Comparative Study of Prebiotics for Infants Using a Fecal Culture System: Insights into Responders and Non-Responders
Abstract
:1. Introduction
2. Materials and Methods
2.1. Oligosaccharides
2.2. Fecal Samples
2.3. In Vitro Fecal Fermentation
2.4. DNA Extraction
2.5. Quantification of Bacterial Cell Numbers
2.6. SCFAs and Lactate Analysis
2.7. Statistical Analysis
3. Results
3.1. Characteristics of the Subjects
3.2. Effects of Various Prebiotic Oligosaccharides and Their Combinations on the Growth of Individual Infant-Type Bifidobacteria
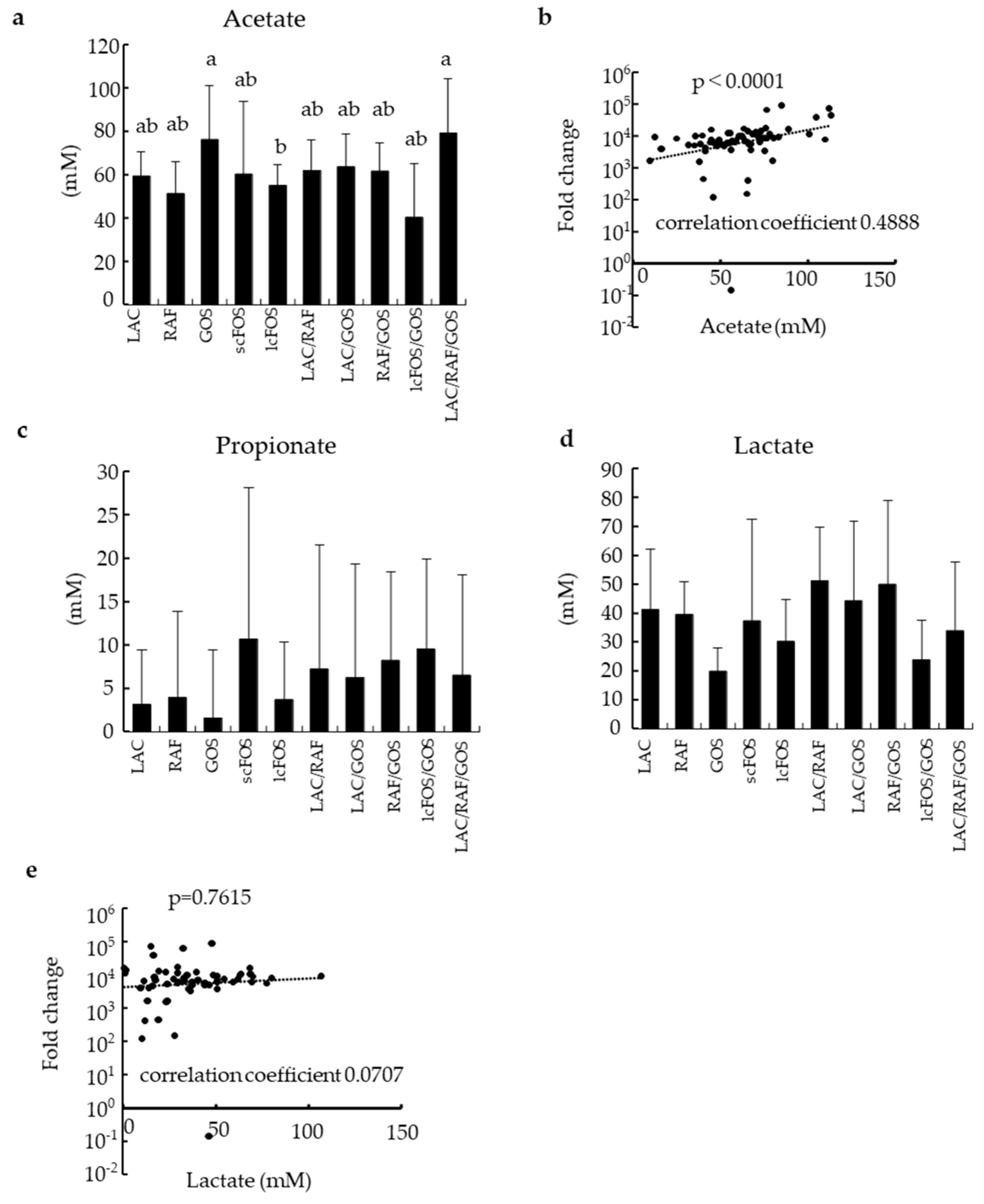
3.3. Effects of Prebiotic Oligosaccharides and Their Combinations on SCFAs and Lactate Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saturio, S.; Nogacka, A.M.; Alvarado-Jasso, G.M.; Salazar, N.; de los Reyes-Gavilán, C.G.; Gueimonde, M.; Arboleya, S. Role of Bifidobacteria on Infant Health. Microorganisms 2021, 9, 2415. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.B.; Sugahara, H.; Odamaki, T.; Xiao, J.Z. Different Physiological Properties of Human-Residential and Non-Human-Residential Bifidobacteria in Human Health. Benef. Microbes 2018, 9, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, H.; Odamaki, T.; Hashikura, N.; Abe, F.; Xiao, J.Z. Differences in Folate Production by Bifidobacteria of Different Origins. Biosci. Microbiota Food Health 2015, 34, 87–93. [Google Scholar] [CrossRef]
- Hiraku, A.; Nakata, S.; Murata, M.; Xu, C.; Mutoh, N.; Arai, S.; Odamaki, T.; Iwabuchi, N.; Tanaka, M.; Tsuno, T.; et al. Early Probiotic Supplementation of Healthy Term Infants with Bifidobacterium Longum Subsp. Infantis M-63 Is Safe and Leads to the Development of Bifidobacterium-Predominant Gut Microbiota: A Double-Blind, Placebo-Controlled Trial. Nutrients 2023, 15, 1402. [Google Scholar] [CrossRef]
- Wong, C.B.; Iwabuchi, N.; Xiao, J.Z. Exploring the Science behind Bifidobacterium Breve M-16V in Infant Health. Nutrients 2019, 11, 1724. [Google Scholar] [CrossRef]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-Mediated Immune System Imprinting Early in Life. Cell 2021, 184, 3884–3898. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Ehara, T.; Sugahara, H.; Matsubara, T.; Mitsuyama, E.; Nakazato, Y.; Tsuda, M.; Shimizu, T.; Odamaki, T.; Xiao, J.Z.; et al. The Combination of Bifidobacterium Breve and Three Prebiotic Oligosaccharides Modifies Gut Immune and Endocrine Functions in Neonatal Mice. J. Nutr. 2019, 149, 344–353. [Google Scholar] [CrossRef]
- Huda, M.N.; Ahmad, S.M.; Alam, M.J.; Khanam, A.; Kalanetra, K.M.; Taft, D.H.; Raqib, R.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Bifidobacterium Abundance in Early Infancy and Vaccine Response at 2 Years of Age. Pediatrics 2019, 143, e20181489. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria Can Protect from Enteropathogenic Infection through Production of Acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Sakurai, T.; Odamaki, T.; Xiao, J.Z. Production of Indole-3-Lactic Acid by Bifidobacterium Strains Isolated Fromhuman Infants. Microorganisms 2019, 7, 340. [Google Scholar] [CrossRef]
- Newburg, D.S.; Walker, W.A. Protection of the Neonate by the Innate Immune System of Developing Gut and of Human Milk. Pediatr. Res. 2007, 61, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Lönnerdal, B. Nutritional and Physiologic Significance of Human Milk Proteins. Am. J. Clin. Nutr. 2003, 77, 1537S–1543S. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Lim, J.Y.; Kim, B.S.; Cho, S.J.; Kim, N.Y.; Kim, O.B.; Kim, Y. Comparison of the Gut Microbiota Profile in Breast-Fed and Formula-Fed Korean Infants Using Pyrosequencing. Nutr. Res. Pract. 2015, 9, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, M.; Gotoh, A.; Yoshida, K.; Odamaki, T.; Koguchi, H.; Xiao, J.Z.; Kitaoka, M.; Katayama, T. Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation with Bifidobacteria-Rich Microbiota Formation. Nutrients 2020, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Soyyilmaz, B.; Mikš, M.H.; Röhrig, C.H.; Matwiejuk, M.; Meszaros-matwiejuk, A.; Vigsnæs, L.K. The Mean of Milk: A Review of Human Milk Oligosaccharide Concentrations throughout Lactation. Nutrients 2021, 13, 2737. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.T.; Chen, C.; Kling, D.E.; Liu, B.; McCoy, J.M.; Merighi, M.; Heidtman, M.; Newburg, D.S. The Principal Fucosylated Oligosaccharides of Human Milk Exhibit Prebiotic Properties on Cultured Infant Microbiota. Glycobiology 2013, 23, 169–177. [Google Scholar] [CrossRef]
- Laursen, M.F.; Roager, H.M. Human Milk Oligosaccharides Modify the Strength of Priority Effects in the Bifidobacterium Community Assembly during Infancy. ISME J. 2023, 17, 2452–2457. [Google Scholar] [CrossRef]
- Schönknecht, Y.B.; Moreno Tovar, M.V.; Jensen, S.R.; Parschat, K. Clinical Studies on the Supplementation of Manufactured Human Milk Oligosaccharides: A Systematic Review. Nutrients 2023, 15, 3622. [Google Scholar] [CrossRef]
- Parschat, K.; Melsaether, C.; Jäpelt, K.R.; Jennewein, S. Clinical Evaluation of 16-Week Supplementation with Tolerability, Safety and Effect on Growth. Nutrients 2021, 13, 2871. [Google Scholar] [CrossRef]
- Bosheva, M.; Tokodi, I.; Krasnow, A.; Pedersen, H.K.; Lukjancenko, O.; Eklund, A.C.; Grathwohl, D.; Sprenger, N.; Berger, B.; Cercamondi, C.I. Infant Formula with a Specific Blend of Five Human Milk Oligosaccharides Drives the Gut Microbiota Development and Improves Gut Maturation Markers: A Randomized Controlled Trial. Front. Nutr. 2022, 9, 920362. [Google Scholar] [CrossRef]
- Kiyosawa, I.; Takase, M.; Yamauchi, K.; Ono, J.; Yaeshima, T.; Okonogi, S. Lactulose and Intestinal Microflora in Infant Nutrition. Bifidobact. Microflora 1986, 5, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Nagendra, R.; Viswanatha, S.; Kumar, S.A.; Murthy, B.K.; Rao, S.V. Effect of Feeding Milk Formula Containing Lactulose to Infants on Faecal Bifidobacterial Flora. Nutr. Res. 1995, 15, 15–24. [Google Scholar] [CrossRef]
- Hattori, K.; Sasai, M.; Yamamoto, A.; Taniuchi, S.; Kojima, T.; Kobayashi, Y.; Iwamoto, H.; Yaeshima, T.; Hayasawa, H. Intestinal Flora of Infants with Cow Milk Hypersensitivity Fed on Casein-Hydrolyzed Formula Supplemented Raffinose. Arerugi 2000, 49, 1146–1155. [Google Scholar] [PubMed]
- Williams, T.; Choe, Y.; Price, P.; Katz, G.; Suarez, F.; Paule, C.; Mackey, A. Tolerance of Formulas Containing Prebiotics in Healthy, Term Infants. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Paineau, D.; Respondek, F.; Menet, V.; Sauvage, R.; Bornet, F.; Wagner, A. Effects of Short-Chain Fructooligosaccharides on Faecal Bifidobacteria and Specific Immune Response in Formula-Fed Term Infants: A Randomized, Double-Blind, Placebo-Cotrolled Trial. J. Nutr. Sci. Vitaminol. 2014, 60, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Veereman, G. Pediatric Applications of Inulin and Oligofructose. J. Nutr. 2007, 137, 2585–2589. [Google Scholar] [CrossRef]
- Ackerman, D.L.; Craft, K.M.; Townsend, S.D. Infant Food Applications of Complex Carbohydrates: Structure, Synthesis, and Function. Carbohydr. Res. 2017, 437, 16–27. [Google Scholar] [CrossRef]
- Scholtens, P.A.M.J.; Alliet, P.; Raes, M.; Alles, M.S.; Kroes, H.; Boehm, G.; Knippels, L.M.J.; Knol, J.; Vandenplas, Y. Fecal Secretory Immunoglobulin A Is Increased in Healthy Infants Who Receive a Formula with Short-Chain Galacto-Oligosaccharides and Long-Chain Fructo-Oligosaccharides. J. Nutr. 2008, 138, 1141–1147. [Google Scholar] [CrossRef]
- Ehara, T.; Izumi, H.; Tsuda, M.; Nakazato, Y.; Iwamoto, H.; Namba, K.; Takeda, Y. Combinational Effects of Prebiotic Oligosaccharides on Bifidobacterial Growth and Host Gene Expression in a Simplified Mixed Culture Model and Neonatal Mice. Br. J. Nutr. 2016, 116, 270–278. [Google Scholar] [CrossRef]
- Ojima, M.N.; Yoshida, K.; Sakanaka, M.; Jiang, L.; Odamaki, T.; Katayama, T. Ecological and Molecular Perspectives on Responders and Non-Responders to Probiotics and Prebiotics. Curr. Opin. Biotechnol. 2022, 73, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hirano, R.; Sakai, Y.; Choi, M.; Sakanaka, M.; Kurihara, S.; Iino, H.; Xiao, J.Z.; Katayama, T.; Odamaki, T. Bifidobacterium Response to Lactulose Ingestion in the Gut Relies on a Solute-Binding Protein-Dependent ABC Transporter. Commun. Biol. 2021, 4, 541. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Odamaki, T.; Namura, M.; Shimizu, T.; Iwatsuki, K.; Nishimoto, M.; Kitaoka, M.; Xiao, J. zhong In Vitro Comparative Evaluation of the Impact of Lacto-N-Biose I, a Major Building Block of Human Milk Oligosaccharides, on the Fecal Microbiota of Infants. Anaerobe 2013, 19, 50–57. [Google Scholar] [CrossRef]
- Murakami, R.; Hashikura, N.; Yoshida, K.; Xiao, J.Z.; Odamaki, T. Growth-Promoting Effect of Alginate on Faecalibacterium Prausnitzii through Cross-Feeding with Bacteroides. Food Res. Int. 2021, 144, 110326. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Xia, X.; Tang, R.; Zhou, J.; Zhao, H.; Wang, K. Development of a Real-Time PCR Method for Firmicutes and Bacteroidetes in Faeces and Its Application to Quantify Intestinal Population of Obese and Lean Pigs. Lett. Appl. Microbiol. 2008, 47, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Takada, T. Use of 16S rRNA Gene-Targeted Group-Specific Primers for Real-Time PCR Analysis of Predominant Bacteria in Human Feces. Appl. Environ. Microbiol. 2004, 70, 7220–7228. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Watanabe, K.; Tanaka, R.; Oyaizu, H. Rapid Identification of Human Intestinal Bifidobacteria by 16S rRNA-Targeted Species- and Group-Specific Primers. FEMS Microbiol. Lett. 1998, 167, 113–121. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Kado, Y.; Takada, T.; Matsumoto, K.; Tanaka, R. Quantitative PCR with 16S rRNA-Gene-Targeted Species-Specific Primers for Analysis of Human Intestinal Bifidobacteria. Appl. Environ. Microbiol. 2004, 70, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Odamaki, T.; Mitsuyama, E.; Sugahara, H.; Xiao, J.Z.; Osawa, R. Age-Related Changes in the Composition of Gut Bifidobacterium Species. Curr. Microbiol. 2017, 74, 987–995. [Google Scholar] [CrossRef]
- Xiao, J.Z.; Takahashi, S.; Nishimoto, M.; Odamaki, T.; Yaeshima, T.; Iwatsuki, K.; Kitaoka, M. Distribution of In Vitro Fermentation Ability of Lacto-TV-Biose I, a Major Building Block of Human Milk Oligosaccharides, in Bifidobacteria! Strains. Appl. Environ. Microbiol. 2010, 76, 54–59. [Google Scholar] [CrossRef]
- Watson, D.; O’Connell Motherway, M.; Schoterman, M.H.C.; van Neerven, R.J.J.; Nauta, A.; Van Sinderen, D. Selective Carbohydrate Utilization by Lactobacilli and Bifidobacteria. J. Appl. Microbiol. 2013, 114, 1132–1146. [Google Scholar] [CrossRef]
- Ojima, M.N.; Jiang, L.; Arzamasov, A.A.; Yoshida, K.; Odamaki, T.; Xiao, J.; Nakajima, A.; Kitaoka, M.; Hirose, J.; Urashima, T.; et al. Priority Effects Shape the Structure of Infant-Type Bifidobacterium Communities on Human Milk Oligosaccharides. ISME J. 2022, 16, 2265–2279. [Google Scholar] [CrossRef] [PubMed]
- Morozumi, M.; Wada, Y.; Tsuda, M.; Tabata, F.; Ehara, T.; Nakamura, H.; Miyaji, K. Cross-Feeding among Bifidobacteria on Glycomacropeptide. J. Funct. Foods 2023, 103, 105463. [Google Scholar] [CrossRef]
- Nishiyama, K.; Nagai, A.; Uribayashi, K.; Yamamoto, Y.; Mukai, T.; Okada, N. Two Extracellular Sialidases from Bifidobacterium Bifidum Promote the Degradation of Sialyl-Oligosaccharides and Support the Growth of Bifidobacterium Breve. Anaerobe 2018, 52, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Nagy, D.U.; Sándor-Bajusz, K.A.; Bódy, B.; Decsi, T.; Van Harsselaar, J.; Theis, S.; Lohner, S. Effect of Chicory-Derived Inulin-Type Fructans on Abundance of Bifidobacterium and on Bowel Function: A Systematic Review with Meta-Analyses. Crit. Rev. Food Sci. Nutr. 2023, 63, 12018–12035. [Google Scholar] [CrossRef]
- Healey, G.; Murphy, R.; Butts, C.; Brough, L.; Whelan, K.; Coad, J. Habitual Dietary Fibre Intake Influences Gut Microbiota Response to an Inulin-Type Fructan Prebiotic: A Randomised, Double-Blind, Placebo-Controlled, Cross-over, Human Intervention Study. Br. J. Nutr. 2018, 119, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Sumiyoshi, W.; Urashima, T.; Nakamura, T.; Arai, I.; Nagasawa, T.; Saito, T.; Tsumura, N.; Wang, B.; Brand-Miller, J.; Watanabe, Y.; et al. Galactosyllactoses in the Milk of Japanese Women: Changes in Concentration during the Course of Lactation. J. Appl. Glycosci. 2004, 51, 341–344. [Google Scholar] [CrossRef]
- Takeuchi, T.; Miyauchi, E.; Kanaya, T.; Kato, T.; Nakanishi, Y.; Watanabe, T.; Kitami, T.; Taida, T.; Sasaki, T.; Negishi, H.; et al. Acetate Differentially Regulates IgA Reactivity to Commensal Bacteria. Nature 2021, 595, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Antunes, K.H.; Fachi, J.L.; de Paula, R.; da Silva, E.F.; Pral, L.P.; dos Santos, A.Á.; Dias, G.B.M.; Vargas, J.E.; Puga, R.; Mayer, F.Q.; et al. Microbiota-Derived Acetate Protects against Respiratory Syncytial Virus Infection through a GPR43-Type 1 Interferon Response. Nat. Commun. 2019, 10, 3273. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T.; et al. The Gut Microbiota Suppresses Insulin-Mediated Fat Accumulation via the Short-Chain Fatty Acid Receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, A.; Allahyar, A.; Greiner, T.U.; Plovier, H.; Lundén, G.Ö.; Larsson, T.; Drucker, D.J.; Delzenne, N.M.; Cani, P.D.; Bäckhed, F. Microbial Modulation of Energy Availability in the Colon Regulates Intestinal Transit. Cell Host Microbe 2013, 14, 582–590. [Google Scholar] [CrossRef]
- Tsukuda, N.; Yahagi, K.; Hara, T.; Watanabe, Y.; Matsumoto, H.; Mori, H.; Higashi, K.; Tsuji, H.; Matsumoto, S.; Kurokawa, K.; et al. Key Bacterial Taxa and Metabolic Pathways Affecting Gut Short-Chain Fatty Acid Profiles in Early Life. ISME J. 2021, 15, 2574–2590. [Google Scholar] [CrossRef] [PubMed]
- Weis, A.M.; Round, J.L. Microbiota-Antibody Interactions That Regulate Gut Homeostasis. Cell Host Microbe 2021, 29, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Masuda, K.; Nakamura, K.; Yoshioka, S.; Fukaya, R.; Sakai, N.; Ayabe, T. Regulation of Microbiota by Antimicrobial Peptides in the Gut. Adv. Otorhinolaryngol. 2011, 72, 97–99. [Google Scholar] [CrossRef]
- Ostaff, M.J.; Stange, E.F.; Wehkamp, J. Antimicrobial Peptides and Gut Microbiota in Homeostasis and Pathology. EMBO Mol. Med. 2013, 5, 1465–1483. [Google Scholar] [CrossRef] [PubMed]
- Donald, K.; Petersen, C.; Turvey, S.E.; Finlay, B.B.; Azad, M.B. Review Secretory IgA: Linking Microbes, Maternal Health, and Infant Health through Human Milk. Cell Host Microbe 2022, 30, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Minami, J.; Odamaki, T.; Hashikura, N.; Abe, F.; Xiao, J.Z. Lysozyme in Breast Milk Is a Selection Factor for Bifidobacterial Colonisation in the Infant Intestine. Benef. Microbes 2016, 7, 53–60. [Google Scholar] [CrossRef]
- Gopalakrishna, K.P.; Hand, T.W. Influence of Maternal Milk on the Neonatal Intestinal Microbiome. Nutrients 2020, 12, 823. [Google Scholar] [CrossRef]

| Subject ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Age (months) * | 1.5 | 3 | 6.4 | 6.4 | 7.0 | 9.1 | 10.2 |
| Breast-fed | + | + | + | + | + | + | + |
| Formula-fed | + | + | + | + | |||
| Solid food | + | + | + |
| Subject ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| B. breve | + | + | + | + | + | + | |
| B. longum subsp. longum | + | + | + | + | + | ||
| B. longum subsp. infantis | + | + | + | + | + | ||
| B. bifidum | + | + |
| Groups | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LAC | RAF | GOS | scFOS | lcFOS | LAC/ RAF | LAC/ GOS | RAF/ GOS | lcFOS/ GOS | LAC /RAF /GOS | |
| Lactulose (LAC) | + | + | + | + | ||||||
| Raffinose (RAF) | + | + | + | + | ||||||
| Galactooligosaccharides (GOS) | + | + | + | + | + | |||||
| Short-chain fructooligosaccharides (scFOS) | + | |||||||||
| Long-chain fructooligosaccharides (lcFOS) | + | + | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mingat, S.; Ehara, T.; Nakamura, H.; Miyaji, K. Comparative Study of Prebiotics for Infants Using a Fecal Culture System: Insights into Responders and Non-Responders. Nutrients 2024, 16, 3347. https://doi.org/10.3390/nu16193347
Mingat S, Ehara T, Nakamura H, Miyaji K. Comparative Study of Prebiotics for Infants Using a Fecal Culture System: Insights into Responders and Non-Responders. Nutrients. 2024; 16(19):3347. https://doi.org/10.3390/nu16193347
Chicago/Turabian StyleMingat, Shijir (Xijier), Tatsuya Ehara, Hirohiko Nakamura, and Kazuhiro Miyaji. 2024. "Comparative Study of Prebiotics for Infants Using a Fecal Culture System: Insights into Responders and Non-Responders" Nutrients 16, no. 19: 3347. https://doi.org/10.3390/nu16193347
APA StyleMingat, S., Ehara, T., Nakamura, H., & Miyaji, K. (2024). Comparative Study of Prebiotics for Infants Using a Fecal Culture System: Insights into Responders and Non-Responders. Nutrients, 16(19), 3347. https://doi.org/10.3390/nu16193347






