Evaluation of the Leaves and Seeds of Cucurbitaceae Plants as a New Source of Bioactive Compounds for Colorectal Cancer Prevention and Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plant Material
2.3. Cold Ethanolic and Reflux Ethanolic Extracts
2.4. Protein Extracts and Protein Hydrolysates
2.5. Cell Viability Assay
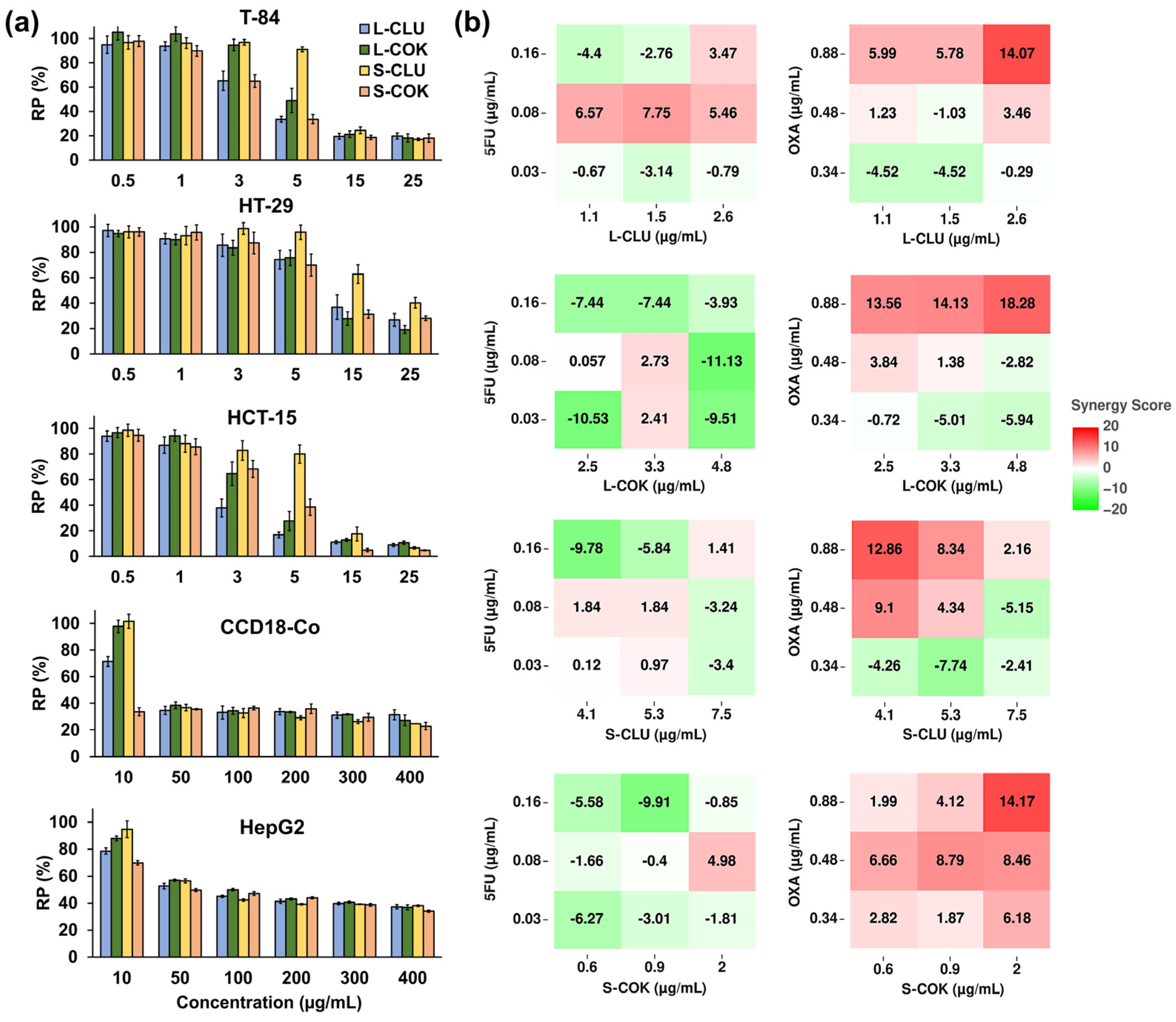
2.6. Synergistic Effect Analysis
2.7. Cell Cycle and Apoptosis Flow Cytometry Analysis
2.8. Lysotracker Labeling
2.9. Immunofluorescence Assay
2.10. Western Blot Analysis
2.11. Clonogenicity Assay
2.12. RT-PCR
2.13. Wound Healing Assay
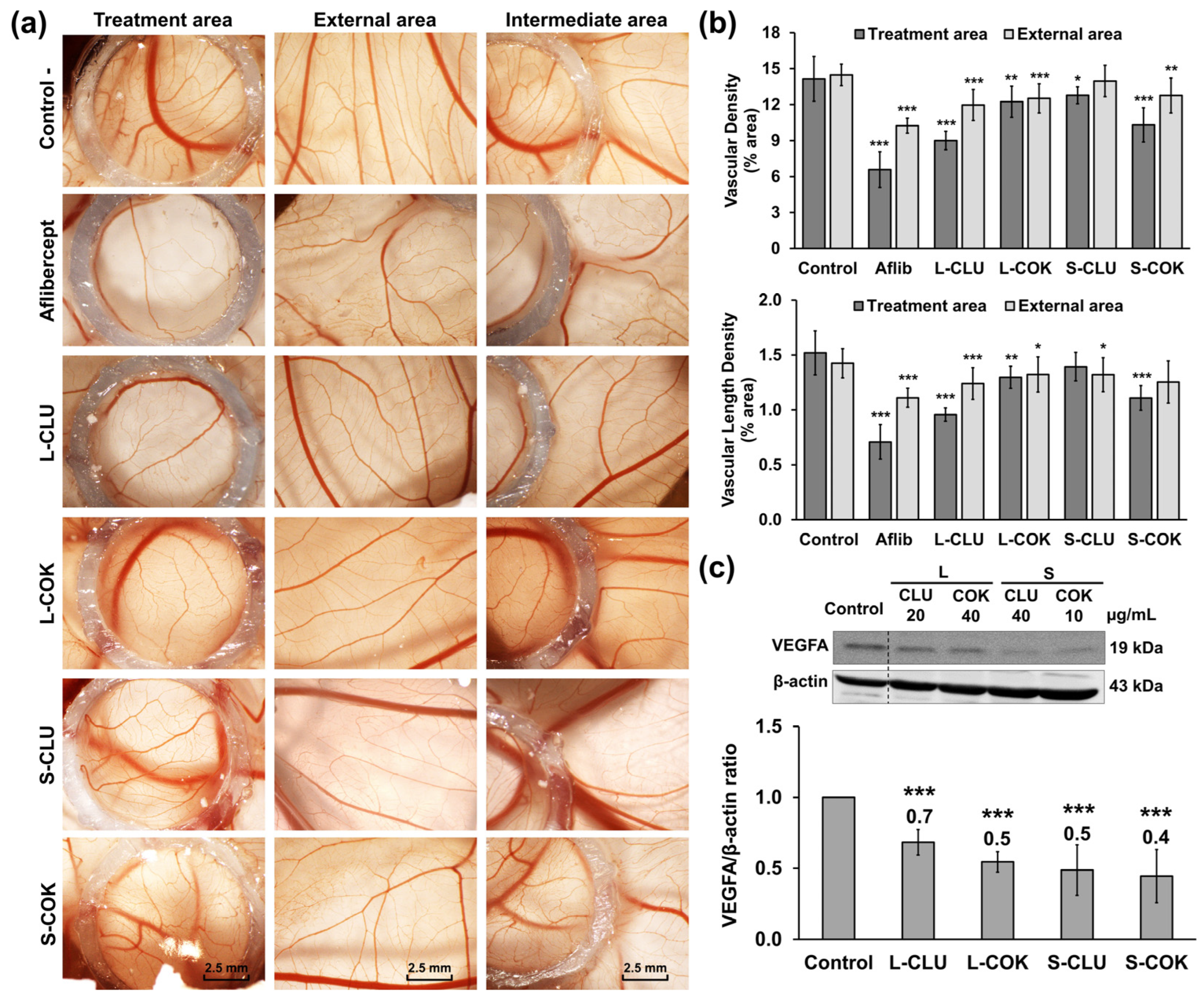
2.14. Angiogenesis Study by CAM Assay
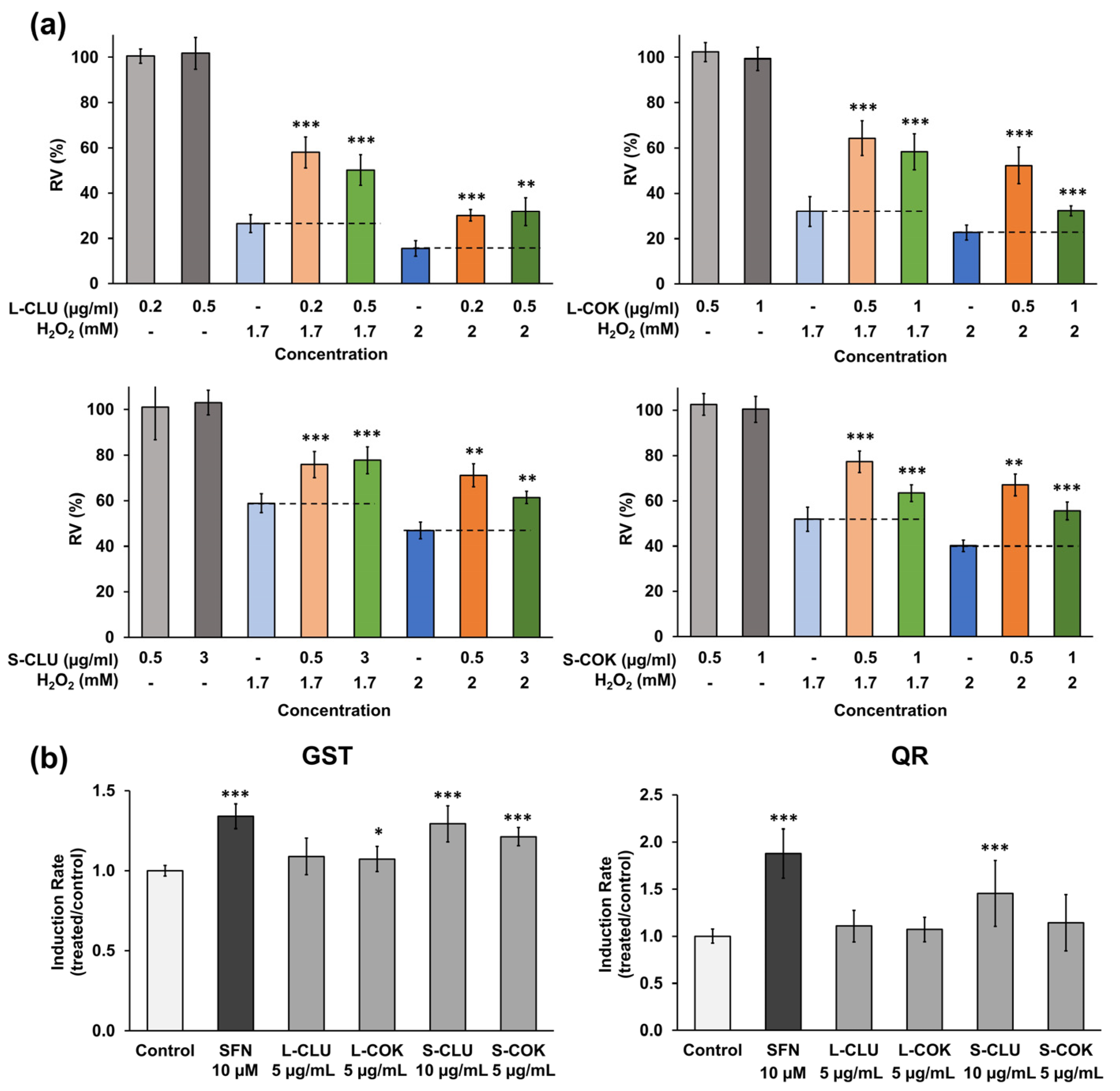
2.15. Antioxidant Activity In Vitro
2.16. Determination of Detoxifying Enzymes Induction Capacity
2.17. Bioguided Fractionation of Active Extracts
2.18. Chromatographic Studies by UPLC-MS Analysis
2.19. Statistical Analysis
3. Results
3.1. Proliferation and Synergy Assay
3.2. Cell Cycle and Apoptosis Assays
3.3. Autophagy and Immunofluorescence Assays
3.4. Clonogenicity, Migration and Angiogenesis Assays
3.5. Expression of CSC Genes by Real Time PCR
3.6. In Vitro Antioxidant and Chemopreventive Activity
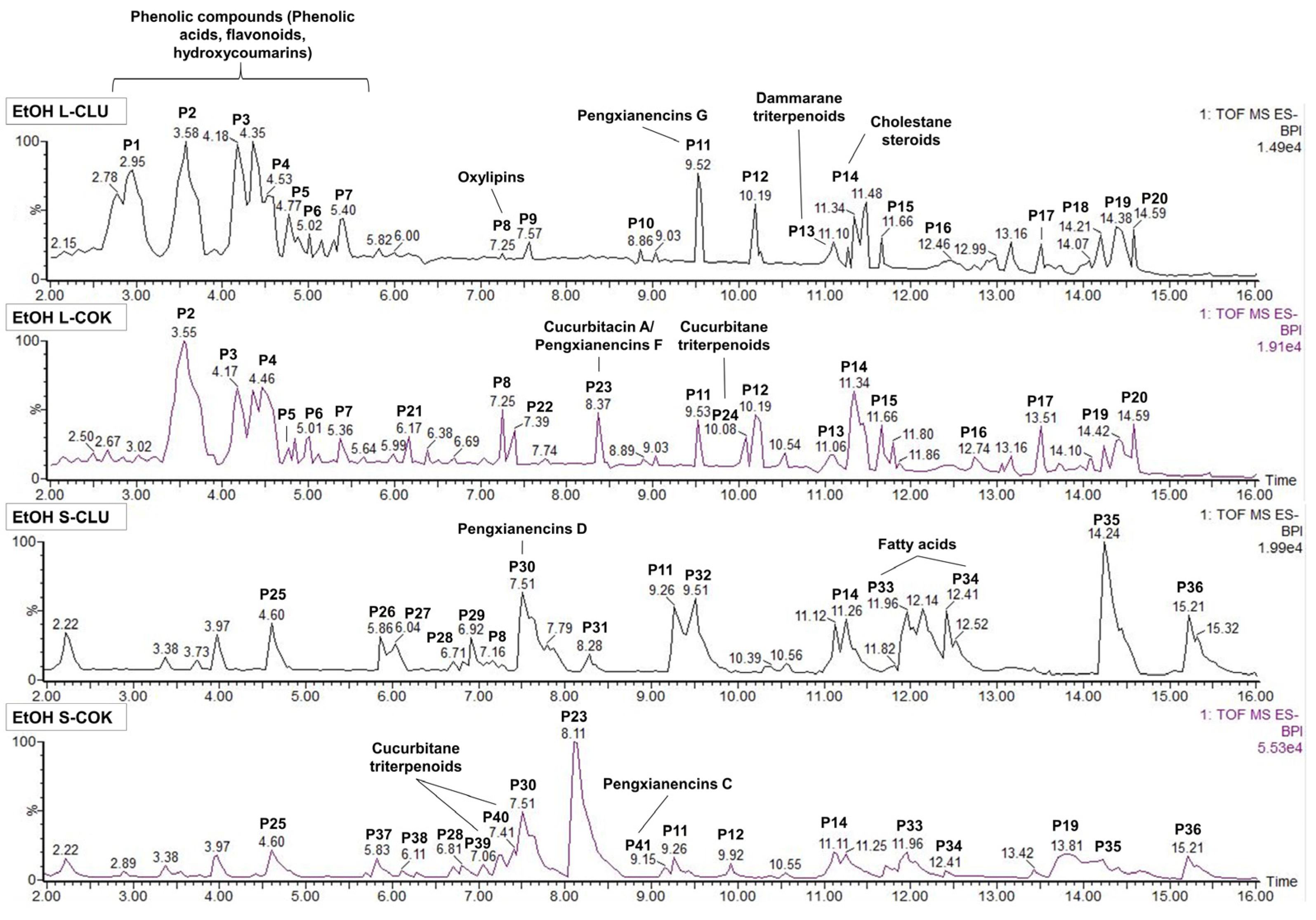
3.7. Analysis of the Bioactive Composition by UPLC-MS
3.8. Bioassay-Guided Fractionation of Seed Extracts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mukherjee, P.K.; Singha, S.; Kar, A.; Chanda, J.; Banerjee, S.; Dasgupta, B.; Haldar, P.K.; Sharma, N. Therapeutic Importance of Cucurbitaceae: A Medicinally Important Family. J. Ethnopharmacol. 2022, 282, 114599. [Google Scholar] [CrossRef] [PubMed]
- Rolnik, A.; Olas, B. A Review of the Effect of Preparations from Vegetables of the Asteraceae Family and Cucurbitaceae Family on the Cardiovascular System and Its Diseases. Nutrients 2022, 14, 3601. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Quispe, C.; Sharifi-Rad, J.; Giri, L.; Suyal, R.; Jugran, A.K.; Zucca, P.; Rescigno, A.; Peddio, S.; Bobiş, O.; et al. Antioxidant Potential of Family Cucurbitaceae with Special Emphasis on Cucurbita Genus: A Key to Alleviate Oxidative Stress-Mediated Disorders. Phytother. Res. 2021, 35, 3533–3557. [Google Scholar] [CrossRef] [PubMed]
- Al-Nablsi, S.; El-Keblawy, A.; Ali, M.A.; Mosa, K.A.; Hamoda, A.M.; Shanableh, A.; Almehdi, A.M.; Soliman, S.S.M. Phenolic Contents and Antioxidant Activity of Citrullus Colocynthis Fruits, Growing in the Hot Arid Desert of the UAE, Influenced by the Fruit Parts, Accessions, and Seasons of Fruit Collection. Antioxidants 2022, 11, 656. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Huang, G.; Huang, H. Preparation, Analysis, Antioxidant Activities In Vivo of Phosphorylated Polysaccharide from Momordica Charantia. Carbohydr. Polym. 2021, 252, 117179. [Google Scholar] [CrossRef]
- Jahan, F.; Islam, M.B.; Moulick, S.P.; Al Bashera, M.; Hasan, M.S.; Tasnim, N.; Saha, T.; Boby, F.; Waliullah, M.; Saha, A.K.; et al. Nutritional Characterization and Antioxidant Properties of Various Edible Portions of Cucurbita Maxima: A Potential Source of Nutraceuticals. Heliyon 2023, 9, e16628. [Google Scholar] [CrossRef]
- Shang, Z.; Li, M.; Zhang, W.; Cai, S.; Hu, X.; Yi, J. Analysis of Phenolic Compounds in Pickled Chayote and Their Effects on Antioxidant Activities and Cell Protection. Food Res. Int. 2022, 157, 111325. [Google Scholar] [CrossRef]
- Psilopatis, I.; Vrettou, K.; Giaginis, C.; Theocharis, S. The Role of Bitter Melon in Breast and Gynecological Cancer Prevention and Therapy. Int. J. Mol. Sci. 2023, 24, 8918. [Google Scholar] [CrossRef]
- Park, H.-J.; Park, S.-H. Root Extract of Trichosanthes Kirilowii Suppresses Metastatic Activity of EGFR TKI-Resistant Human Lung Cancer Cells by Inhibiting Src-Mediated EMT. Nutr. Cancer 2023, 75, 1945–1957. [Google Scholar] [CrossRef]
- Pitchakarn, P.; Ogawa, K.; Suzuki, S.; Takahashi, S.; Asamoto, M.; Chewonarin, T.; Limtrakul, P.; Shirai, T. Momordica Charantia Leaf Extract Suppresses Rat Prostate Cancer Progression In Vitro and In Vivo. Cancer Sci. 2010, 101, 2234–2240. [Google Scholar] [CrossRef]
- Ai, Z.; Ma, C.; Wan, R.; Yin, J.; Li, G.; Li, Y.; Chen, L. Anticancer Activity and Molecular Mechanism of Momordica Cochinchinensis Seed Extract in Chronic Myeloid Leukemia Cells. Nutr. Cancer 2021, 74, 2644–2656. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.L.; Lin, B.J.; Lin, Y.C.; Tu, C.C.; Nguyen, N.L.; Wang, C.C.; Chen, M.C.; Chen, C.H. Cucurbitacin E Exerts Anti-Proliferative Activity via Promoting P62-Dependent Apoptosis in Human Non-Small-Cell Lung Cancer A549 Cells. Curr. Issues Mol. Biol. 2023, 45, 8138–8151. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Lee, K.; Ku, J.M.; Choi, Y.J.; Mok, K.; Kim, D.; Cheon, C.; Ko, S.G. Cucurbitacin D Induces G2/M Phase Arrest and Apoptosis via the ROS/P38 Pathway in Capan-1 Pancreatic Cancer Cell Line. Evid. Based Complement. Alternat Med. 2020, 2020, 6571674. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Y.; Yao, Y.; Li, H.; Gao, C.; Sun, C.; Zhuang, J. Potential of Cucurbitacin as an Anticancer Drug. Biomed. Pharmacother. 2023, 168, 115707. [Google Scholar] [CrossRef]
- Xu, D.; Shen, H.; Tian, M.; Chen, W.; Zhang, X. Cucurbitacin I Inhibits the Proliferation of Pancreatic Cancer through the JAK2/STAT3 Signalling Pathway In Vivo and In Vitro. J. Cancer 2022, 13, 2050–2060. [Google Scholar] [CrossRef]
- Yuan, R.; Zhao, W.; Wang, Q.-Q.; He, J.; Han, S.; Gao, H.; Feng, Y.; Yang, S. Cucurbitacin B Inhibits Non-Small Cell Lung Cancer In Vivo and In Vitro by Triggering TLR4/NLRP3/GSDMD-Dependent Pyroptosis. Pharmacol. Res. 2021, 170, 105748. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, J.; Huang, Q.; Ren, Y.; Li, T.; Zhang, X.; Yao, R.; Sun, J. Cucurbitacin IIa: A Review of Phytochemistry and Pharmacology. Phytother. Res. 2021, 35, 4155–4170. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Abdulridha, M.K.; Al-Marzoqi, A.H.; Ghasemian, A. The Anticancer Efficiency of Citrullus Colocynthis Toward the Colorectal Cancer Therapy. J. Gastrointest. Cancer 2020, 51, 439–444. [Google Scholar] [CrossRef]
- Dandawate, P.; Subramaniam, D.; Panovich, P.; Standing, D.; Krishnamachary, B.; Kaushik, G.; Thomas, S.M.; Dhar, A.; Weir, S.J.; Jensen, R.A.; et al. Cucurbitacin B and I Inhibits Colon Cancer Growth by Targeting the Notch Signaling Pathway. Sci. Rep. 2020, 10, 1290. [Google Scholar] [CrossRef]
- Mesas, C.; Fuel, M.; Martínez, R.; Prados, J.; Melguizo, C.; Porres, J.M. In Vitro Evidence of the Antitumor Capacity of Solanaceae and Cucurbitaceae in Colon Cancer: A Systematic Review. Crit. Rev. Food Sci. Nutr. 2021, 62, 6293–6314. [Google Scholar] [CrossRef] [PubMed]
- Di Napoli, R.; Balzano, N.; Mascolo, A.; Cimmino, C.; Vitiello, A.; Zovi, A.; Capuano, A.; Boccellino, M. What Is the Role of Nutraceutical Products in Cancer Patients? A Systematic Review of Randomized Clinical Trials. Nutrients 2023, 15, 3249. [Google Scholar] [CrossRef] [PubMed]
- Maiuolo, J.; Gliozzi, M.; Carresi, C.; Musolino, V.; Oppedisano, F.; Scarano, F.; Nucera, S.; Scicchitano, M.; Bosco, F.; Macri, R.; et al. Nutraceuticals and Cancer: Potential for Natural Polyphenols. Nutrients 2021, 13, 3834. [Google Scholar] [CrossRef] [PubMed]
- Telang, N. Drug-Resistant Stem Cells: Novel Approach for Colon Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 2519. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.K.; Yin, T.C.; Su, W.C.; Tsai, H.L.; Huang, C.W.; Chen, Y.-C.; Li, C.-C.; Chen, P.-J.; Ma, C.-J.; Chuang, K.-H.; et al. A Pilot Study of Silymarin as Supplementation to Reduce Toxicities in Metastatic Colorectal Cancer Patients Treated With First-Line FOLFIRI Plus Bevacizumab. Oncol. Res. 2021, 28, 801–809. [Google Scholar] [CrossRef]
- Islam, M.R.; Akash, S.; Rahman, M.M.; Nowrin, F.T.; Akter, T.; Shohag, S.; Rauf, A.; Aljohani, A.S.M.; Simal-Gandara, J. Colon Cancer and Colorectal Cancer: Prevention and Treatment by Potential Natural Products. Chem. Biol. Interact. 2022, 368, 110170. [Google Scholar] [CrossRef]
- Jeon, Y.; Sym, S.J.; Yoo, B.K.; Baek, J.H. Long-Term Survival, Tolerability, and Safety of First-Line Bevacizumab and FOLFIRI in Combination With Ginsenoside-Modified Nanostructured Lipid Carrier Containing Curcumin in Patients With Unresectable Metastatic Colorectal Cancer. Integr. Cancer Ther. 2022, 21, 1–7. [Google Scholar] [CrossRef]
- Cione, E.; La Torre, C.; Cannataro, R.; Caroleo, M.C.; Plastina, P.; Gallelli, L. Quercetin, Epigallocatechin Gallate, Curcumin, and Resveratrol: From Dietary Sources to Human MicroRNA Modulation. Molecules 2020, 25, 63. [Google Scholar] [CrossRef]
- Kapravelou, G.; Martínez, R.; Andrade, A.M.; López Chaves, C.; López-Jurado, M.; Aranda, P.; Arrebola, F.; Cañizares, F.J.; Galisteo, M.; Porres, J.M. Improvement of the Antioxidant and Hypolipidaemic Effects of Cowpea Flours (Vigna unguiculata) by Fermentation: Results of In Vitro and In Vivo Experiments. J. Sci. Food Agric. 2015, 95, 1207–1216. [Google Scholar] [CrossRef]
- Martínez, R.; García Beltrán, A.; Kapravelou, G.; Guzmán, A.; Lozano, A.; Gómez-Villegas, P.; León, R.; Vigara, J.; Galisteo, M.; Aranda, P.; et al. Nutritional and Functional Assessment of Haloarchaea and Microalgae from the Andalusian Shoreline: Promising Functional Foods with a High Nutritional Value. J. Funct. Foods 2024, 116, 106194. [Google Scholar] [CrossRef]
- Kapravelou, G.; Martínez, R.; Andrade, A.M.; Sánchez, C.; Chaves, C.L.; López-Jurado, M.; Aranda, P.; Cantarero, S.; Arrebola, F.; Fernández-Segura, E.; et al. Health Promoting Effects of Lupin (Lupinus albus Var. Multolupa) Protein Hydrolyzate and Insoluble Fiber in a Diet-Induced Animal Experimental Model of Hypercholesterolemia. Food Res. Int. 2013, 54, 1471–1481. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B Colorimetric Assay for Cytotoxicity Screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.; Wennerberg, K.; Aittokallio, T.; Tang, J. Searching for Drug Synergy in Complex Dose–Response Landscapes Using an Interaction Potency Model. Comput. Struct. Biotechnol. J. 2015, 13, 504–513. [Google Scholar] [CrossRef]
- Zheng, S.; Wang, W.; Aldahdooh, J.; Malyutina, A.; Shadbahr, T.; Tanoli, Z.; Pessia, A.; Tang, J. SynergyFinder Plus: Toward Better Interpretation and Annotation of Drug Combination Screening Datasets. Genom. Proteom. Bioinform. 2022, 20, 587–596. [Google Scholar] [CrossRef]
- Mesas, C.; Martínez, R.; Ortíz, R.; Galisteo, M.; López-Jurado, M.; Cabeza, L.; Perazzoli, G.; Melguizo, C.; Porres, J.M.; Prados, J. Antitumor Effect of the Ethanolic Extract from Seeds of Euphorbia Lathyris in Colorectal Cancer. Nutrients 2021, 13, 566. [Google Scholar] [CrossRef]
- Peña, M.; Mesas, C.; Perazzoli, G.; Martínez, R.; Porres, J.M.; Doello, K.; Prados, J.; Melguizo, C.; Cabeza, L. Antiproliferative, Antioxidant, Chemopreventive and Antiangiogenic Potential of Chromatographic Fractions from Anemonia Sulcata with and without Its Symbiont Symbiodinium in Colorectal Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 11249. [Google Scholar] [CrossRef]
- Cabeza, L.; Peña, M.; Martínez, R.; Mesas, C.; Galisteo, M.; Perazzoli, G.; Prados, J.; Porres, J.M.; Melguizo, C. Anemonia Sulcata and Its Symbiont Symbiodinium as a Source of Anti-Tumor and Anti-Oxidant Compounds for Colon Cancer Therapy: A Preliminary In Vitro Study. Biology 2021, 10, 134. [Google Scholar] [CrossRef]
- Fuel, M.; Mesas, C.; Martínez, R.; Ortiz, R.; Quiñonero, F.; Prados, J.; Porres, J.M.; Melguizo, C. Antioxidant and Antiproliferative Potential of Ethanolic Extracts from Moringa Oleifera, Tropaeolum Tuberosum and Annona Cherimola in Colorrectal Cancer Cells. Biomed. Pharmacother. 2021, 143, 112248. [Google Scholar] [CrossRef]
- Madhu, C.S.; Balaji, K.S.; Shankar, J.; Sharada, A.C. Antitumor Effects of Chitin Specific Lectin from Praecitrullus Fistulosus by Targeting Angiogenesis and Apoptosis. Biochem. Biophys. Res. Commun. 2019, 518, 381–387. [Google Scholar] [CrossRef]
- Stein, U.; Walther, W.; Shoemaker, R.H. Modulation of Mdr1 Expression by Cytokines in Human Colon Carcinoma Cells: An Approach for Reversal of Multidrug Resistance. Br. J. Cancer 1996, 74, 1384–1391. [Google Scholar] [CrossRef]
- Polidoro, M.A.; Ferrari, E.; Marzorati, S.; Lleo, A.; Rasponi, M. Experimental Liver Models: From Cell Culture Techniques to Microfluidic Organs-on-Chip. Liver Int. 2021, 41, 1744–1761. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.B.; Attar, U.A.; Sakate, D.M.; Ghane, S.G. Efficient Extraction of Cucurbitacins from Diplocyclos palmatus (L.) C. Jeffrey: Optimization Using Response Surface Methodology, Extraction Methods and Study of Some Important Bioactivities. Sci. Rep. 2020, 10, 2109. [Google Scholar] [CrossRef]
- Serala, K.; Steenkamp, P.; Mampuru, L.; Prince, S.; Poopedi, K.; Mbazima, V. In Vitro Antimetastatic Activity of Momordica Balsamina Crude Acetone Extract in HT-29 Human Colon Cancer Cells. Environ. Toxicol. 2021, 36, 2196–2205. [Google Scholar] [CrossRef]
- Yang, P.; Liu, W.; Fu, R.; Ding, G.-B.; Amin, S.; Li, Z. Cucurbitacin E Chemosensitizes Colorectal Cancer Cells via Mitigating TFAP4/Wnt/β-Catenin Signaling. J. Agric. Food Chem. 2020, 68, 14148–14160. [Google Scholar] [CrossRef]
- Kwatra, D.; Venugopal, A.; Standing, D.; Ponnurangam, S.; Dhar, A.; Mitra, A.; Anant, S. Bitter Melon Extracts Enhance the Activity of Chemotherapeutic Agents Through the Modulation of Multiple Drug Resistance. J. Pharm. Sci. 2013, 102, 4444–4454. [Google Scholar] [CrossRef]
- Haciosmanoglu Aldogan, E.; Önsü, K.A.; Saylan, C.C.; Günçer, B.; Baday, S.; Bektaş, M. Depolymerization of Actin Filaments by Cucurbitacin I through Binding G-Actin. Food Sci. Nutr. 2024, 12, 881–889. [Google Scholar] [CrossRef]
- Chaitanya, G.V.; Alexander, J.S.; Babu, P.P. PARP-1 Cleavage Fragments: Signatures of Cell-Death Proteases in Neurodegeneration. Cell Commun. Signal. 2010, 8, 31. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Nieves-Neira, W.; Boon, C.; Pommier, Y.; Bonner, W.M. Initiation of DNA Fragmentation during Apoptosis Induces Phosphorylation of H2AX Histone at Serine 139. J. Biol. Chem. 2000, 275, 9390–9395. [Google Scholar] [CrossRef]
- Liu, X.; Duan, C.; Ji, J.; Zhang, T.; Yuan, X.; Zhang, Y.; Ma, W.; Yang, J.; Yang, L.; Jiang, Z.; et al. Cucurbitacin B Induces Autophagy and Apoptosis by Suppressing CIP2A/PP2A/mTORC1 Signaling Axis in Human Cisplatin Resistant Gastric Cancer Cells. Oncol. Rep. 2017, 38, 271–278. [Google Scholar] [CrossRef]
- Song, H.; Sui, H.; Zhang, Q.; Wang, P.; Wang, F. Cucurbitacin E Induces Autophagy-Involved Apoptosis in Intestinal Epithelial Cells. Front. Physiol. 2020, 11, 1020. [Google Scholar] [CrossRef]
- Huang, J.; Pan, W.; Ou, D.; Dai, W.; Lin, Y.; Chen, Y.; Chen, X. LC3B, a Protein That Serves as an Autophagic Marker, Modulates Angiotensin II-Induced Myocardial Hypertrophy. J. Cardiovasc. Pharmacol. 2015, 66, 576. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Liu, Y.; Liang, K.; Chen, Z.; Ding, K.; Qiu, L.; Wei, J.; Du, H. Cucurbitacin I Reverses Tumor-Associated Macrophage Polarization to Affect Cancer Cell Metastasis. Int. J. Mol. Sci. 2023, 24, 15920. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, X.; Yang, H.; Zhu, C.; Fan, D.; Deng, J. Preliminary Investigation of the Anti-Colon Cancer Activity of Cucurbitacin C from Cucumber: A Network Pharmacological Study and Experimental Validation. Food Front. 2024, 5, 142–159. [Google Scholar] [CrossRef]
- Lopes-Coelho, F.; Martins, F.; Pereira, S.A.; Serpa, J. Anti-Angiogenic Therapy: Current Challenges and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 3765. [Google Scholar] [CrossRef]
- Bielenberg, D.R.; Zetter, B.R. The Contribution of Angiogenesis to the Process of Metastasis. Cancer J. 2015, 21, 267–273. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, J.-K. Antiangiogenic Effects of Cucurbitacin-I. Arch. Pharm. Res. 2015, 38, 290–298. [Google Scholar] [CrossRef]
- Piao, X.M.; Gao, F.; Zhu, J.X.; Wang, L.J.; Zhao, X.; Li, X.; Sheng, M.M.; Zhang, Y. Cucurbitacin B Inhibits Tumor Angiogenesis by Triggering the Mitochondrial Signaling Pathway in Endothelial Cells. Int. J. Mol. Med. 2018, 42, 1018–1025. [Google Scholar] [CrossRef]
- Huang, J.L.; Oshi, M.; Endo, I.; Takabe, K. Clinical Relevance of Stem Cell Surface Markers CD133, CD24, and CD44 in Colorectal Cancer. Am. J. Cancer Res. 2021, 11, 5141–5154. [Google Scholar]
- Roudi, R.; Barodabi, M.; Madjd, Z.; Roviello, G.; Corona, S.P.; Panahi, M. Expression Patterns and Clinical Significance of the Potential Cancer Stem Cell Markers OCT4 and NANOG in Colorectal Cancer Patients. Mol. Cell. Oncol. 2020, 7, 1788366. [Google Scholar] [CrossRef]
- Zhang, Y. Phase II Enzymes. In Encyclopedia of Cancer; Schwab, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 2853–2855. ISBN 978-3-642-16483-5. [Google Scholar]
- Moreno-Quiroga, G.; Alba-Jiménez, J.E.; Aquino-Bolaños, E.N.; Chávez-Servia, J.L. Phenolic Compounds and Antioxidant Activity in Cucurbita Ficifolia Fruits, an Underrated Fruit. Front. Nutr. 2023, 9, 1029826. [Google Scholar] [CrossRef]
- Li, P.; Zhu, N.; Hu, M.; Wu, H.; Yu, T.; Wu, T.; Zhang, D.; Sun, Z.; Yang, J.; Ma, G.; et al. New Cucurbitane Triterpenoids with Cytotoxic Activities from Hemsleya Penxianensis. Fitoterapia 2017, 120, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Varela, C.; Melim, C.; Neves, B.G.; Sharifi-Rad, J.; Calina, D.; Mamurova, A.; Cabral, C. Cucurbitacins as Potential Anticancer Agents: New Insights on Molecular Mechanisms. J. Transl. Med. 2022, 20, 630. [Google Scholar] [CrossRef] [PubMed]
- Zieniuk, B.; Pawełkowicz, M. Recent Advances in the Application of Cucurbitacins as Anticancer Agents. Metabolites 2023, 13, 1081. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zheng, J.; Li, Y.; Xu, D.P.; Li, S.; Chen, Y.M.; Li, H.B. Natural Polyphenols for Prevention and Treatment of Cancer. Nutrients 2016, 8, 515. [Google Scholar] [CrossRef] [PubMed]



| Genus | Specie | Sample Code | cEtOH | rEtOH | PE | PH |
|---|---|---|---|---|---|---|
| Cucurbita | C. pepo | 151SQ | 784.2 ± 128.9 | >1000 | >1000 | 224.9 ± 49.3 |
| MURCIA | 731.1 ± 10.5 | >1000 | >1000 | 388.6 ± 6.3 | ||
| C. moshata | BTN18010 | >1000 | >1000 | >1000 | 367.5 ± 4.8 | |
| C. maxima | ZP180235 | >1000 | >1000 | >1000 | 372.1 ± 2.2 | |
| ZP180301 | >1000 | >1000 | 534.7 ± 108.7 | 472.6 ± 67.4 | ||
| Interspecific | CLU01002 | 3.9 ± 0.3 | 5.5 ± 1.0 | 8.4 ± 0.7 | 42.1 ± 13.9 | |
| COK01001 | 8.8 ± 1.2 | 10.4 ± 1.4 | 9.8 ± 0.3 | 30.4 ± 0.9 | ||
| Sechium | S. edule | CHAYOTE | >1000 | >1000 | 512.7 ± 101.6 | 251.0 ± 29.2 |
| Sample | Plant Part | T-84 | HCT-15 | HT-29 | CCD-18Co | HepG2 |
|---|---|---|---|---|---|---|
| CLU | Leaves | 3.90 ± 0.30 | 2.51 ± 0.27 | 11.36 ± 2.21 | 33.33 ± 0.53 | 66.11 ± 2.66 |
| Seeds | 11.10 ± 0.31 | 9.71 ± 0.98 | 20.37 ± 2.29 | 41.86 ± 1.36 | 72.65 ± 3.50 | |
| COK | Leaves | 8.84 ± 1.18 | 3.76 ± 0.44 | 10.33 ± 1.08 | 42.20 ± 2.04 | 99.12 ± 5.74 |
| Seeds | 3.92 ± 0.28 | 4.18 ± 0.44 | 10.29 ± 1.35 | <10 | 51.0 ± 5.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peña, M.; Guzmán, A.; Mesas, C.; Porres, J.M.; Martínez, R.; Bermúdez, F.; Melguizo, C.; Cabeza, L.; Prados, J. Evaluation of the Leaves and Seeds of Cucurbitaceae Plants as a New Source of Bioactive Compounds for Colorectal Cancer Prevention and Treatment. Nutrients 2024, 16, 4233. https://doi.org/10.3390/nu16234233
Peña M, Guzmán A, Mesas C, Porres JM, Martínez R, Bermúdez F, Melguizo C, Cabeza L, Prados J. Evaluation of the Leaves and Seeds of Cucurbitaceae Plants as a New Source of Bioactive Compounds for Colorectal Cancer Prevention and Treatment. Nutrients. 2024; 16(23):4233. https://doi.org/10.3390/nu16234233
Chicago/Turabian StylePeña, Mercedes, Ana Guzmán, Cristina Mesas, Jesús M. Porres, Rosario Martínez, Francisco Bermúdez, Consolación Melguizo, Laura Cabeza, and Jose Prados. 2024. "Evaluation of the Leaves and Seeds of Cucurbitaceae Plants as a New Source of Bioactive Compounds for Colorectal Cancer Prevention and Treatment" Nutrients 16, no. 23: 4233. https://doi.org/10.3390/nu16234233
APA StylePeña, M., Guzmán, A., Mesas, C., Porres, J. M., Martínez, R., Bermúdez, F., Melguizo, C., Cabeza, L., & Prados, J. (2024). Evaluation of the Leaves and Seeds of Cucurbitaceae Plants as a New Source of Bioactive Compounds for Colorectal Cancer Prevention and Treatment. Nutrients, 16(23), 4233. https://doi.org/10.3390/nu16234233






