Circadian Regulation of Vitamin D Target Genes Reveals a Network Shaped by Individual Responsiveness
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptome Data Sources
2.2. Differential Gene Expression Analysis
2.3. Characterization of Target Genes
2.4. Analysis of Genomic Regions of Vitamin D Target Genes
2.5. Statistical Tests
3. Results
3.1. Circadian Expression Profile of In Vivo Vitamin D Target Genes
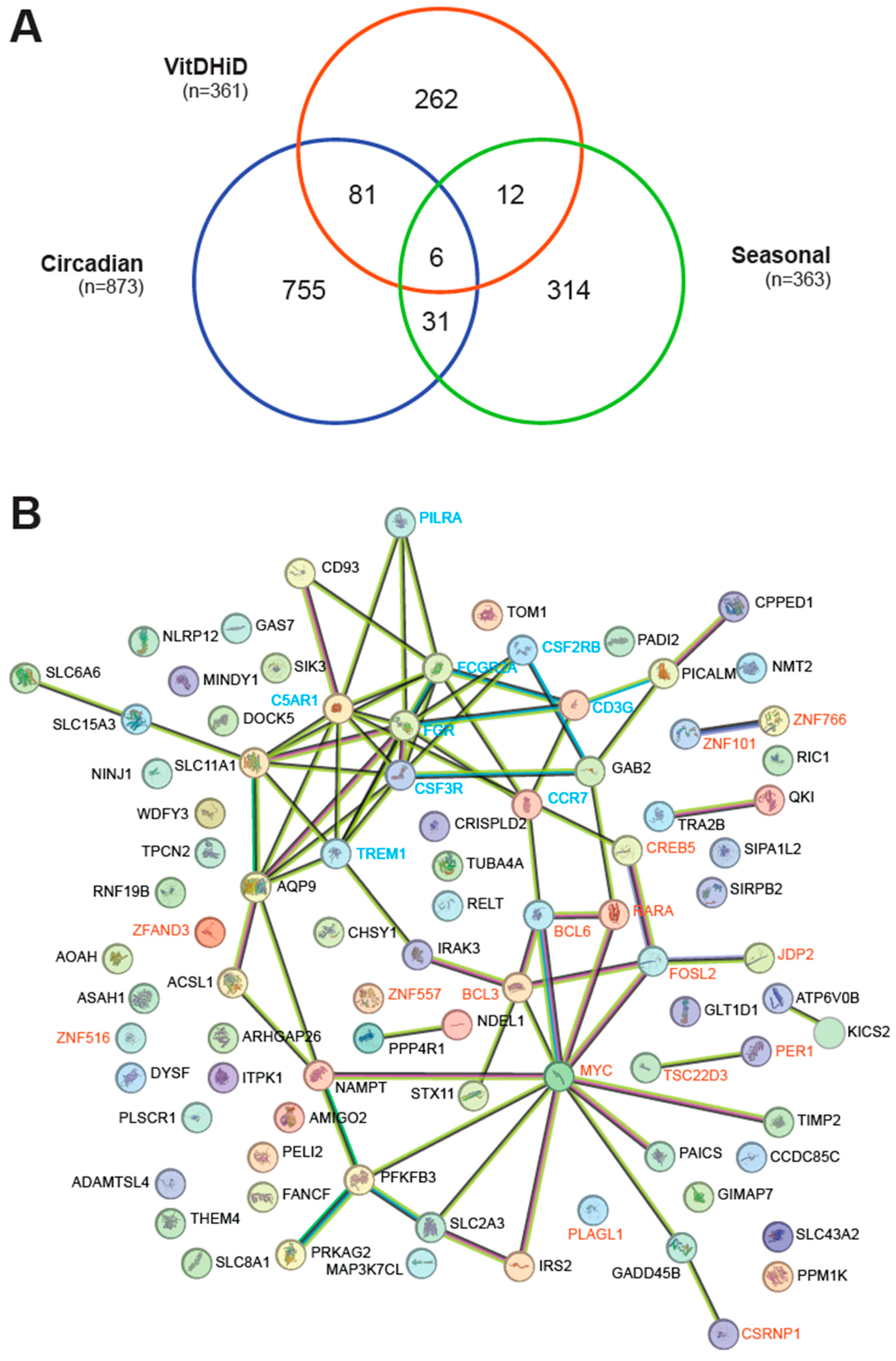
3.2. Characterization of Vitamin D Targets with Circadian Behavior
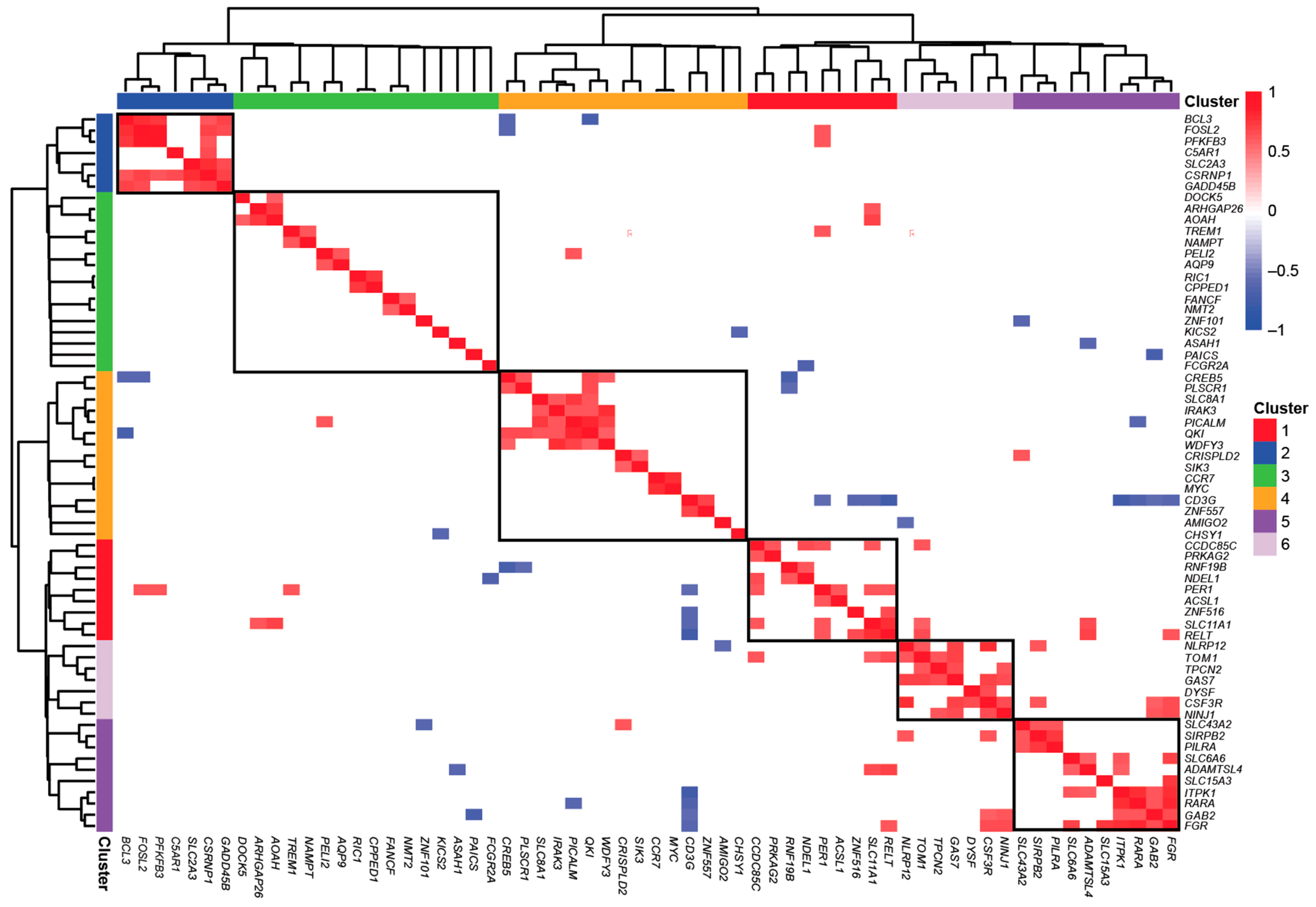
3.3. Individual-Specific Responses of Circadian Vitamin D Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 1,25(OH)2D3 | 1α,25-dihydroxyvitamin D3 |
| AMIGO2 | adhesion molecule with Ig-like domain 2 |
| AMPK | AMP-activated protein kinase |
| AQP9 | aquaporin 9 |
| ASCL1 | acyl-CoA synthetase long chain family member 1 |
| BCL | BCL transcription coactivator |
| C5AR1 | complement C5a receptor 1 |
| CCR7 | C-C motif chemokine receptor 7 |
| CD3G | CD3 gamma subunit of T-cell receptor complex |
| CD93 | CD93 molecule |
| ChIP-seq | chromatin immunoprecipitation sequencing |
| CPM | counts per million |
| CPPED1 | calcineurin-like phosphoesterase domain containing 1 |
| CREB5 | CAMP responsive element binding protein 5 |
| CRISPLD2 | cysteine-rich secretory protein LCCL domain containing 2 |
| CSF2RB | colony stimulating factor 2 receptor subunit beta |
| CSF3R | colony stimulating factor 3 receptor |
| CSRNP1 | cysteine- and serine-rich nuclear protein 1 |
| DYSF | dysferlin |
| FAIRE-seq | formaldehyde-assisted isolation of regulatory elements followed by sequencing |
| FC | fold change |
| FCGR2A | Fc gamma receptor IIa |
| FDR | false discovery rate |
| FGR | FGR proto-oncogene, Src family tyrosine kinase |
| FOSL2 | FOS-like 2, AP1 transcription factor subunit |
| GADD45B | growth arrest and DNA damage inducible beta |
| GAS7 | growth arrest specific 7 |
| IRAK3 | interleukin 1 receptor-associated kinase 3 |
| IRS2 | insulin receptor substrate 2 |
| JDP2 | Jun dimerization protein 2 |
| KLF11 | KLF transcription factor 11 |
| MAP3K7CL | MAP3K7 C-terminal like |
| MYC | MYC proto-oncogene, BHLH transcription factor |
| NAMPT | nicotinamide phosphoribosyltransferase |
| NDEL1 | NudE neurodevelopment protein 1 like 1 |
| NINJ1 | ninjurin 1 |
| NLRP12 | NLR family pyrin domain containing 12 |
| PADI2 | peptidyl arginine deiminase |
| PBMC | peripheral blood mononuclear cell |
| PER1 | period circadian regulator 1 |
| PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| PILRA | paired immunoglobin-like type 2 receptor alpha |
| PRKAG2 | protein kinase AMP-activated non-catalytic subunit gamma 2 |
| RARA | retinoic acid receptor alpha |
| RNA-seq | RNA sequencing |
| RNF19B | ring finger protein 19B |
| SIK3 | SIK family kinase 3 |
| SIPA1L2 | signal-induced proliferation-associated 1 like 2 |
| SIRPB2 | signal regulatory protein beta 2 |
| SLC | solute carrier family |
| STX11 | syntaxin 11 |
| TAD | topologically associating domain |
| TIMP2 | TIMP metallopeptidase inhibitor 2 |
| TOM1 | target of Myb1 membrane trafficking protein |
| TPCN2 | two pore segment channel 2 |
| TREM1 | triggering receptor expressed on myeloid cells 1 |
| TSC22D3 | TSC22 domain family member 3 |
| TSS | transcription start site |
| VDR | vitamin D receptor |
| WDFY3 | WD repeat and FYVE domain containing 3 |
| ZNF | zinc finger protein |
References
- Holick, M.F.; MacLaughlin, J.A.; Doppelt, S.H. Regulation of cutaneous previtamin D3 photosynthesis in man: Skin pigment is not an essential regulator. Science 1981, 211, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Holick, M.F. The cutaneous photosynthesis of previtamin D3: A unique photoendocrine system. J. Investig. Dermatol. 1981, 77, 51–58. [Google Scholar] [CrossRef]
- Hanel, A.; Carlberg, C. Skin color and vitamin D: An update. Exp. Dermatol. 2020, 29, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Pagani, L.; Lawson, D.J.; Jagoda, E.; Morseburg, A.; Eriksson, A.; Mitt, M.; Clemente, F.; Hudjashov, G.; DeGiorgio, M.; Saag, L.; et al. Genomic analyses inform on migration events during the peopling of Eurasia. Nature 2016, 538, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Skoglund, P.; Mathieson, I. Ancient genomics of modern humans: The first decade. Annu. Rev. Genom. Hum. Genet. 2018, 19, 381–404. [Google Scholar] [CrossRef]
- O’Neill, C.M.; Kazantzidis, A.; Ryan, M.J.; Barber, N.; Sempos, C.T.; Durazo-Arvizu, R.A.; Jorde, R.; Grimnes, G.; Eiriksdottir, G.; Gudnason, V.; et al. Seasonal changes in vitamin D-effective UVB availability in Europe and associations with population serum 25-hydroxyvitamin D. Nutrients 2016, 8, 533. [Google Scholar] [CrossRef]
- Bouillon, R.; Suda, T. Vitamin D: Calcium and bone homeostasis during evolution. BoneKEy Rep. 2014, 3, 480. [Google Scholar] [CrossRef]
- Levine, M.A. Diagnosis and management of vitamin D dependent rickets. Front. Pediatr. 2020, 8, 315. [Google Scholar] [CrossRef]
- Bouillon, R.; Carmeliet, G.; Lieben, L.; Watanabe, M.; Perino, A.; Auwerx, J.; Schoonjans, K.; Verstuyf, A. Vitamin D and energy homeostasis-of mice and men. Nat. Rev. Endocrinol. 2014, 10, 79–87. [Google Scholar] [CrossRef]
- Sunyecz, J.A. The use of calcium and vitamin D in the management of osteoporosis. Ther. Clin. Risk Manag. 2008, 4, 827–836. [Google Scholar] [CrossRef]
- Uchitomi, R.; Oyabu, M.; Kamei, Y. Vitamin D and sarcopenia: Potential of vitamin D supplementation in sarcopenia prevention and treatment. Nutrients 2020, 12, 3189. [Google Scholar] [CrossRef]
- Feige, J.; Moser, T.; Bieler, L.; Schwenker, K.; Hauer, L.; Sellner, J. Vitamin D supplementation in multiple sclerosis: A critical analysis of potentials and threats. Nutrients 2020, 12, 783. [Google Scholar] [CrossRef] [PubMed]
- Vaughan-Shaw, P.G.; Buijs, L.F.; Blackmur, J.P.; Theodoratou, E.; Zgaga, L.; Din, F.V.N.; Farrington, S.M.; Dunlop, M.G. The effect of vitamin D supplementation on survival in patients with colorectal cancer: Systematic review and meta-analysis of randomised controlled trials. Br. J. Cancer 2020, 123, 1705–1712. [Google Scholar] [CrossRef] [PubMed]
- Bouillon, R.; Manousaki, D.; Rosen, C.; Trajanoska, K.; Rivadeneira, F.; Richards, J.B. The health effects of vitamin D supplementation: Evidence from human studies. Nat. Rev. Endocrinol. 2022, 18, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Gospodarska, E.; Ghosh Dastidar, R.; Carlberg, C. Intervention approaches in studying the response to vitamin D3 supplementation. Nutrients 2023, 15, 3382. [Google Scholar] [CrossRef]
- Bischoff-Ferrari, H.A.; Vellas, B.; Rizzoli, R.; Kressig, R.W.; da Silva, J.A.P.; Blauth, M.; Felson, D.T.; McCloskey, E.V.; Watzl, B.; Hofbauer, L.C.; et al. Effect of vitamin D supplementation, omega-3 fatty acid supplementation, or a strength-training exercise program on clinical outcomes in older adults: The DO-HEALTH randomized clinical trial. JAMA 2020, 324, 1855–1868. [Google Scholar] [CrossRef]
- Whitfield, G.K.; Dang, H.T.; Schluter, S.F.; Bernstein, R.M.; Bunag, T.; Manzon, L.A.; Hsieh, G.; Dominguez, C.E.; Youson, J.H.; Haussler, M.R.; et al. Cloning of a functional vitamin D receptor from the lamprey (Petromyzon marinus), an ancient vertebrate lacking a calcified skeleton and teeth. Endocrinology 2003, 144, 2704–2716. [Google Scholar] [CrossRef]
- Haussler, M.R.; Haussler, C.A.; Bartik, L.; Whitfield, G.K.; Hsieh, J.C.; Slater, S.; Jurutka, P.W. Vitamin D receptor: Molecular signaling and actions of nutritional ligands in disease prevention. Nutr. Rev. 2008, 66, S98–S112. [Google Scholar] [CrossRef]
- Chun, R.F.; Liu, P.T.; Modlin, R.L.; Adams, J.S.; Hewison, M. Impact of vitamin D on immune function: Lessons learned from genome-wide analysis. Front. Physiol. 2014, 5, 151. [Google Scholar] [CrossRef]
- Pike, J.W.; Meyer, M.B.; Lee, S.M.; Onal, M.; Benkusky, N.A. The vitamin D receptor: Contemporary genomic approaches reveal new basic and translational insights. J. Clin. Investig. 2017, 127, 1146–1154. [Google Scholar] [CrossRef]
- Carlberg, C.; Haq, A. The concept of the personal vitamin D response index. J. Steroid Biochem. Mol. Biol. 2018, 175, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Vukic, M.; Neme, A.; Seuter, S.; Saksa, N.; de Mello, V.D.; Nurmi, T.; Uusitupa, M.; Tuomainen, T.P.; Virtanen, J.K.; Carlberg, C. Relevance of vitamin D receptor target genes for monitoring the vitamin D responsiveness of primary human cells. PLoS ONE 2015, 10, e0124339. [Google Scholar] [CrossRef]
- Seuter, S.; Virtanen, J.K.; Nurmi, T.; Pihlajamäki, J.; Mursu, J.; Voutilainen, S.; Tuomainen, T.P.; Neme, A.; Carlberg, C. Molecular evaluation of vitamin D responsiveness of healthy young adults. J. Steroid Biochem. Mol. Biol. 2017, 174, 314–321. [Google Scholar] [CrossRef]
- Hanel, A.; Neme, A.; Malinen, M.; Hamalainen, E.; Malmberg, H.R.; Etheve, S.; Tuomainen, T.P.; Virtanen, J.K.; Bendik, I.; Carlberg, C. Common and personal target genes of the micronutrient vitamin D in primary immune cells from human peripheral blood. Sci. Rep. 2020, 10, 21051. [Google Scholar] [CrossRef]
- Ghosh Dastidar, R.; Jaroslawska, J.; Malinen, M.; Tuomainen, T.P.; Virtanen, J.K.; Bendik, I.; Carlberg, C. In vivo vitamin D targets reveal the upregulation of focal adhesion-related genes in primary immune cells of healthy individuals. Sci. Rep. 2024, 14, 17552. [Google Scholar] [CrossRef]
- Moller-Levet, C.S.; Archer, S.N.; Bucca, G.; Laing, E.E.; Slak, A.; Kabiljo, R.; Lo, J.C.; Santhi, N.; von Schantz, M.; Smith, C.P.; et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc. Natl. Acad. Sci. USA 2013, 110, E1132–E1141. [Google Scholar] [CrossRef]
- Sailani, M.R.; Metwally, A.A.; Zhou, W.; Rose, S.M.S.; Ahadi, S.; Contrepois, K.; Mishra, T.; Zhang, M.J.; Kidzinski, L.; Chu, T.J.; et al. Deep longitudinal multiomics profiling reveals two biological seasonal patterns in California. Nat. Commun. 2020, 11, 4933. [Google Scholar] [CrossRef]
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.; Bruford, E.A. Genenames.org: The HGNC resources in 2023. Nucleic Acids Res. 2023, 51, D1003–D1009. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Sjostedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Neme, A.; Seuter, S.; Carlberg, C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim. Biophys. Acta 2017, 1860, 952–961. [Google Scholar] [CrossRef]
- Seuter, S.; Neme, A.; Carlberg, C. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res. 2016, 44, 4090–4104. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Hanel, A.; Carlberg, C. Time-resolved gene expression analysis monitors the regulation of inflammatory mediators and attenuation of adaptive immune response by vitamin D. Int. J. Mol. Sci. 2022, 23, 911. [Google Scholar] [CrossRef]
- Malmberg, H.R.; Hanel, A.; Taipale, M.; Heikkinen, S.; Carlberg, C. Vitamin D treatment sequence is critical for transcriptome modulation of immune challenged primary human cells. Front. Immunol. 2021, 12, 754056. [Google Scholar] [CrossRef]
- Scheiermann, C.; Kunisaki, Y.; Frenette, P.S. Circadian control of the immune system. Nat. Rev. Immunol. 2013, 13, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Chen, P.; Qi, C. Circadian rhythm regulation in the immune system. Immunology 2024, 171, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Teboul, M.; Barrat-Petit, M.A.; Li, X.M.; Claustrat, B.; Formento, J.L.; Delaunay, F.; Levi, F.; Milano, G. Atypical patterns of circadian clock gene expression in human peripheral blood mononuclear cells. J. Mol. Med. 2005, 83, 693–699. [Google Scholar] [CrossRef]
- Tahara, Y.; Aoyama, S.; Shibata, S. The mammalian circadian clock and its entrainment by stress and exercise. J. Physiol. Sci. 2017, 67, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Luxwolda, M.F.; Kuipers, R.S.; Kema, I.P.; Dijck-Brouwer, D.A.; Muskiet, F.A. Traditionally living populations in East Africa have a mean serum 25-hydroxyvitamin D concentration of 115 nmol/l. Br. J. Nutr. 2012, 108, 1557–1561. [Google Scholar] [CrossRef]
- Mylopotamitaki, D.; Weiss, M.; Fewlass, H.; Zavala, E.I.; Rougier, H.; Sumer, A.P.; Hajdinjak, M.; Smith, G.M.; Ruebens, K.; Sinet-Mathiot, V.; et al. Homo sapiens reached the higher latitudes of Europe by 45,000 years ago. Nature 2024, 626, 341–346. [Google Scholar] [CrossRef]
- Macdonald, C.D.; Falconer, A.M.D.; Chan, C.M.; Wilkinson, D.J.; Skelton, A.; Reynard, L.; Litherland, G.J.; Europe-Finner, G.N.; Rowan, A.D. Cytokine-induced cysteine-serine-rich nuclear protein-1 (CSRNP1) selectively contributes to MMP1 expression in human chondrocytes. PLoS ONE 2018, 13, e0207240. [Google Scholar] [CrossRef]
- Niyonzima, N.; Rahman, J.; Kunz, N.; West, E.E.; Freiwald, T.; Desai, J.V.; Merle, N.S.; Gidon, A.; Sporsheim, B.; Lionakis, M.S.; et al. Mitochondrial C5aR1 activity in macrophages controls IL-1beta production underlying sterile inflammation. Sci. Immunol. 2021, 6, eabf2489. [Google Scholar] [CrossRef]
- She, B.R.; Liou, G.G.; Lin-Chao, S. Association of the growth-arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth. Exp. Cell Res. 2002, 273, 34–44. [Google Scholar] [CrossRef]
- Roach, T.G.; Lang, H.K.M.; Xiong, W.; Ryhanen, S.J.; Capelluto, D.G.S. Protein trafficking or cell signaling: A dilemma for the adaptor protein TOM1. Front. Cell Dev. Biol. 2021, 9, 643769. [Google Scholar] [CrossRef]
- Zong, X.; Schieder, M.; Cuny, H.; Fenske, S.; Gruner, C.; Rotzer, K.; Griesbeck, O.; Harz, H.; Biel, M.; Wahl-Schott, C. The two-pore channel TPCN2 mediates NAADP-dependent Ca(2+)-release from lysosomal stores. Pflug. Arch. Eur. J. Physiol. 2009, 458, 891–899. [Google Scholar] [CrossRef]
- Sundaram, B.; Pandian, N.; Mall, R.; Wang, Y.; Sarkar, R.; Kim, H.J.; Malireddi, R.K.S.; Karki, R.; Janke, L.J.; Vogel, P.; et al. NLRP12-PANoptosome activates PANoptosis and pathology in response to heme and PAMPs. Cell 2023, 186, 2783–2801.e20. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Shaw, R.J. AMPK: Mechanisms of cellular energy sensing and restoration of metabolic balance. Mol. Cell 2017, 66, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, C. Vitamin D in the context of evolution. Nutrients 2022, 14, 3018. [Google Scholar] [CrossRef]
- Lowrey, P.L.; Takahashi, J.S. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu. Rev. Genom. Hum. Genet. 2004, 5, 407–441. [Google Scholar] [CrossRef]
- Zimmet, P.; Alberti, K.; Stern, N.; Bilu, C.; El-Osta, A.; Einat, H.; Kronfeld-Schor, N. The circadian syndrome: Is the metabolic syndrome and much more! J. Intern. Med. 2019, 286, 181–191. [Google Scholar] [CrossRef]
- Abboud, M. Vitamin D supplementation and sleep: A systematic review and meta-analysis of intervention studies. Nutrients 2022, 14, 1076. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maissan, P.; Carlberg, C. Circadian Regulation of Vitamin D Target Genes Reveals a Network Shaped by Individual Responsiveness. Nutrients 2025, 17, 1204. https://doi.org/10.3390/nu17071204
Maissan P, Carlberg C. Circadian Regulation of Vitamin D Target Genes Reveals a Network Shaped by Individual Responsiveness. Nutrients. 2025; 17(7):1204. https://doi.org/10.3390/nu17071204
Chicago/Turabian StyleMaissan, Parcival, and Carsten Carlberg. 2025. "Circadian Regulation of Vitamin D Target Genes Reveals a Network Shaped by Individual Responsiveness" Nutrients 17, no. 7: 1204. https://doi.org/10.3390/nu17071204
APA StyleMaissan, P., & Carlberg, C. (2025). Circadian Regulation of Vitamin D Target Genes Reveals a Network Shaped by Individual Responsiveness. Nutrients, 17(7), 1204. https://doi.org/10.3390/nu17071204






