Assessment of the Effect of High or Low Protein Diet on the Human Urine Metabolome as Measured by NMR
Abstract
:1. Introduction
2. Experimental Section
2.1. Study Design
2.2. Subjects

2.3. Experimental Design
2.4. 24-h Urine Collection and Storage
2.5. Urine Sample Handling
2.6. 1H NMR Analyses
2.7. Pre-Processing of the NMR Spectra

2.8. Statistics and Multivariate Data Analyses
3. Results and Discussion
3.1. Dietary Intake
| HP diet | LP diet | P 2 | P 2 | P 2 | |
|---|---|---|---|---|---|
| (n = 42) | (n = 35) | Main effect of time | Main effect of treatment | Time × treatment interaction | |
| Men/women | 21/21 | 12/23 | |||
| Age (year) | 43.9 ± 4.9 3 | 42 ± 5.1 | |||
| BMI | <0.0001 | NS 5 | NS | ||
| Baseline (kg/m2) | 33.9 ± 1.1 | 34.6 ± 1.2 | |||
| Month 1 (kg/m2) | 28.0 ± 1.1 | 28.0 ± 1.3 | |||
| Month 3 (kg/m2) | 27.9 ± 1.1 | 29.6 ± 1.3 | |||
| Month 6 (kg/m2) | 30.6 ± 1.2 | 30.9 ± 1.3 | |||
| Energy intake | <0.0001 | NS | NS | ||
| Baseline (MJ) | 10.1 ± 0.5 | 10.3 ± 0.6 | |||
| Month 1 (MJ) | 6.8 ± 0.5 | 5.6 ± 0.6 | |||
| Month 6 (MJ) | 5.6 ± 0.5 | 5.4 ± 0.6 | |||
| Protein | - | - | <0.0001 | ||
| Aim | 23–28 | 10–15 | |||
| Baseline (% E) | 15.8 ± 0.6 | 17.5 ± 0.7 | |||
| Month 1 (% E) | 24.1 ± 0.6 | 17.3 ± 0.4 | |||
| Month 6 (% E) | 25.1 ± 0.7 | 18.1 ± 0.4 | |||
| Supermarket | 26.6 ± 0.3 | 13.9 ± 0.3 | |||
| Months 0–6 (% E) | <0.0001 | ||||
| Carbohydrate | - | - | <0.0001 | ||
| Aim | 45–50 | 57–62 | |||
| Baseline (% E) | 46.9 ± 1.1 | 47.5 ± 1.3 | |||
| Month 1 (% E) | 44.7 ± 1.1 | 56.3 ± 1.2 | |||
| Month 6 (% E) | 46.9 ± 1.3 | 55.2 ± 1.4 | |||
| Supermarket | 43.6 ± 0.3 | 56.2 ± 0.4 | |||
| Months 0–6 (% E) | <0.0001 | ||||
| Fat | <0.0001 | NS | NS | ||
| Aim | 25–30 | 25–30 | |||
| Baseline (% E) | 33.9 ± 1.0 | 32.8 ± 1.2 | |||
| Month 1 (% E) | 28.9 ± 1.1 | 26.3 ± 1.2 | |||
| Month 6 (% E) | 26.1 ± 1.2 | 25.3 ± 1.3 | |||
| Supermarket | 25.8 ± 0.4 | 25.3 ± 0.4 | |||
| Months 0–6 (% E) | NS | ||||
| U-Nitrogen 4 | NS | 0.001 | NS | ||
| Baseline (g/24 h) | 14.8 ± 0.6 | 14.8 ± 0.7 | |||
| Month 1 (g/24 h) | 15.8 ± 0.6 | 11.4 ± 0.6 | |||
| Month 3 (g/24 h) | 17.5 ± 0.6 | 12.8 ± 0.7 | |||
| Month 6 (g/24 h) | 16.6 ± 0.6 | 13.8 ± 0.6 | |||
| U-Creatinine 4 | NS | 0.078 | NS | ||
| Baseline (mmol/24 h) | 14.8 ± 0.5 | 14.4 ± 0.6 | |||
| Month 6 (mmol/24 h) | 16.1 ± 0.7 | 13.9 ± 0.6 |

3.2. Validation of NMR Data by Means of Creatinine and Nitrogen Calibration


3.3. 1H NMR Spectroscopy of Human Urine


3.4. Associations Between Dietary Patterns Related to Protein Intake and Human Urinary Metabolites
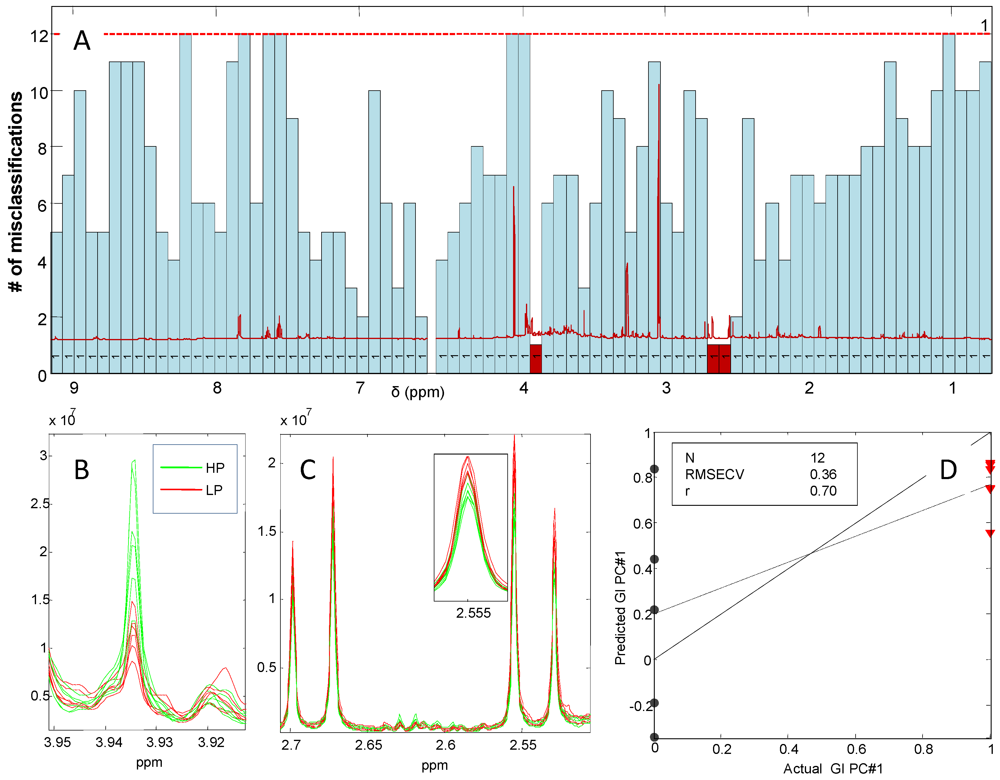
3.5. Associations Between Dietary Patterns Related to Protein Intake and Gender-Specific Metabolites
| δ ppm | Female | Male | Chemical assignment a |
|---|---|---|---|
| 9.13 | HP | Trigonelline (from Caffeine) | |
| 8.84 (triplet) | HP | Trigonelline (from Caffeine) | |
| 8.46 | HP | Formate | |
| 4.34 | LP | Tartaric acid (grape/wine) | |
| 3.93 | HP | HP | Creatine |
| 3.88 (doublet) | LP | Mannitol | |
| 3.84 (doublet) | LP | Mannitol | |
| 3.75-3.79 | LP | Alanine | |
| 3.57 | LP | Glycine | |
| 3.43 (triplet) | HP | HP | Taurine |
| 3.27 | HP | TMAO (trimethylamine-N-oxide) | |
| 3.21 | HP | Carnitine | |
| 3.04 | HP | HP | Creatine |
| 2.92 | LP | (dimethyl-glycine) | |
| 2.88 | HP | TMA (trimethylamine) | |
| 2.73 | LP | DMA (dimethylamine) | |
| 2.68 (doublet) | LP | LP | Citric acid |
| 2.54 (doublet) | LP | LP | Citric acid |
| 2.41 | LP | Succinate | |
| 2.32 | HP | Taurine | |
| 2.20 | HP | Carnitine | |
| 2.27 (triplet) | HP | Taurine | |
| 2.22 | LP | Ribose | |
| 2.06 | LP | N-acethyl glycosamine (NAG) | |
| 1.48 (doublet) | LP | Alanine |
4. Conclusions
Acknowledgements
Conflict of Interest
References
- Bingham, S.A. Biomarkers in nutritional epidemiology. Public Health Nutr. 2002, 5, 821–827. [Google Scholar]
- Schoeller, D.A. How accurate is self-reported dietary energy intake? Nutr. Rev. 1990, 48, 373–379. [Google Scholar] [CrossRef]
- Briefel, R.R.; McDowell, M.A.; Alaimo, K.; Caughman, C.R.; Bischof, A.L.; Carroll, M.D.; Johnson, C.L. Total energy intake of the US population: The third National Health and Nutrition Examination Survey, 1988-1991. Am. J. Clin. Nutr. 1995, 62, 1072S–1080S. [Google Scholar]
- Heitmann, B.L.; Lissner, L. Dietary underreporting by obese individuals-is it specific or non-specific? BMJ 1995, 311, 986–989. [Google Scholar] [CrossRef]
- Cross, A.J.; Ward, M.H.; Schenk, M.; Kulldorff, M.; Cozen, W.; Davis, S.; Colt, J.S.; Hartge, P.; Cerhan, J.R.; Sinha, R. Meat and meat-mutagen intake and risk of non-Hodgkin lymphoma: results from a NCI-SEER case-control study. Carcinogenesis 2006, 27, 293–297. [Google Scholar] [CrossRef]
- Cross, A.J.; Leitzmann, M.F.; Gail, M.H.; Hollenbeck, A.R.; Schatzkin, A.; Sinha, R. A prospective study of red and processed meat intake in relation to cancer risk. PLoS Med. 2007, 4, e325. [Google Scholar]
- Verhoef, P.; van Vliet, T.; Olthof, M.R.; Katan, M.B. A high-protein diet increases postprandial but not fasting plasma total homocysteine concentrations: A dietary controlled, crossover trial in healthy volunteers. Am. J. Clin. Nutr. 2005, 82, 553–558. [Google Scholar]
- Stella, C.; Beckwith-Hall, B.; Cloarec, O.; Holmes, E.; Lindon, J.C.; Powell, J.; van der Ouderaa, F.; Bingham, S.; Cross, A.J.; Nicholson, J.K. Susceptibility of human metabolic phenotypes to dietary modulation. J. Proteome Res. 2006, 5, 2780–2788. [Google Scholar]
- Xu, J.; Yang, S.; Cai, S.; Dong, J.; Li, X.; Chen, Z. Identification of biochemical changes in lactovegetarian urine using 1H NMR spectroscopy and pattern recognition. Anal. Bioanal. Chem. 2010, 396, 1451–1463. [Google Scholar]
- Larsen, T.M.; Dalskov, S.; van Baak, M.; Jebb, S.; Kafatos, A.; Pfeiffer, A.; Martinez, J.A.; Handjieva-Darlenska, T.; Kunesová, M.; Holst, C.; et al. The diet, obesity and genes (diogenes) dietary study in eight European countries-A comprehensive design for long—term intervention. Obes. Rev. 2010, 11, 76–91. [Google Scholar]
- Moore, C.S.; Lindroos, A.K.; Kreutzer, M.; Larsen, T.M.; Astrup, A.; van Baak, M.A.; Handjieva-Darlenska, T.; Hlavaty, P.; Kafatos, A.; Kohl, A.; et al. Dietary strategy to manipulate ad libitum macronutrient intake, and glycaemic index, across eight European countries in the Diogenes Studies. Obes. Rev. 2010, 11, 67–75. [Google Scholar]
- Rasmussen, L.G.; Larsen, T.M.; Mortensen, P.K.; Due, A.; Astrup, A. Effect on 24-h energy expenditure of a moderate-fat diet high in monounsaturated fatty acids compared with that of a low-fat, carbohydrate-rich diet: A 6-mo controlled dietary intervention trial. Am. J. Clin. Nutr. 2007, 85, 1014–1022. [Google Scholar]
- Rasmussen, L.G.; Savorani, F.; Larsen, T.M.; Dragsted, L.O.; Astrup, A.; Engelsen, S.B. Standardization of factors that influence human urine metabolomics. Metabolomics 2011, 7, 71–83. [Google Scholar]
- Bingham, S.A. Urine nitrogen as a biomarker for the validation of dietary protein intake. J. Nutr. 2003, 133, 921S–924S. [Google Scholar]
- Savorani, F.; Tomasi, G.; Engelsen, S.B. icoshift: A versatile tool for the rapid alignment of 1D NMR spectra. J. Magn. Reson. 2010, 202, 190–202. [Google Scholar]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933, 24, 498–520. [Google Scholar]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar]
- Nørgaard, L.; Saudland, A.; Wagner, J.; Nielsen, J.P.; Munck, L.; Engelsen, S.B. Interval partial least squares regression (iPLS): A comparative chemometric study with an example from near infrared spectroscopy. Appl. Spectrosc. 2000, 54, 413–419. [Google Scholar]
- Savorani, F.; Kristensen, M.; Larsen, F.H.; Astrup, A.; Engelsen, S.B. High throughput prediction of chylomicron triglycerides in human plasma by nuclear magnetic resonance and chemometrics. Nutr. Metab. 2010, 7, 43. [Google Scholar]
- Kristensen, M.; Savorani, F.; Ravn-Haren, G.; Poulsen, M.; Markowski, J.; Larsen, F.H.; Dragsted, L.O.; Engelsen, S.B. NMR and interval PLS as reliable methods for determination of cholesterol in rodent lipoprotein fractions. Metabolomics 2010, 6, 129–136. [Google Scholar]
- Skov, A.R.; Toubro, S.; Raben, A.; Astrup, A. A method to achieve control of dietary macronutrient composition in ad libitum diets consumed by free-living subjects. Eur. J. Clin. Nutr. 1997, 51, 667–672. [Google Scholar]
- Day, N.; McKeown, N.; Wong, M.; Welch, A.; Bingham, S. Epidemiological assessment of diet: A comparison of a 7-day diary with a food frequency questionnaire using urinary markers of nitrogen, potassium and sodium. Int. J. Epidemiol. 2001, 30, 309–317. [Google Scholar]
- Lenz, E.M.; Bright, J.; Wilson, I.D.; Hughes, A.; Morrisson, J.; Lindberg, H.; Lockton, A. Metabonomics, dietary influences and cultural differences: A 1H NMR-based study of urine samples obtained from healthy British and Swedish subjects. J. Pharm. Biomed. Anal. 2004, 36, 841–849. [Google Scholar]
- Walsh, M.C.; Brennan, L.; Pujos-Guillot, E.; Sébédio, J.L.; Scalbert, A.; Fagan, A.; Higgins, D.G.; Gibney, M.J. Influence of acute phytochemical intake on human urinary metabolomic profiles. Am. J. Clin. Nutr. 2007, 86, 1687–1693. [Google Scholar]
- Holmes, E.; Loo, R.L.; Stamler, J.; Bictash, M.; Yap, I.K.; Chan, Q.; Ebbels, T.; de Iorio, M.; Brown, I.J.; Veselkov, K.A.; et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008, 453, 396–400. [Google Scholar]
- Dumas, M.E.; Maibaum, E.C.; Teague, C.; Ueshima, H.; Zhou, B.; Lindon, J.C.; Nicholson, J.K.; Stamler, J.; Elliott, P.; Chan, Q.; Holmes, E. Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: The INTERMAP Study. Anal. Chem. 2006, 78, 2199–2208. [Google Scholar]
- Yancey, P.H.; Rhea, M.D.; Kemp, K.M.; Bailey, D.M. Trimethylamine oxide, betaine and other osmolytes in deep-sea animals: Depth trends and effects on enzymes under hydrostatic pressure. Cell. Mol. Biol. 2004, 50, 371–376. [Google Scholar]
- Nicholson, J.K.; Foxall, P.J.; Spraul, M.; Farrant, R.D.; Lindon, J.C. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal. Chem. 1995, 67, 793–811. [Google Scholar]
- Holmes, E.; Foxall, P.J.; Spraul, M.; Farrant, R.D.; Nicholson, J.K.; Lindon, J.C. 750 MHz 1H NMR spectroscopy characterisation of the complex metabolic pattern of urine from patients with inborn errors of metabolism: 2-Hydroxyglutaric aciduria and maple syrup urine disease. J. Pharm. Biomed. Anal. 1997, 15, 1647–1659. [Google Scholar]
- Nicholls, A.W.; Mortishire-Smith, R.J.; Nicholson, J.K. NMR spectroscopic-based metabonomic studies of urinary metabolite variation in acclimatizing germ-free rats. Chem. Res. Toxicol. 2003, 16, 1395–1404. [Google Scholar]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar]
- Siener, R.; Hesse, A. The effect of different diets on urine composition and the risk of calcium oxalate crystallisation in healthy subjects. Eur. Urol. 2002, 42, 289–296. [Google Scholar]
- Yang, W.J.; Wang, Y.W.; Zhou, Q.F.; Tang, H.R. Analysis of human urine metabolites using SPE and NMR spectroscopy. Sci. China Ser. B Chem. 2008, 51, 218–225. [Google Scholar]
- Dragsted, L.O. Biomarkers of meat intake and the application of nutrigenomics. Meat Sci. 2009, 84, 301–307. [Google Scholar]
- Bingham, S.A.; Gill, C.; Welch, A.; Cassidy, A.; Runswick, S.A.; Oakes, S.; Lubin, R.; Thurnham, D.I.; Key, T.J.; Roe, L.et al. Validation of dietary assessment methods in the UK arm of EPIC using weighed records, and 24-hour urinary nitrogen and potassium and serum vitamin C and carotenoids as biomarkers. Int. J. Epidemiol. 1997, 26, S137–S151. [Google Scholar] [CrossRef]
- Weber, J.L.; Reid, P.M.; Greaves, K.A.; DeLany, J.P.; Stanford, V.A.; Going, S.B.; Howell, W.H.; Houtkooper, L.B. Validity of self-reported energy intake in lean and obese young women, using two nutrient databases, compared with total energy expenditure assessed by doubly labeled water. Eur. J. Clin. Nutr. 2001, 55, 940–950. [Google Scholar] [CrossRef]
- The Diogenes project—Sponsor list. Available online: http://www.diogenes-eu.org/sponsors (accessed on 15 April 2010).
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Rasmussen, L.G.; Winning, H.; Savorani, F.; Toft, H.; Larsen, T.M.; Dragsted, L.O.; Astrup, A.; Engelsen, S.B. Assessment of the Effect of High or Low Protein Diet on the Human Urine Metabolome as Measured by NMR. Nutrients 2012, 4, 112-131. https://doi.org/10.3390/nu4020112
Rasmussen LG, Winning H, Savorani F, Toft H, Larsen TM, Dragsted LO, Astrup A, Engelsen SB. Assessment of the Effect of High or Low Protein Diet on the Human Urine Metabolome as Measured by NMR. Nutrients. 2012; 4(2):112-131. https://doi.org/10.3390/nu4020112
Chicago/Turabian StyleRasmussen, Lone G., Hanne Winning, Francesco Savorani, Henrik Toft, Thomas M. Larsen, Lars O. Dragsted, Arne Astrup, and Søren B. Engelsen. 2012. "Assessment of the Effect of High or Low Protein Diet on the Human Urine Metabolome as Measured by NMR" Nutrients 4, no. 2: 112-131. https://doi.org/10.3390/nu4020112
APA StyleRasmussen, L. G., Winning, H., Savorani, F., Toft, H., Larsen, T. M., Dragsted, L. O., Astrup, A., & Engelsen, S. B. (2012). Assessment of the Effect of High or Low Protein Diet on the Human Urine Metabolome as Measured by NMR. Nutrients, 4(2), 112-131. https://doi.org/10.3390/nu4020112






