Proteomic Profiling of Plasma- and Gut-Derived Extracellular Vesicles in Obesity
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse Models
2.2. Intra-Peritoneal Glucose Tolerance Test (ipGTT)
2.3. Hematoxylin–Eosin Staining
2.4. Extracellular Vesicles Isolation
2.5. Nanoparticle Tracking Analysis (Nanosight)
2.6. Protein Quantification
2.7. Nano-LC-MS/MS Analysis
2.8. Database Search
2.9. Proteomic Functional Enrichment Analysis
2.10. Statistical Analysis
3. Results
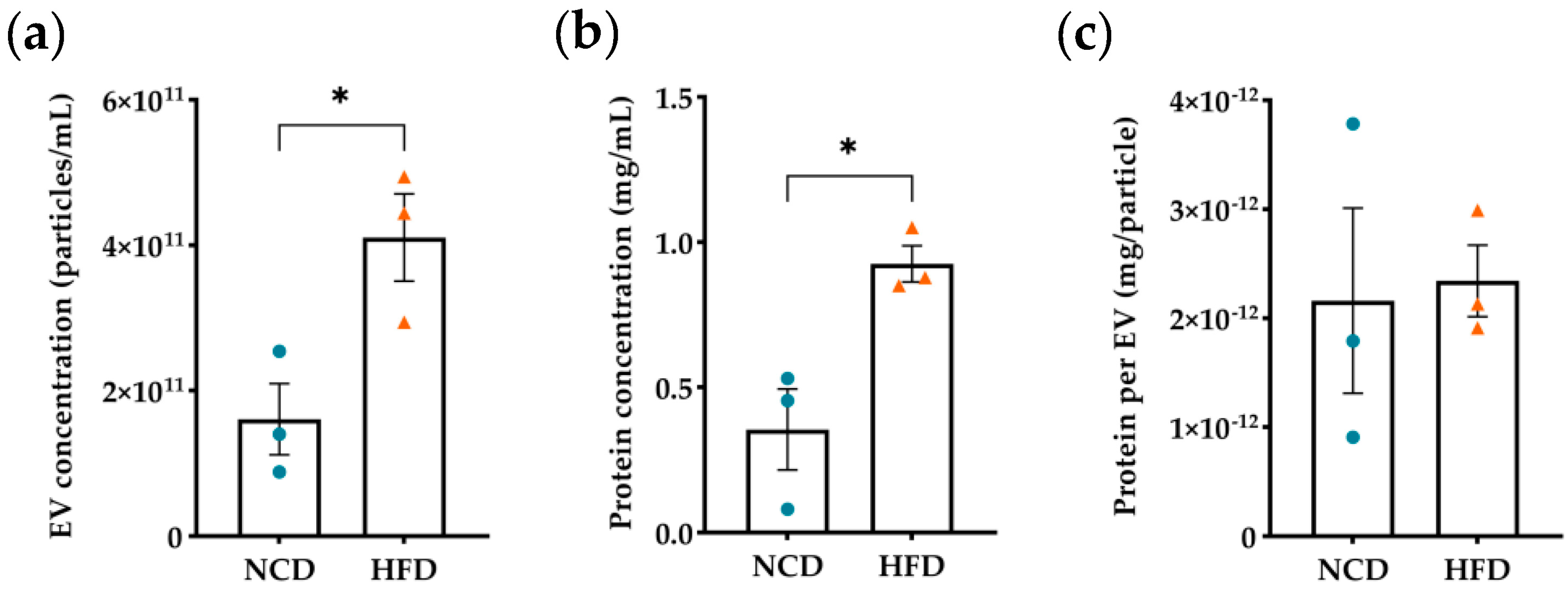
3.1. Evaluation of Diet-Induced Obesity’s Effects on Plasma Derived EVs
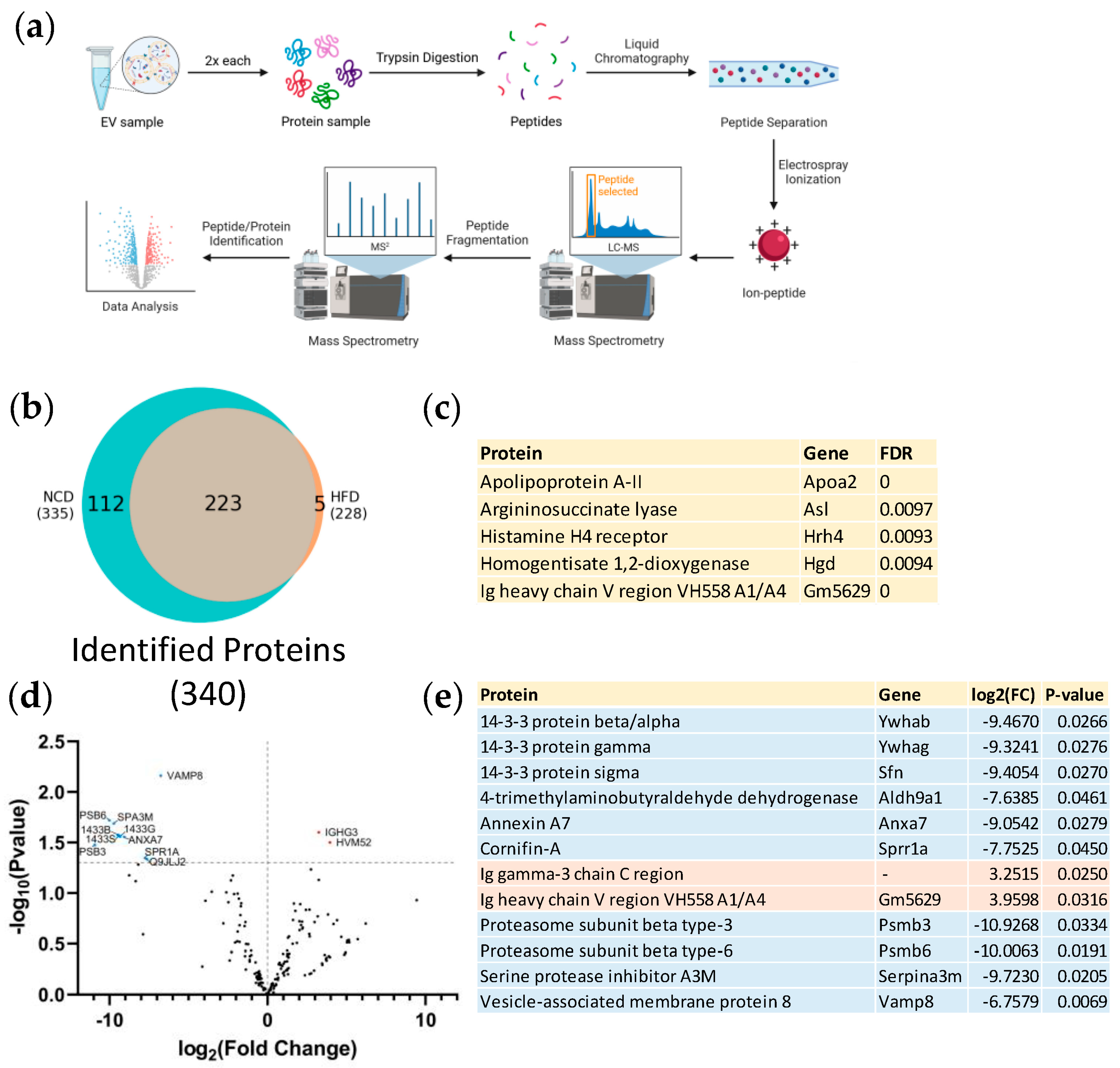
3.2. Proteomic Analysis of Plasma Derived EVs
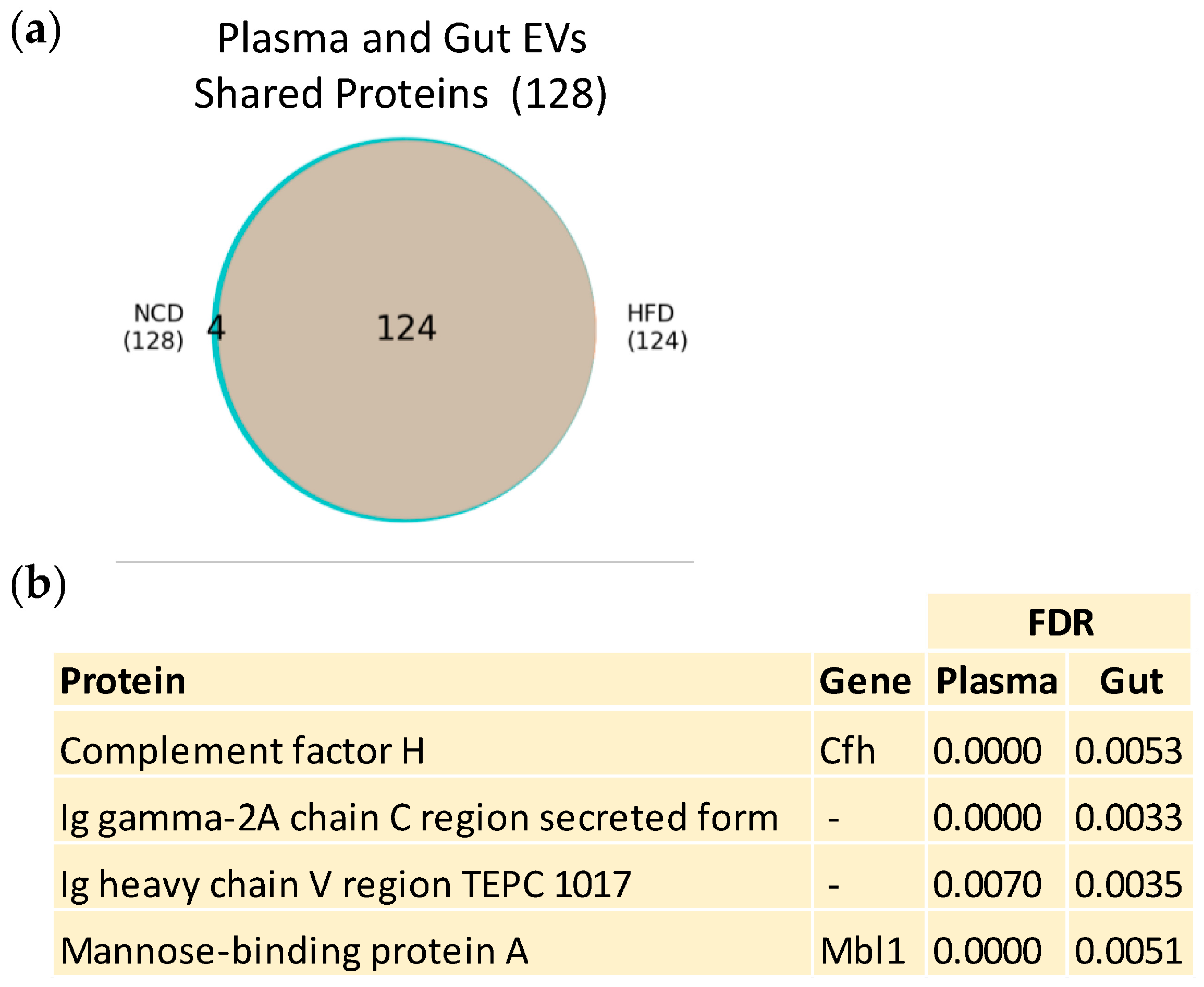
3.3. Plasma and Gut EVs Crosstalk
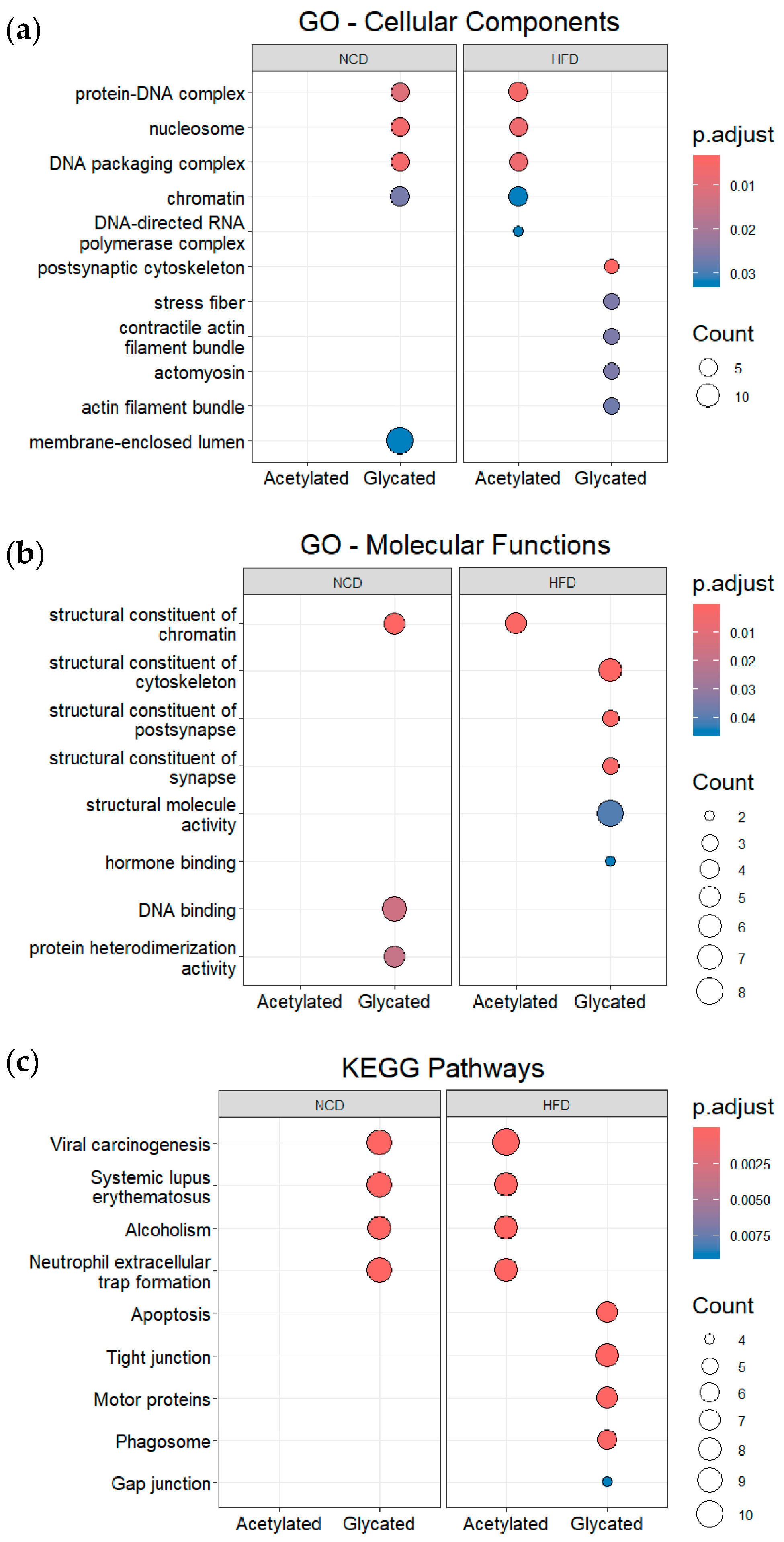
3.4. Post-Translational Modifications (PTMs) in Plasma and Gut EVs Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. World Obesity Day 2022—Accelerating Action to Stop Obesity; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- Bastías-Pérez, M.; Serra, D.; Herrero, L. Dietary Options for Rodents in the Study of Obesity. Nutrients 2020, 12, 3234. [Google Scholar] [CrossRef]
- Hill, J.O.; Melanson, E.L.; Wyatt, H.T. Dietary Fat Intake and Regulation of Energy Balance: Implications for Obesity. J. Nutr. 2000, 130, 284S–288S. [Google Scholar] [CrossRef]
- Jéquier, E. Pathways to Obesity. Int. J. Obes. 2002, 26, S12–S17. [Google Scholar] [CrossRef]
- French, S.; Robinson, T. Fats and Food Intake. Curr. Opin. Clin. Nutr. Metab. Care 2003, 6, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Losacco, M.C.; de Almeida, C.F.T.; Hijo, A.H.T.; Bargi-Souza, P.; Gama, P.; Nunes, M.T.; Goulart-Silva, F. High-Fat Diet Affects Gut Nutrients Transporters in Hypo and Hyperthyroid Mice by PPAR-a Independent Mechanism. Life Sci. 2018, 202, 35–43. [Google Scholar] [CrossRef]
- Hu, F.B. Globalization of Diabetes. Diabetes Care 2011, 34, 1249–1257. [Google Scholar] [CrossRef]
- Shoelson, S.E.; Herrero, L.; Naaz, A. Obesity, Inflammation, and Insulin Resistance. Gastroenterology 2007, 132, 2169–2180. [Google Scholar] [CrossRef]
- Fernández-Sánchez, A.; Madrigal-Santillán, E.; Bautista, M.; Esquivel-Soto, J.; Morales-González, Á.; Esquivel-Chirino, C.; Durante-Montiel, I.; Sánchez-Rivera, G.; Valadez-Vega, C.; Morales-González, J.A. Inflammation, Oxidative Stress, and Obesity. Int. J. Mol. Sci. 2011, 12, 3117–3132. [Google Scholar] [CrossRef]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in Inflammation and Metabolic Disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef]
- Pardo, F.; Villalobos-Labra, R.; Sobrevia, B.; Toledo, F.; Sobrevia, L. Extracellular Vesicles in Obesity and Diabetes Mellitus. Mol. Aspects Med. 2018, 60, 81–91. [Google Scholar] [CrossRef]
- Garcia-Contreras, M.; Shah, S.H.; Tamayo, A.; Robbins, P.D.; Golberg, R.B.; Mendez, A.J.; Ricordi, C. Plasma-Derived Exosome Characterization Reveals a Distinct MicroRNA Signature in Long Duration Type 1 Diabetes. Sci. Rep. 2017, 7, 5998. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.C.; Andriantsitohaina, R. Extracellular Vesicles in Metabolic Syndrome. Circ. Res. 2017, 120, 1674–1686. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Kalra, H.; Drummen, G.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Akbar, N.; Azzimato, V.; Choudhury, R.P.; Aouadi, M. Extracellular Vesicles in Metabolic Disease. Diabetologia 2019, 62, 2179–2187. [Google Scholar] [CrossRef]
- Huang-Doran, I.; Zhang, C.-Y.; Vidal-Puig, A. Extracellular Vesicles: Novel Mediators of Cell Communication in Metabolic Disease. Trends Endocrinol. Metab. 2017, 28, 3–18. [Google Scholar] [CrossRef]
- Li, C.-J.; Fang, Q.-H.; Liu, M.-L.; Lin, J.-N. Current Understanding of the Role of Adipose-Derived Extracellular Vesicles in Metabolic Homeostasis and Diseases: Communication from the Distance between Cells/Tissues. Theranostics 2020, 10, 7422–7435. [Google Scholar] [CrossRef]
- Aswad, H.; Forterre, A.; Wiklander, O.P.B.; Vial, G.; Danty-Berger, E.; Jalabert, A.; Lamazière, A.; Meugnier, E.; Pesenti, S.; Ott, C.; et al. Exosomes Participate in the Alteration of Muscle Homeostasis during Lipid-Induced Insulin Resistance in Mice. Diabetologia 2014, 57, 2155–2164. [Google Scholar] [CrossRef]
- Dang, S.-Y.; Leng, Y.; Wang, Z.-X.; Xiao, X.; Zhang, X.; Wen, T.; Gong, H.-Z.; Hong, A.; Ma, Y. Exosomal Transfer of Obesity Adipose Tissue for Decreased MiR-141-3p Mediate Insulin Resistance of Hepatocytes. Int. J. Biol. Sci. 2019, 15, 351–368. [Google Scholar] [CrossRef]
- Jalabert, A.; Vial, G.; Guay, C.; Wiklander, O.P.B.; Nordin, J.Z.; Aswad, H.; Forterre, A.; Meugnier, E.; Pesenti, S.; Regazzi, R.; et al. Exosome-like Vesicles Released from Lipid-Induced Insulin-Resistant Muscles Modulate Gene Expression and Proliferation of Beta Recipient Cells in Mice. Diabetologia 2016, 59, 1049–1058. [Google Scholar] [CrossRef]
- Wu, J.; Dong, T.; Chen, T.; Sun, J.; Luo, J.; He, J.; Wei, L.; Zeng, B.; Zhang, H.; Li, W.; et al. Hepatic Exosome-Derived MiR-130a-3p Attenuates Glucose Intolerance via Suppressing PHLPP2 Gene in Adipocyte. Metabolism 2020, 103, 154006. [Google Scholar] [CrossRef]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-Derived Exosomal MiR-27a Induces Insulin Resistance in Skeletal Muscle Through Repression of PPARγ. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef]
- Deng, Z.; Poliakov, A.; Hardy, R.W.; Clements, R.; Liu, C.; Liu, Y.; Wang, J.; Xiang, X.; Zhang, S.; Zhuang, X.; et al. Adipose Tissue Exosome-Like Vesicles Mediate Activation of Macrophage-Induced Insulin Resistance. Diabetes 2009, 58, 2498–2505. [Google Scholar] [CrossRef]
- Kranendonk, M.E.G.; Visseren, F.L.J.; van Balkom, B.W.M.; Nolte-’t Hoen, E.N.M.; van Herwaarden, J.A.; de Jager, W.; Schipper, H.S.; Brenkman, A.B.; Verhaar, M.C.; Wauben, M.H.M.; et al. Human Adipocyte Extracellular Vesicles in Reciprocal Signaling between Adipocytes and Macrophages. Obesity 2014, 22, 1296–1308. [Google Scholar] [CrossRef]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-Associated Exosomal MiRNAs Modulate Glucose and Lipid Metabolism in Mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal MiRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.; Machado de Oliveira, R.; Carvalho, A.S.; Teshima, A.; Beck, H.C.; Matthiesen, R.; Costa-Silva, B.; Macedo, M.P. Messages from the Small Intestine Carried by Extracellular Vesicles in Prediabetes: A Proteomic Portrait. J. Proteome Res. 2022, 21, 910–920. [Google Scholar] [CrossRef]
- Breen, D.M.; Rasmussen, B.A.; Côté, C.D.; Jackson, V.M.; Lam, T.K.T. Nutrient-Sensing Mechanisms in the Gut as Therapeutic Targets for Diabetes. Diabetes 2013, 62, 3005–3013. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of Gut Microbiota in Type 2 Diabetes Pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Evers, S.S.; Sandoval, D.A.; Seeley, R.J. The Physiology and Molecular Underpinnings of the Effects of Bariatric Surgery on Obesity and Diabetes. Annu. Rev. Physiol. 2017, 79, 313–334. [Google Scholar] [CrossRef]
- Pareek, M.; Schauer, P.R.; Kaplan, L.M.; Leiter, L.A.; Rubino, F.; Bhatt, D.L. Metabolic Surgery. J. Am. Coll. Cardiol. 2018, 71, 670–687. [Google Scholar] [CrossRef]
- Hillyer, R.; Sullivan, T.; Christ, K.; Hodarkar, A.; Hodge, M.B. 2117-P: Biomarkers Isolated from Extracellular Vesicles Prior to Bariatric Surgery May Be Associated with Postoperative Resolution of Type 2 Diabetes. Diabetes 2019, 68, 2117-P. [Google Scholar] [CrossRef]
- Duarte, N.; Coelho, I.C.; Patarrão, R.S.; Almeida, J.I.; Penha-Gonçalves, C.; Macedo, M.P. How Inflammation Impinges on NAFLD: A Role for Kupffer Cells. BioMed Res. Int. 2015, 2015, 984578. [Google Scholar] [CrossRef] [PubMed]
- Freeman, D.W.; Noren Hooten, N.; Eitan, E.; Green, J.; Mode, N.A.; Bodogai, M.; Zhang, Y.; Lehrmann, E.; Zonderman, A.B.; Biragyn, A.; et al. Altered Extracellular Vesicle Concentration, Cargo, and Function in Diabetes. Diabetes 2018, 67, 2377–2388. [Google Scholar] [CrossRef]
- Eguchi, A.; Lazic, M.; Armando, A.M.; Phillips, S.A.; Katebian, R.; Maraka, S.; Quehenberger, O.; Sears, D.D.; Feldstein, A.E. Circulating Adipocyte-Derived Extracellular Vesicles Are Novel Markers of Metabolic Stress. J. Mol. Med. 2016, 94, 1241–1253. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Horigome, H.; Tanaka, K.; Nakata, Y.; Ohkawara, K.; Katayama, Y.; Matsui, A. Impact of Weight Reduction on Production of Platelet-Derived Microparticles and Fibrinolytic Parameters in Obesity. Thromb. Res. 2007, 119, 45–53. [Google Scholar] [CrossRef]
- Stepanian, A.; Bourguignat, L.; Hennou, S.; Coupaye, M.; Hajage, D.; Salomon, L.; Alessi, M.-C.; Msika, S.; de Prost, D. Microparticle Increase in Severe Obesity: Not Related to Metabolic Syndrome and Unchanged after Massive Weight Loss. Obesity 2013, 21, 2236–2243. [Google Scholar] [CrossRef]
- Campello, E.; Zabeo, E.; Radu, C.M.; Spiezia, L.; Foletto, M.; Prevedello, L.; Gavasso, S.; Bulato, C.; Vettor, R.; Simioni, P. Dynamics of Circulating Microparticles in Obesity after Weight Loss. Intern. Emerg. Med. 2016, 11, 695–702. [Google Scholar] [CrossRef]
- Stocks, B.; Zierath, J.R. Post-Translational Modifications: The Signals at the Intersection of Exercise, Glucose Uptake, and Insulin Sensitivity. Endocr. Rev. 2022, 43, 654–677. [Google Scholar] [CrossRef]
- Wu, X.; Xu, M.; Geng, M.; Chen, S.; Little, P.J.; Xu, S.; Weng, J. Targeting Protein Modifications in Metabolic Diseases: Molecular Mechanisms and Targeted Therapies. Signal Transduct. Target. Ther. 2023, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wei, M.; Zhao, Y.; Yang, Z.; Song, M.; Mi, J.; Yang, X.; Tian, G. Regulation of Insulin Secretion by the Post-Translational Modifications. Front. Cell Dev. Biol. 2023, 11, 1217189. [Google Scholar] [CrossRef]
- Dieterich, I.A.; Lawton, A.J.; Peng, Y.; Yu, Q.; Rhoads, T.W.; Overmyer, K.A.; Cui, Y.; Armstrong, E.A.; Howell, P.R.; Burhans, M.S.; et al. Acetyl-CoA Flux Regulates the Proteome and Acetyl-Proteome to Maintain Intracellular Metabolic Crosstalk. Nat. Commun. 2019, 10, 3929. [Google Scholar] [CrossRef]
- Maessen, D.E.M.; Stehouwer, C.D.A.; Schalkwijk, C.G. The Role of Methylglyoxal and the Glyoxalase System in Diabetes and Other Age-Related Diseases. Clin. Sci. 2015, 128, 839–861. [Google Scholar] [CrossRef]
- Menzies, K.J.; Zhang, H.; Katsyuba, E.; Auwerx, J. Protein Acetylation in Metabolism—Metabolites and Cofactors. Nat. Rev. Endocrinol. 2016, 12, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.T.; Workman, J.L. Introducing the Acetylome. Nat. Biotechnol. 2009, 27, 917–919. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Suttapitugsakul, S.; Xiao, H.; Wu, R. Comprehensive Analysis of Protein Glycation Reveals Its Potential Impacts on Protein Degradation and Gene Expression in Human Cells. J. Am. Soc. Mass Spectrom. 2019, 30, 2480–2490. [Google Scholar] [CrossRef]
- Pessoa Rodrigues, C.; Chatterjee, A.; Wiese, M.; Stehle, T.; Szymanski, W.; Shvedunova, M.; Akhtar, A. Histone H4 Lysine 16 Acetylation Controls Central Carbon Metabolism and Diet-Induced Obesity in Mice. Nat. Commun. 2021, 12, 6212. [Google Scholar] [CrossRef]
- Li, Z.; Zhuang, M.; Zhang, L.; Zheng, X.; Yang, P.; Li, Z. Acetylation Modification Regulates GRP78 Secretion in Colon Cancer Cells. Sci. Rep. 2016, 6, 30406. [Google Scholar] [CrossRef]
- Iyer, A.; Fairlie, D.P.; Brown, L. Lysine Acetylation in Obesity, Diabetes and Metabolic Disease. Immunol. Cell Biol. 2012, 90, 39–46. [Google Scholar] [CrossRef]
- Rabbani, N.; Thornalley, P.J. Glycation Research in Amino Acids: A Place to Call Home. Amino Acids 2012, 42, 1087–1096. [Google Scholar] [CrossRef]
- Twarda-Clapa, A.; Olczak, A.; Białkowska, A.M.; Koziołkiewicz, M. Advanced Glycation End-Products (AGEs): Formation, Chemistry, Classification, Receptors, and Diseases Related to AGEs. Cells 2022, 11, 1312. [Google Scholar] [CrossRef]
- Mengstie, M.A.; Chekol Abebe, E.; Behaile Teklemariam, A.; Tilahun Mulu, A.; Agidew, M.M.; Teshome Azezew, M.; Zewde, E.A.; Agegnehu Teshome, A. Endogenous Advanced Glycation End Products in the Pathogenesis of Chronic Diabetic Complications. Front. Mol. Biosci. 2022, 9, 1002710. [Google Scholar] [CrossRef]
- Negre-Salvayre, A.; Salvayre, R.; Augé, N.; Pamplona, R.; Portero-Otín, M. Hyperglycemia and Glycation in Diabetic Complications. Antioxid. Redox Signal. 2009, 11, 3071–3109. [Google Scholar] [CrossRef]
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021. [Google Scholar]
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Gaglia, J.L.; Hilliard, M.E.; Isaacs, D.; et al. 2. Classification and Diagnosis of Diabetes: Standards of Care in Diabetes—2023. Diabetes Care 2023, 46, S19–S40. [Google Scholar] [CrossRef]
- Carvalho, A.S.; Baeta, H.; Henriques, A.F.A.; Ejtehadifar, M.; Tranfield, E.M.; Sousa, A.L.; Farinho, A.; Silva, B.C.; Cabeçadas, J.; Gameiro, P.; et al. Proteomic Landscape of Extracellular Vesicles for Diffuse Large B-Cell Lymphoma Subtyping. Int. J. Mol. Sci. 2021, 22, 1004. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Carvalho, A.S.; Ribeiro, H.; Voabil, P.; Penque, D.; Jensen, O.N.; Molina, H.; Matthiesen, R. Global Mass Spectrometry and Transcriptomics Array Based Drug Profiling Provides Novel Insight into Glucosamine Induced Endoplasmic Reticulum Stress. Mol. Cell. Proteom. 2014, 13, 3294–3307. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, D.H.; Hwang, S.-I.; Wu, L.; Han, D.K. Role of Spectral Counting in Quantitative Proteomics. Expert Rev. Proteom. 2010, 7, 39–53. [Google Scholar] [CrossRef]
- Schwanhüusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global Quantification of Mammalian Gene Expression Control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO::TermFinder—Open Source Software for Accessing Gene Ontology Information and Finding Significantly Enriched Gene Ontology Terms Associated with a List of Genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; Hill, D.P.; et al. The Gene Ontology Knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for Taxonomy-Based Analysis of Pathways and Genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Evans, M.K. Extracellular Vesicles as Signaling Mediators in Type 2 Diabetes Mellitus. Am. J. Physiol.-Cell Physiol. 2020, 318, C1189–C1199. [Google Scholar] [CrossRef]
- Sandau, U.S.; McFarland, T.J.; Smith, S.J.; Galasko, D.R.; Quinn, J.F.; Saugstad, J.A. Differential Effects of APOE Genotype on MicroRNA Cargo of Cerebrospinal Fluid Extracellular Vesicles in Females with Alzheimer’s Disease Compared to Males. Front. Cell Dev. Biol. 2022, 10, 864022. [Google Scholar] [CrossRef]
- Chung, D.D.; Mahnke, A.H.; Pinson, M.R.; Salem, N.A.; Lai, M.S.; Collins, N.P.; Hillhouse, A.E.; Miranda, R.C. Sex Differences in the Transcriptome of Extracellular Vesicles Secreted by Fetal Neural Stem Cells and Effects of Chronic Alcohol Exposure. Biol. Sex Differ. 2023, 14, 19. [Google Scholar] [CrossRef]
- Perpiñá-Clérigues, C.; Mellado, S.; Galiana-Roselló, C.; Fernández-Regueras, M.; Marcos, M.; García-García, F.; Pascual, M. Novel Insight into the Lipid Network of Plasma Extracellular Vesicles Reveal Sex-Based Differences in the Lipidomic Profile of Alcohol Use Disorder Patients. Biol. Sex Differ. 2024, 15, 10. [Google Scholar] [CrossRef]
- Perpiñá-Clérigues, C.; Mellado, S.; Català-Senent, J.F.; Ibáñez, F.; Costa, P.; Marcos, M.; Guerri, C.; García-García, F.; Pascual, M. Lipidomic Landscape of Circulating Extracellular Vesicles Isolated from Adolescents Exposed to Ethanol Intoxication: A Sex Difference Study. Biol. Sex Differ. 2023, 14, 22. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Byappanahalli, A.M.; Vannoy, M.; Omoniyi, V.; Evans, M.K. Influences of Age, Race, and Sex on Extracellular Vesicle Characteristics. Theranostics 2022, 12, 4459–4476. [Google Scholar] [CrossRef]
- Ter-Ovanesyan, D.; Gilboa, T.; Budnik, B.; Nikitina, A.; Whiteman, S.; Lazarovits, R.; Trieu, W.; Kalish, D.; Church, G.M.; Walt, D.R. Improved Isolation of Extracellular Vesicles by Removal of Both Free Proteins and Lipoproteins. Elife 2023, 12, e86394. [Google Scholar] [CrossRef]
- Sódar, B.W.; Kittel, Á.; Pálóczi, K.; Vukman, K.V.; Osteikoetxea, X.; Szabó-Taylor, K.; Németh, A.; Sperlágh, B.; Baranyai, T.; Giricz, Z.; et al. Low-Density Lipoprotein Mimics Blood Plasma-Derived Exosomes and Microvesicles during Isolation and Detection. Sci. Rep. 2016, 6, 24316. [Google Scholar] [CrossRef]
- Smolarz, M.; Pietrowska, M.; Matysiak, N.; Mielańczyk, Ł.; Widłak, P. Proteome Profiling of Exosomes Purified from a Small Amount of Human Serum: The Problem of Co-Purified Serum Components. Proteomes 2019, 7, 18. [Google Scholar] [CrossRef]
- Thomas, S.E. Unravelling the Story of Protein Misfolding in Diabetes Mellitus. World J. Diabetes 2011, 2, 114. [Google Scholar] [CrossRef]
- Mukherjee, A.; Morales-Scheihing, D.; Butler, P.C.; Soto, C. Type 2 Diabetes as a Protein Misfolding Disease. Trends Mol. Med. 2015, 21, 439–449. [Google Scholar] [CrossRef]
- Mukherjee, A.; Morales-Scheihing, D.; Salvadores, N.; Moreno-Gonzalez, I.; Gonzalez, C.; Taylor-Presse, K.; Mendez, N.; Shahnawaz, M.; Gaber, A.O.; Sabek, O.M.; et al. Induction of IAPP Amyloid Deposition and Associated Diabetic Abnormalities by a Prion-like Mechanism. J. Exp. Med. 2017, 214, 2591–2610. [Google Scholar] [CrossRef]
- Klyosova, E.; Azarova, I.; Polonikov, A. A Polymorphism in the Gene Encoding Heat Shock Factor 1 (HSF1) Increases the Risk of Type 2 Diabetes: A Pilot Study Supports a Role for Impaired Protein Folding in Disease Pathogenesis. Life 2022, 12, 1936. [Google Scholar] [CrossRef]
- Lim, G.E.; Piske, M.; Johnson, J.D. 14-3-3 Proteins Are Essential Signalling Hubs for Beta Cell Survival. Diabetologia 2013, 56, 825–837. [Google Scholar] [CrossRef]
- Lontchi-Yimagou, E.; Sobngwi, E.; Matsha, T.E.; Kengne, A.P. Diabetes Mellitus and Inflammation. Curr. Diabetes Rep. 2013, 13, 435–444. [Google Scholar] [CrossRef]
- Lin, H.; Du, J.; Jiang, H. Post-Translational Modifications to Regulate Protein Function. In Wiley Encyclopedia of Chemical Biology; Wiley: Hoboken, NJ, USA, 2008; pp. 1–31. [Google Scholar]
- Bischoff, R.; Schlüter, H. Amino Acids: Chemistry, Functionality and Selected Non-Enzymatic Post-Translational Modifications. J. Proteom. 2012, 75, 2275–2296. [Google Scholar] [CrossRef]
- Campos, E.I.; Reinberg, D. Histones: Annotating Chromatin. Annu. Rev. Genet. 2009, 43, 559–599. [Google Scholar] [CrossRef]
- Ren, J.; Panther, E.; Liao, X.; Grammer, A.; Lipsky, P.; Reilly, C. The Impact of Protein Acetylation/Deacetylation on Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2018, 19, 4007. [Google Scholar] [CrossRef]
- Pandey, S.C.; Bohnsack, J.P. Alcohol Makes Its Epigenetic Marks. Cell Metab. 2020, 31, 213–214. [Google Scholar] [CrossRef]
- Mews, P.; Egervari, G.; Nativio, R.; Sidoli, S.; Donahue, G.; Lombroso, S.I.; Alexander, D.C.; Riesche, S.L.; Heller, E.A.; Nestler, E.J.; et al. Alcohol Metabolism Contributes to Brain Histone Acetylation. Nature 2019, 574, 717–721. [Google Scholar] [CrossRef]
- Hamam, H.; Khan, M.; Palaniyar, N. Histone Acetylation Promotes Neutrophil Extracellular Trap Formation. Biomolecules 2019, 9, 32. [Google Scholar] [CrossRef]
- Mirzaei, H.; Ghorbani, S.; Khanizadeh, S.; Namdari, H.; Faghihloo, E.; Akbari, A. Histone Deacetylases in Virus-associated Cancers. Rev. Med. Virol. 2020, 30, e2085. [Google Scholar] [CrossRef]
- Csizmok, V.; Forman-Kay, J.D. Complex Regulatory Mechanisms Mediated by the Interplay of Multiple Post-Translational Modifications. Curr. Opin. Struct. Biol. 2018, 48, 58–67. [Google Scholar] [CrossRef]
- Zheng, Q.; Omans, N.D.; Leicher, R.; Osunsade, A.; Agustinus, A.S.; Finkin-Groner, E.; D’Ambrosio, H.; Liu, B.; Chandarlapaty, S.; Liu, S.; et al. Reversible Histone Glycation Is Associated with Disease-Related Changes in Chromatin Architecture. Nat. Commun. 2019, 10, 1289. [Google Scholar] [CrossRef]
- Smith, B.C.; Denu, J.M. Chemical Mechanisms of Histone Lysine and Arginine Modifications. Biochim. Biophys. Acta BBA—Gene Regul. Mech. 2009, 1789, 45–57. [Google Scholar] [CrossRef]
- Song, Y.; Brady, S.T. Post-Translational Modifications of Tubulin: Pathways to Functional Diversity of Microtubules. Trends Cell Biol. 2015, 25, 125–136. [Google Scholar] [CrossRef]
- Terman, J.R.; Kashina, A. Post-Translational Modification and Regulation of Actin. Curr. Opin. Cell Biol. 2013, 25, 30–38. [Google Scholar] [CrossRef]
- Horton, F.; Wright, J.; Smith, L.; Hinton, P.J.; Robertson, M.D. Increased Intestinal Permeability to Oral Chromium (51Cr)—EDTA in Human Type 2 Diabetes. Diabet. Med. 2014, 31, 559–563. [Google Scholar] [CrossRef]
- Monsour, M.; Garbuzova-Davis, S.; Borlongan, C.V. Patching Up the Permeability: The Role of Stem Cells in Lessening Neurovascular Damage in Amyotrophic Lateral Sclerosis. Stem Cells Transl. Med. 2022, 11, 1196–1209. [Google Scholar] [CrossRef]
- Kawada, T.; Nakazawa, M.; Nakauchi, S.; Yamazaki, K.; Shimamoto, R.; Urabe, M.; Nakata, J.; Hemmi, C.; Masui, F.; Nakajima, T.; et al. Rescue of Hereditary Form of Dilated Cardiomyopathy by RAAV-Mediated Somatic Gene Therapy: Amelioration of Morphological Findings, Sarcolemmal Permeability, Cardiac Performances, and the Prognosis of TO-2 Hamsters. Proc. Natl. Acad. Sci. USA 2002, 99, 901–906. [Google Scholar] [CrossRef]
- Kaye, D.; Pimental, D.; Prasad, S.; Mäki, T.; Berger, H.J.; McNeil, P.L.; Smith, T.W.; Kelly, R.A. Role of Transiently Altered Sarcolemmal Membrane Permeability and Basic Fibroblast Growth Factor Release in the Hypertrophic Response of Adult Rat Ventricular Myocytes to Increased Mechanical Activity in Vitro. J. Clin. Investig. 1996, 97, 281–291. [Google Scholar] [CrossRef]
- Zmora, N.; Bashiardes, S.; Levy, M.; Elinav, E. The Role of the Immune System in Metabolic Health and Disease. Cell Metab. 2017, 25, 506–521. [Google Scholar] [CrossRef]
- Ghosh, P.; Sahoo, R.; Vaidya, A.; Chorev, M.; Halperin, J.A. Role of Complement and Complement Regulatory Proteins in the Complications of Diabetes. Endocr. Rev. 2015, 36, 272–288. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baptista Pereira, P.; Torrejón, E.; Ferreira, I.; Carvalho, A.S.; Teshima, A.; Sousa-Lima, I.; Beck, H.C.; Costa-Silva, B.; Matthiesen, R.; Macedo, M.P.; et al. Proteomic Profiling of Plasma- and Gut-Derived Extracellular Vesicles in Obesity. Nutrients 2024, 16, 736. https://doi.org/10.3390/nu16050736
Baptista Pereira P, Torrejón E, Ferreira I, Carvalho AS, Teshima A, Sousa-Lima I, Beck HC, Costa-Silva B, Matthiesen R, Macedo MP, et al. Proteomic Profiling of Plasma- and Gut-Derived Extracellular Vesicles in Obesity. Nutrients. 2024; 16(5):736. https://doi.org/10.3390/nu16050736
Chicago/Turabian StyleBaptista Pereira, Pedro, Estefania Torrejón, Inês Ferreira, Ana Sofia Carvalho, Akiko Teshima, Inês Sousa-Lima, Hans Christian Beck, Bruno Costa-Silva, Rune Matthiesen, Maria Paula Macedo, and et al. 2024. "Proteomic Profiling of Plasma- and Gut-Derived Extracellular Vesicles in Obesity" Nutrients 16, no. 5: 736. https://doi.org/10.3390/nu16050736
APA StyleBaptista Pereira, P., Torrejón, E., Ferreira, I., Carvalho, A. S., Teshima, A., Sousa-Lima, I., Beck, H. C., Costa-Silva, B., Matthiesen, R., Macedo, M. P., & de Oliveira, R. M. (2024). Proteomic Profiling of Plasma- and Gut-Derived Extracellular Vesicles in Obesity. Nutrients, 16(5), 736. https://doi.org/10.3390/nu16050736







