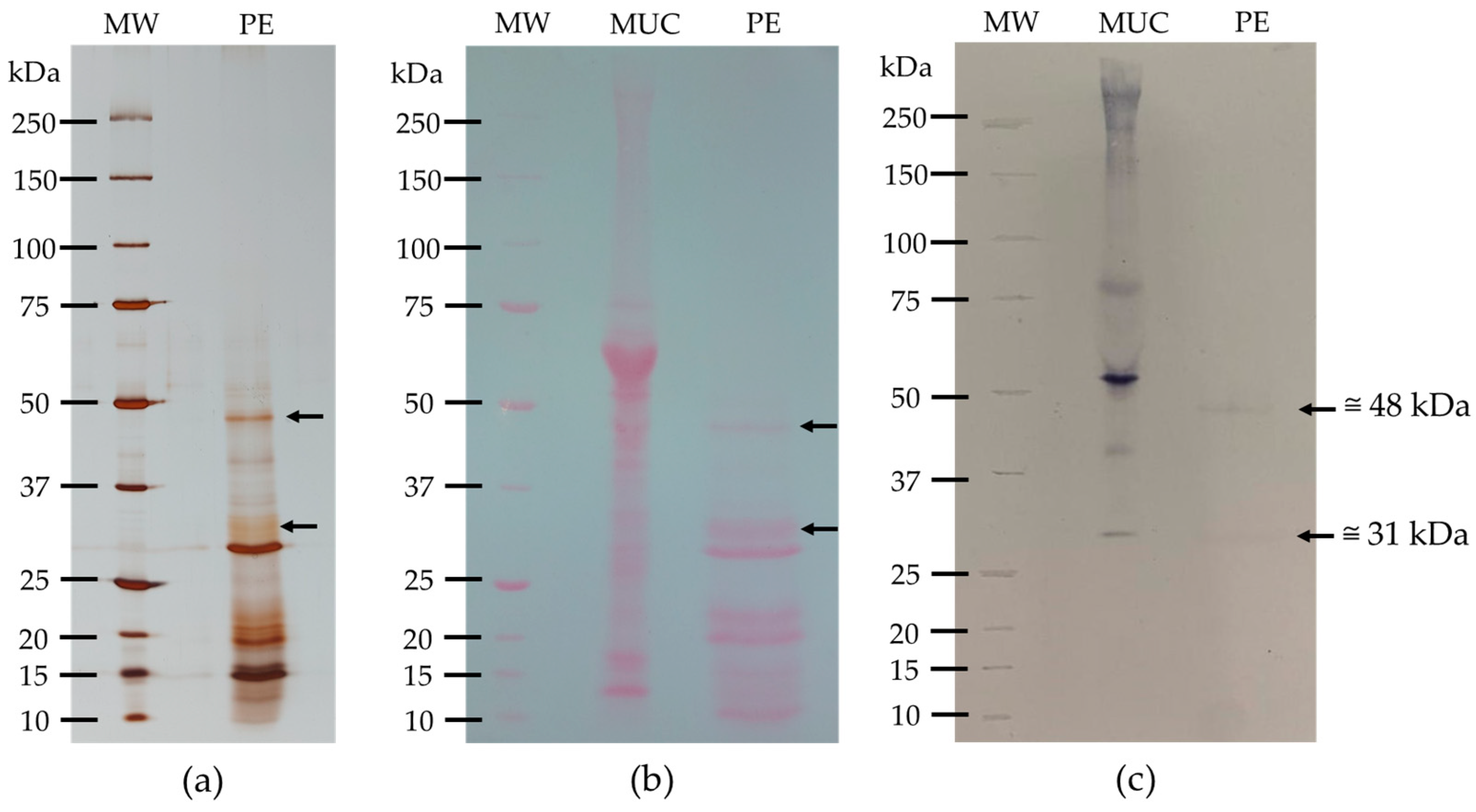
3.3.2. Glycoprotein Detection
3.4. Lectin Characterization
3.4.1. Hemagglutination Activity and Inhibition
3.4.2. Purification of Sialic Acid Lectins
3.5. Lectin Binding to HT-29 and MIA PaCa-2
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Associação dos Jovens Agricultores de Portugal. Manual Boas Práticas para Culturas Emergentes—A Cultura do Kiwi; Associação dos Jovens Agricultores de Portugal: Lisboa, Portugal, 2017; pp. 10–18. [Google Scholar]
- Richardson, D.P.; Ansell, J.; Drummond, L.N. The nutritional and health attributes of kiwifruit: A review. Eur. J. Nutr. 2018, 57, 2659–2676. [Google Scholar] [CrossRef] [PubMed]
- Franco, J. História e Desenvolvimento Comercial. In Kiwi da Produção à Comercialização; Antunes, M.D., Ed.; Ciências da Terra, Universidade do Algarve: Faro, Portugal, 2008; pp. 13–19. [Google Scholar]
- Wang, J.; Qiu, Y.; Zhu, F. Kiwifruit (Actinidia spp.): A review of chemical diversity and biological activities. Food Chem. 2021, 350, 128469. [Google Scholar] [CrossRef] [PubMed]
- McGhie, T. Secondary Metabolite Components of Kiwifruit. In Advances in Food and Nutrition Research; Boland, M., Moughan, P.J., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 68, pp. 101–124. [Google Scholar]
- Direito, R.; Rocha, J.; Sepodes, B.; Eduardo-Figueira, M. Phenolic Compounds Impact on Rheumatoid Arthritis, Inflammatory Bowel Disease and Microbiota Modulation. Pharmaceutics 2021, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Mattiuzzi, C. Kiwifruit and Cancer: An Overview of Biological Evidence. Nutr. Cancer 2020, 72, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Pisoschi, A.M.; Pop, A. The role of antioxidants in the chemistry of oxidative stress: A review. Eur. J. Med. Chem. 2015, 97, 55–74. [Google Scholar] [CrossRef]
- Chamorro, F.; Carpena, M.; Fraga-Corral, M.; Echave, J.; Riaz Rajoka, M.S.; Barba, F.J.; Cao, H.; Xiao, J.; Prieto, M.A.; Simal-Gandara, J. Valorization of kiwi agricultural waste and industry by-products by recovering bioactive compounds and applications as food additives: A circular economy model. Food Chem. 2022, 370, 131315. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, N.; Shu, C.; Cheng, S.; Sun, X.; Liu, C.; Xin, G.; Li, B.; Tian, J. Phenolics Profile and Antioxidant Activity Analysis of Kiwi Berry (Actinidia arguta) Flesh and Peel Extracts from Four Regions in China. Front. Plant. Sci. 2021, 12, 689038. [Google Scholar] [CrossRef]
- Stonehouse, W.; Gammon, C.S.; Beck, K.L.; Conlon, C.A.; Von Hurst, P.R.; Kruger, R. Kiwifruit: Our daily prescription for health. Can. J. Physiol. Pharmacol. 2013, 91, 442–447. [Google Scholar] [CrossRef]
- Zhou, Y.; Fei, G.; Faridul Hasan, K.M.; Kang, Y.; Wu, Y.; Li, H.; Zhou, S. Cultivar difference characterization of kiwifruit wines on phenolic profiles, volatiles and antioxidant activity. Food Chem. 2023, 18, 100691. [Google Scholar] [CrossRef]
- Lampe, J.W. Health effects of vegetables and fruit: Assessing mechanisms of action in human experimental studies. Am. J. Clin. Nutr. 1999, 70, 475S–490S. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, K.; Yan, C.; Yin, Y.; He, S.; Qiu, L.; Li, G. Natural Polyphenols for Treatment of Colorectal Cancer. Molecules 2022, 27, 8810. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, I.; Nunes, A.; Lima, A.; Borralho, P.; Rodrigues, C.; Ferreira, R.B.; Ribeiro, A.C. New lectins from mediterranean flora. Activity against HT29 colon cancer cells. Int. J. Mol. Sci. 2019, 20, 3059. [Google Scholar] [CrossRef] [PubMed]
- De Mejia, E.G.; Prisecaru, V.I. Lectins as bioactive plant proteins: A potential for anti-cancer therapy. Crit. Rev. Food Sci. Nutr. 2005, 45, 425–445. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.-L.; Zhou, C.-C.; Yao, S.; Yu, J.-Y.; Liu, B.; Bao, J.-K. Plant lectins: Targeting programmed cell death pathways as antitumor agents. Int. J. Biochem. Cell Biol. 2011, 43, 1442–1449. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Sun, R.; Yu, T.; Liu, R.; Cheng, L.-J.; Bao, J.-K.; Zou, L.; Tang, Y. Identification of Novel Pathways in Plant Lectin-Induced Cancer Cell Apoptosis. Int. J. Mol. Sci. 2016, 17, 228. [Google Scholar] [CrossRef]
- Lei, H.Y.; Chang, C.P. Induction of autophagy by concanavalin A and its application in anti-tumor therapy. Autophagy 2007, 3, 402–404. [Google Scholar] [CrossRef]
- Folin, O.; Ciocalteu, V. On tyrosine and tryptophane determinations in proteins. J. Biol. Chem. 1927, 73, 627–650. [Google Scholar] [CrossRef]
- Direito, R.; Reis, C.; Roque, L.; Gonçalves, M.; Sanches-Silva, A.; Gaspar, M.M.; Pinto, R.; Rocha, J.; Sepodes, B.; Bronze, M.R.; et al. Phytosomes with persimmon (Diospyros kaki L.) extract: Preparation and preliminary demonstration of in vivo tolerability. Pharmaceutics 2019, 11, 296. [Google Scholar] [CrossRef]
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Çam, M.; Hişil, Y. Pressurized water extraction of polyphenols from pomegranate peels. Food Chem. 2010, 123, 878–885. [Google Scholar] [CrossRef]
- Bento-Silva, A.; Duarte, N.; Mecha, E.; Belo, M.; Patto, M.C.V.; Bronze, M.R. Hydroxycinnamic Acids and Their Derivatives in Broa, a Traditional Ethnic Maize Bread. Foods 2020, 9, 1471. [Google Scholar] [CrossRef] [PubMed]
- Sucupira, N.R.; Silva, A.B.; Pereira, G.; Costa, J.N. Methods for measuring antioxidant activity of fruits. UNOPAR Cient. Ciênc. Biol. Saúde 2012, 14, 263–269. [Google Scholar]
- Hamiaux, C.; Maddumage, R.; Middleditch, M.J.; Prakash, R.; Brummell, D.A.; Baker, E.N.; Atkinson, R.G. Crystal structure of kiwellin, a major cell-wall protein from kiwifruit. J. Struct. Biol. 2014, 187, 276–281. [Google Scholar] [CrossRef]
- Grozdanovic, M.M.; Burazer, L.; Gavrovic-Jankulovic, M. Kiwifruit (Actinidia deliciosa) extract shows potential as a low-cost and efficient milk-clotting agent. Int. Dairy J. 2013, 32, 46–52. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Blum, H.; Beier, H.; Gross, H.J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 1987, 8, 93–99. [Google Scholar] [CrossRef]
- Faye, L.; Chrispeels, M.J. Characterization of N-linked Oligosaccharides by Affinoblotting with Concanavalin A-Peroxidase and Treatment of the Blots with Glycosidases. Anal. Biochem. 1985, 149, 218–224. [Google Scholar] [CrossRef]
- Ribeiro, A.C.; Catarino, S.; Freitas, R. Multiple lectin detection by cell membrane affinity binding. Carbohydr. Res. 2012, 352, 206–210. [Google Scholar] [CrossRef]
- Vercoutter-Edouart, A.S.; Slomianny, M.C.; Dekeyzer-Beseme, O.; Haeuw, J.F.; Michalski, J.C. Glycoproteomics and glycomics investigation of membrane N-glycosylproteins from human colon carcinoma cells. Proteomics 2008, 8, 3236–3256. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Chem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Liu, Z.; Shi, L.; Qi, Y.; Barrow, C.; Dunshea, F.; Suleria, H. Antioxidative Properties and Phenolic Profile of the Core, Pulp and Peel of Commercialized Kiwifruit by LC-ESI-QTOF-MS/MS. Processes 2022, 10, 1811. [Google Scholar] [CrossRef]
- Byland, D.; Norström, S.H.; Essén, S.A.; Lundström, U.S. Analysis of low molecular mass organic acids in natural waters by ion exclusion chromatography tandem mass spectrometry. J. Chromatogr. A 2007, 1176, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Rombolà, A.D.; Brüggemann, W.; López-Millán, A.F.; Tagliavini, M.; Abadía, J.; Marangoni, B.; Moog, P.R. Biochemical responses to iron deficiency in kiwifruit (Actinidia deliciosa). Tree Physiol. 2002, 22, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life science. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Dias, M.; Caleja, C.; Pereira, C.; Calhelha, R.C.; Kostic, M.; Sokovic, M.; Tavares, D.; Baraldi, I.J.; Barros, L.; Ferreira, I.C.F.R. Chemical composition and bioactive properties of byproducts from two different kiwi varieties. Int. Food Res. 2020, 127, 108753. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Ren, Y.; Wang, Y.; Han, M.; Yue, T.; Wang, Z.; Gao, Z. Physicochemical, nutritional, and bioactive properties of pulp and peel from 15 kiwifruit cultivars. Food Biosci. 2021, 42, 101157. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Q.; Lan, T.; Geng, T.; Gao, C.; Tuan, Q.; Zhang, Q.; Xu, P.; Sun, X.; Liu, X.; et al. Comparative Analysis of Physicochemical Characteristics, Nutritional and Functional Components and Antioxidant Capacity of Fifteen Kiwifruit (Actinidia) Cultivars—Comparative Analysis of Fifteen Kiwifruit (Actinidia) Cultivars. Foods 2020, 9, 1267. [Google Scholar] [CrossRef]
- Fiorentino, A.; D’Abrosca, B.; Pacifico, S.; Mastellone, C.; Scognamiglio, M.; Monaco, P. Identification and Assessment of Antioxidant Capacity of Phytochemicals from Kiwi Fruits. J. Agric. Food Chem. 2009, 57, 4148–4155. [Google Scholar] [CrossRef]
- Wu, X.; Beecher, G.R.; Holden, J.M.; Haytowitz, D.B.; Gebhardt, S.E.; Prior, R.L. Lipophilic and Hydrophilic Antioxidant Capacities of Common Foods in the United States. J. Agric. Food Chem. 2004, 52, 4026–4037. [Google Scholar] [CrossRef]
- Taofiq, O.; González-Paramás, A.M.; Barreiro, M.F.; Ferreira, I.C.F.R. Hydroxycinnamic Acids and Their Derivatives: Cosmeceutical Significance, Challenges and Future Perspectives, a Review. Molecules 2017, 22, 281. [Google Scholar] [CrossRef]
- Seifried, H.E.; Anderson, D.E.; Fisher, E.I.; Milner, J.A. A review of the interaction among dietary antioxidants and reactive oxygen species. J. Nutr. Biochem. 2007, 18, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Dartsch, P.C.; Kler, A.; Kriesl, E. Antioxidative and anti-inflammatory potential of different functional drink concepts in vitro. Phytother. Res. 2009, 23, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Su, M.S.; Chein, P.J. Antioxidant activity, anthocyanins, and phenolics of rabbiteye blueberry (Vaccinium ashei) fluid products as affected by fermentation. Food Chem. 2007, 104, 182–187. [Google Scholar] [CrossRef]
- Sousa, M.; Machado, V.; Costa, R.; Figueira, M.E.; Sepodes, B.; Barata, P.; Ribeiro, L.; Soares, R. Red Raspberry Phenols Inhibit Angiogenesis: A Morphological and Subcellular Analysis Upon Human Endothelial Cells. J. Cell. Biochem. 2016, 117, 1604–1612. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef]
- Golawska, S.; Lukasik, I.; Chojnacki, A.A.; Chrzanowski, G. Flavonoids and Phenolic Acids Content in Cultivation and Wild Collection of European Cranberry Bush Viburnum opulus L. Molecules 2023, 28, 2285. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Liu, H.; Zhao, T.; Meng, C.; Liu, Z.; Liu, X. Bioactive compounds and in vitro antioxidant activities of peel, flesh and seed powder of kiwi fruit. Int. J. Food Sci. Technol. 2018, 53, 2239–2245. [Google Scholar] [CrossRef]
- Bernardes, N.R.; Talma, S.V.; Sampaio, S.H.; Nunes, C.R.; Almeida, J.; Oliveira, D. Atividade antioxidante e fenóis totais de frutas de Campos dos Goytacazes RJ. Seção Artig. 2011, 1, 53–59. [Google Scholar] [CrossRef]
- Rajashekar, C.B. Dual Role of Plant Phenolic Compounds as Antioxidants and Prooxidants. Am. J. Plant Sci. 2023, 14, 15–28. [Google Scholar] [CrossRef]
- Carmo, M.A.V.; Granato, D.; Azevedo, L. Antioxidant/pro-oxidant and antiproliferative activities of phenolic-rich foods and extracts. In Advances in Food and Nutrition Research; Granato, D., Ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 98, pp. 253–280. [Google Scholar]
- Dobie, C.; Skropeta, D. Insights into the role of sialylation in cancer progression and metastasis. Br. J. Cancer 2021, 124, 76–90. [Google Scholar] [CrossRef]
- Munkley, J. Aberrant Sialylation in Cancer: Therapeutic Opportunities. Cancers 2022, 14, 4248. [Google Scholar] [CrossRef] [PubMed]
- Rastmanesh, R.; Bowirrat, A.; Gupta, A.; Gilley, E.; Blum, K. Anti(angiogenic) food components: Can be a major source of bias in the investigation of angiogenesis inhibitors. Ann. Transl. Med. 2023, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.; Sarkar, S.; Natoni, A.; Stark, J.C.; Riley, N.M.; Bertozzi, C.R.; Carlsten, M.; O’Dwyer, M.E. Targeting hipersialylation in multiple myeloma represents a novel approach to enhance NK cell-mediated tumor responses. Blood Adv. 2022, 6, 3352–3366. [Google Scholar] [CrossRef] [PubMed]


| No. | RT (min) | Putative Identification | λmax (nm) | Ionization (ESI+/ESI−) | m/z | MS2 Product Ions | References |
|---|---|---|---|---|---|---|---|
| 1 | 6.36 | Caffeoyl glucose | 269 | [M − H]− | 341 | 59, 89, 101, 119, 161, 179 | [34] |
| 2 | 7.23 | 2-Hydroxybenzoic acid | 274 | [M + H]+ | 139 | 65, 77, 93, 95 | [34] |
| 3 | 7.95 | Malic acid | 240 | [M − H]− | 133 | 71, 73, 115 | [5,35] |
| 4 | 8.29 | cis-Aconitic acid | 254 | [M − H]− | 173 | 67, 111 | [35,36] |
| 5 | 10.36 | Citric acid | 253 | [M − H]− | 191 | 85, 87, 111 | [5,35] |
| 6 | 29.84 | 2-Methylquinoline | 283 | [M + H]+ | 144 | 91, 115, 117, 143, 144 | [37] |
| 7 | 35.63 | Caffeic acid hexoside isomer 1 | 316 | [M − H]− | 341 | 71, 135, 179 | [38] |
| 8 | 40.11 | Caffeic acid hexoside isomer 2 | 283 | [M − H]− | 341 | 118, 135, 179, 194, 364 | [38] |
| No. | MW (kDa) | pI |
|---|---|---|
| 1 | 10 | 5.7 |
| 2 (A–E) | 11 | 4.3; 4.5; 4.7; 4.9; 8.4–8.9 |
| 3 (A–C) | 12 | 3.9; 5.9; 7.4 |
| 4 (A–D) | 13 | 3.9; 4.2; 5.8; 6.4 |
| 5 (A–B) | 14 | 4.8; 5 |
| 6 (A–C) | 15 | 5; 5.4; 5.7 |
| 7 (A–B) | 16 | 4.3–4.6; 10 |
| 8 (A–B) | 17 | 4.2; 5.9 |
| 9 | 19 | 3.7 |
| 10 | 20 | 3.8 |
| 11 (A–C) | 22 | 3.9; 4.4; 9.7 |
| 12 (A–C) | 24 | 3.8; 6.4; 9.6 |
| 13 (A–B) | 25 | 4.8; 5.4 |
| 14 (A–B) | 26 | 3.9; 6.2–6.7 |
| 15 | 27 | 5.4 |
| 16 (A–C) | 29 | 3.7; 3.9; 4 |
| 17 | 31 | 3.6–4.3 |
| 18 | 32 | 9.9 |
| 19 | 37 | 5.1 |
| Sugars | Kiwifruit Protein Extract | |
|---|---|---|
| No. | (0.1 M) | Sugar Minimal Inhibitory Concentration (MIC) (M) |
| 1 | d-Glucose | UD |
| 2 | d-Glucosamine | 0.1 |
| 3 | N-Acetyl-d-glucosamine | UD |
| 4 | d-Galactose | UD |
| 5 | d-Galactosamine | 0.1 |
| 6 | N-Acetyl-d-galactosamine | UD |
| 7 | Lactose | UD |
| 8 | d-Mannose | UD |
| 9 | Raffinose | UD |
| 10 | l-Fucose * | UD |
| 11 | Melezitose | UD |
| 12 | α-Methyl-d-glucopyranoside | UD |
| 13 | α-Methyl-d-mannopyranoside | UD |
| 14 | Sucrose | UD |
| 15 | Maltose | UD |
| 16 | Sialic acid | 1.56 × 10−3 |
| 17 | l(−)-Fucose * | UD |
| Hemagglutination Activity (H.U.) | |
|---|---|
| Peak 1 | Until 1:512 dilution |
| Peak 2 | Until 1:1024 dilution |
| Peak 3 | <1:2048 dilution |
| Peak 4 | <1:2048 dilution |
| Molecular Weight (kDa) | 2D Spots Analysis | Glycoprotein Detection | Lectin Binding to HT-29 Membranes | Lectin Binding to MIA PaCa-2 Membranes | Sialic Acid Specific Lectin Purification |
|---|---|---|---|---|---|
| 10 | 1 | + | |||
| 12 | 2 | + | |||
| 13 | 3 | + | + | ||
| 15 | 4 | ? | ? | + | |
| 20 | 6 | + | + | + | |
| 22 | 10 | + | ? | ||
| 24 | 11 | + | ? | + | |
| 25 | 12 | ? | |||
| 31 | 13 | + | + | + | + |
| 37 | 17 | + | |||
| 42 | 19 | + | + | ||
| 48 | - | + | + | ||
| 50 | - | + | |||
| 52 | - | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, R.; Figueira, M.E.; Direito, R.; Bento-Silva, A.; Ferreira, R.B.; Ribeiro, A.C. Exploring Lectin Bioactivity and Total Phenolic Compounds in Kiwifruit (Actinidia deliciosa var. Hayward). Nutrients 2024, 16, 3292. https://doi.org/10.3390/nu16193292
Rodrigues R, Figueira ME, Direito R, Bento-Silva A, Ferreira RB, Ribeiro AC. Exploring Lectin Bioactivity and Total Phenolic Compounds in Kiwifruit (Actinidia deliciosa var. Hayward). Nutrients. 2024; 16(19):3292. https://doi.org/10.3390/nu16193292
Chicago/Turabian StyleRodrigues, Raquel, Maria Eduardo Figueira, Rosa Direito, Andreia Bento-Silva, Ricardo Boavida Ferreira, and Ana Cristina Ribeiro. 2024. "Exploring Lectin Bioactivity and Total Phenolic Compounds in Kiwifruit (Actinidia deliciosa var. Hayward)" Nutrients 16, no. 19: 3292. https://doi.org/10.3390/nu16193292
APA StyleRodrigues, R., Figueira, M. E., Direito, R., Bento-Silva, A., Ferreira, R. B., & Ribeiro, A. C. (2024). Exploring Lectin Bioactivity and Total Phenolic Compounds in Kiwifruit (Actinidia deliciosa var. Hayward). Nutrients, 16(19), 3292. https://doi.org/10.3390/nu16193292







