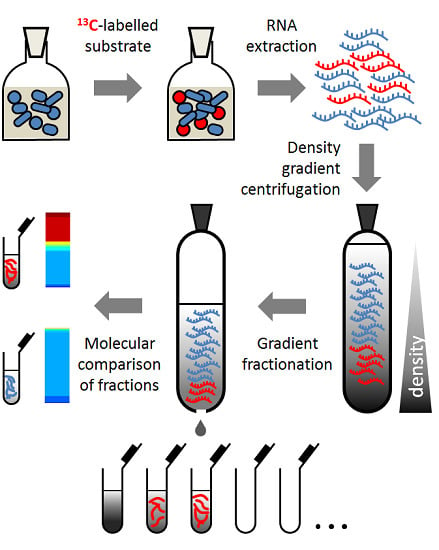
Detection of Sialic Acid-Utilising Bacteria in a Caecal Community Batch Culture Using RNA-Based Stable Isotope Probing
Abstract
:1. Introduction
2. Experimental Section
2.1. Animal Information
2.2. Caecal Cultures
2.3. Nucleic Acid Extraction
2.4. RNA-SIP
2.5. Reverse-Transcription and PCR
2.6. Sequence Analysis
2.7. Statistics
3. Results
3.1. Characterisation of Fresh and Cultured Piglet Caecal Microbiota Composition

3.2. Detection of Piglet Caecal Bacteria Able to Utilise 13C-sialic Acid

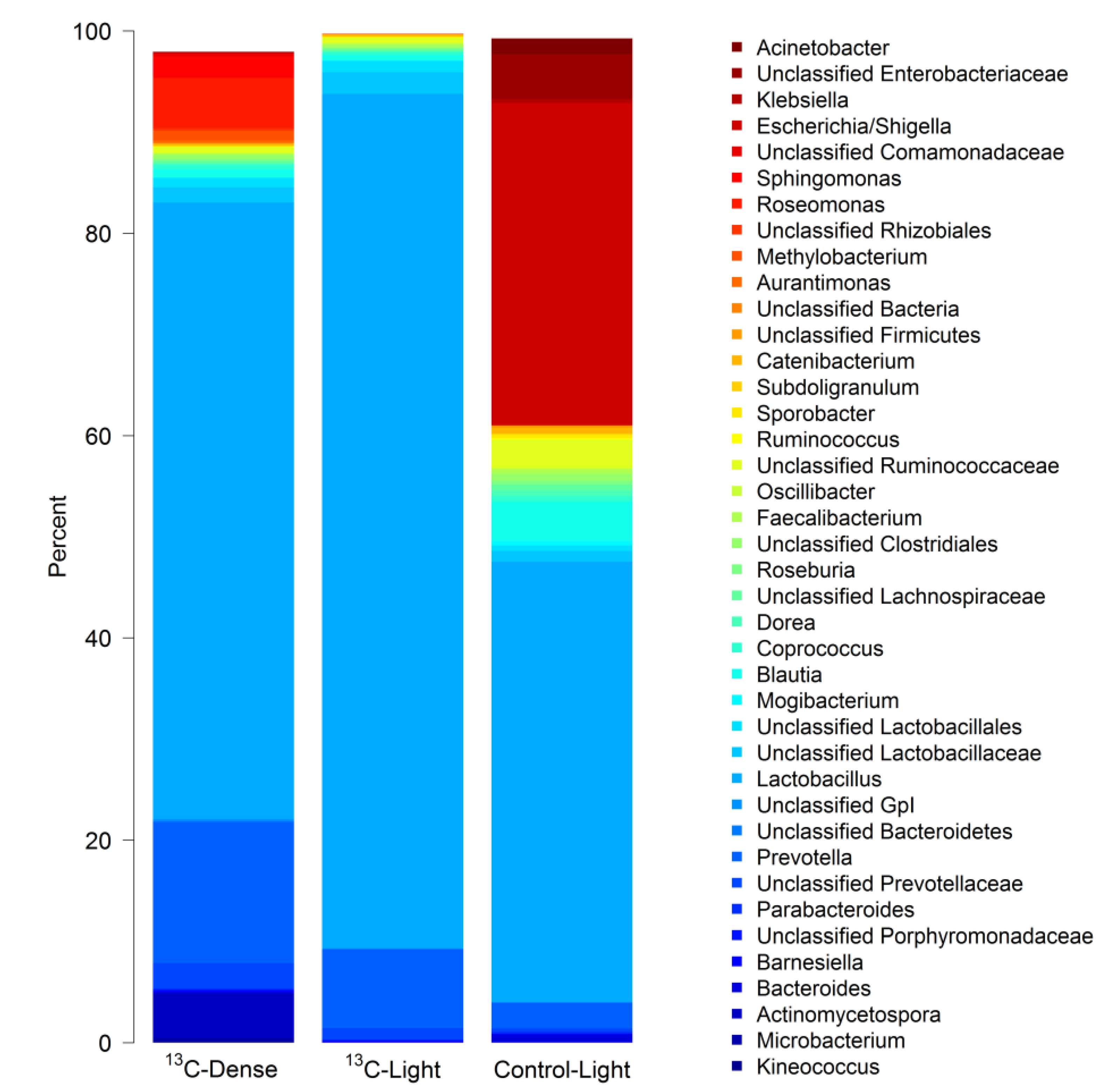


4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Schnaar, R.L.; Gerardy-Schahn, R.; Hildebrandt, H. Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014, 94, 461–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Miller, J.B.; McNeil, Y.; McVeagh, P. Sialic acid concentration of brain gangliosides: Variation among eight mammalian species. Compar. Biochem. Physiol. Part A Mol. Integr. Physiol. 1998, 119, 435–439. [Google Scholar] [CrossRef]
- Kracun, I.; Rosner, H.; Drnovsek, V.; Heffer-Lauc, M.; Cosovic, C.; Lauc, G. Human brain gangliosides in development, aging and disease. Int. J. Dev. Biol. 1991, 35, 289–295. [Google Scholar] [PubMed]
- Morgan, B.L.; Winick, M. Effects of administration of N-acetylneuraminic acid (NANA) on brain NANA content and behavior. J. Nutr. 1980, 110, 416–424. [Google Scholar] [PubMed]
- Wang, B.; Yu, B.; Karim, M.; Hu, H.; Sun, Y.; McGreevy, P.; Petocz, P.; Held, S.; Brand-Miller, J. Dietary sialic acid supplementation improves learning and memory in piglets. Am. J. Clin. Nutr. 2007, 85, 561–569. [Google Scholar] [PubMed]
- Wang, B.; Brand-Miller, J.; McVeagh, P.; Petocz, P. Concentration and distribution of sialic acid in human milk and infant formulas. Am. J. Clin. Nutr. 2001, 74, 510–515. [Google Scholar] [PubMed]
- Zivkovic, A.M.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. USA 2011, 108, 4653–4658. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.T.; Chen, C.; Newburg, D.S. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 2013, 23, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Morrow, A.L.; Ruiz-Palacios, G.M.; Jiang, X.; Newburg, D.S. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J. Nutr. 2005, 135, 1304–1307. [Google Scholar] [PubMed]
- Gurnida, D.A.; Rowan, A.M.; Idjradinata, P.; Muchtadi, D.; Sekarwana, N. Association of complex lipids containing gangliosides with cognitive development of 6-month-old infants. Early Hum. Dev. 2012, 88, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Rueda, R.; Sabatel, J.L.; Maldonado, J.; Molina-Font, J.A.; Gil, A. Addition of gangliosides to an adapted milk formula modifies levels of fecal Escherichia coli in preterm newborn infants. J. Pediatr. 1998, 133, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, L.; Nilsson, Å.; Lundgren, P.; Duan, R.-D. Localization and capacity of sphingomyelin digestion in the rat intestinal tract. J. Nutr. Biochem. 1997, 8, 112–118. [Google Scholar] [CrossRef]
- Gil, A.; Rueda, R. Modulation of intestinal microflora by specific dietary components. Microb. Ecol. Health Dis. 2000, Suppl 2, 31–39. [Google Scholar]
- Radajewski, S.; Ineson, P.; Parekh, N.R.; Murrell, J.C. Stable-isotope probing as a tool in microbial ecology. Nature 2000, 403, 646–649. [Google Scholar] [CrossRef] [PubMed]
- Boschker, H.T.S.; Nold, S.C.; Wellsbury, P.; Bos, D.; de Graaf, W.; Pel, R.; Parkes, R.J.; Cappenberg, T.E. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 1998, 392, 801–805. [Google Scholar] [CrossRef]
- Abraham, W.R. Applications and impacts of stable isotope probing for analysis of microbial interactions. Appl. Microbiol. Biotechnol. 2014, 98, 4817–4828. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, J.D.; Wagner, M.; Murrell, J.C. Who eats what, where and when? Isotope-labelling experiments are coming of age. ISME J. 2007, 1, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Egert, M.; de Graaf, A.A.; Maathuis, A.; de Waard, P.; Plugge, C.M.; Smidt, H.; Deutz, N.E.; Dijkema, C.; de Vos, W.M.; Venema, K. Identification of glucose-fermenting bacteria present in an in vitro model of the human intestine by RNA-stable isotope probing. FEMS Microbiol. Ecol. 2007, 60, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva-Datchary, P.; Egert, M.; Maathuis, A.; Rajilic-Stojanovic, M.; de Graaf, A.A.; Smidt, H.; de Vos, W.M.; Venema, K. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ. Microbiol. 2009, 11, 914–926. [Google Scholar] [CrossRef] [PubMed]
- Tannock, G.W.; Lawley, B.; Munro, K.; Sims, I.M.; Lee, J.; Butts, C.A.; Roy, N. RNA-stable-isotope probing shows utilization of carbon from inulin by specific bacterial populations in the rat large bowel. Appl. Environ. Microbiol. 2014, 80, 2240–2247. [Google Scholar] [CrossRef] [PubMed]
- Falk, P.; Hoskins, L.C.; Larson, G. Bacteria of the human intestinal microbiota produce glycosidases specific for lacto-series glycosphingolipids. J. Biochem. 1990, 108, 466–474. [Google Scholar] [PubMed]
- Calder, P.C.; Krauss-Etschmann, S.; de Jong, E.C.; Dupont, C.; Frick, J.S.; Frokiaer, H.; Heinrich, J.; Garn, H.; Koletzko, S.; Lack, G.; et al. Early nutrition and immunity—Progress and perspectives. Br. J. Nutr. 2006, 96, 774–790. [Google Scholar] [CrossRef] [PubMed]
- Kohler, H.; Donarski, S.; Stocks, B.; Parret, A.; Edwards, C.; Schroten, H. Antibacterial characteristics in the feces of breast-fed and formula-fed infants during the first year of life. J. Pediatr. Gastroenterol. Nutr. 2002, 34, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Tannock, G.W.; Munro, K.; Bibiloni, R.; Simon, M.A.; Hargreaves, P.; Gopal, P.; Harmsen, H.; Welling, G. Impact of consumption of oligosaccharide-containing biscuits on the fecal microbiota of humans. Appl. Environ. Microbiol. 2004, 70, 2129–2136. [Google Scholar] [CrossRef] [PubMed]
- Karl, D.M.; Bossard, P. Measurement of microbial nucleic acid synthesis and specific growth rate by 32PO4 and [3H]adenine: Field comparison. Appl. Environ. Microbiol. 1985, 50, 706–709. [Google Scholar] [PubMed]
- Furet, J.P.; Firmesse, O.; Gourmelon, M.; Bridonneau, C.; Tap, J.; Mondot, S.; Dore, J.; Corthier, G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 2009, 68, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Claus, S.P.; Ellero, S.L.; Berger, B.; Krause, L.; Bruttin, A.; Molina, J.; Paris, A.; Want, E.J.; de Waziers, I.; Cloarec, O.; et al. Colonization-induced host-gut microbial metabolic interaction. MBio 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Whiteley, A.S.; Thomson, B.; Lueders, T.; Manefield, M. RNA stable-isotope probing. Nat. Protocols 2007, 2, 838–844. [Google Scholar] [CrossRef]
- Whiteley, A.S.; Manefield, M.; Lueders, T. Unlocking the ‘microbial black box’ using RNA-based stable isotope probing technologies. Curr. Opin. Biotechnol. 2006, 17, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, B. Sialic acid is an essential nutrient for brain development and cognition. Annu. Rev. Nutr. 2009, 29, 177–222. [Google Scholar] [CrossRef] [PubMed]
- Severi, E.; Hood, D.W.; Thomas, G.H. Sialic acid utilization by bacterial pathogens. Microbiology (Reading, Engl.) 2007, 153, 2817–2822. [Google Scholar] [CrossRef]
- Wright, D.P.; Rosendale, D.I.; Robertson, A.M. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 2000, 190, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; van Passel, M.W.; van de Bovenkamp, J.H.; Schipper, R.G.; de Vos, W.M.; Dekker, J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 2010, 1, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.A.; Li, Y.; Lerno, L.; Wu, S.; Marcobal, A.M.; German, J.B.; Chen, X.; Lebrilla, C.B.; Mills, D.A. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J. Biol. Chem. 2011, 286, 11909–11918. [Google Scholar] [CrossRef] [PubMed]
- Mahowald, M.A.; Rey, F.E.; Seedorf, H.; Turnbaugh, P.J.; Fulton, R.S.; Wollam, A.; Shah, N.; Wang, C.; Magrini, V.; Wilson, R.K.; et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA 2009, 106, 5859–5864. [Google Scholar] [CrossRef] [PubMed]
- Crost, E.H.; Tailford, L.E.; Le Gall, G.; Fons, M.; Henrissat, B.; Juge, N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS ONE 2013, 8, e76341. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Dassa, B.; Borovok, I.; Ruimy-Israeli, V.; Lamed, R.; Flint, H.J.; Duncan, S.H.; Henrissat, B.; Coutinho, P.; Morrison, M.; Mosoni, P.; et al. Rumen cellulosomics: Divergent fiber-degrading strategies revealed by comparative genome-wide analysis of six ruminococcal strains. PLoS ONE 2014, 9, e99221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, D.E.; Smalley, D.J.; Tucker, D.L.; Leatham, M.P.; Norris, W.E.; Stevenson, S.J.; Anderson, A.B.; Grissom, J.E.; Laux, D.C.; Cohen, P.S.; et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 2004, 101, 7427–7432. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.M.; Ferreyra, J.A.; Higginbottom, S.K.; Lynch, J.B.; Kashyap, P.C.; Gopinath, S.; Naidu, N.; Choudhury, B.; Weimer, B.C.; Monack, D.M.; et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013, 502, 96–99. [Google Scholar] [CrossRef] [PubMed]
- El-Labany, S.; Sohanpal, B.K.; Lahooti, M.; Akerman, R.; Blomfield, I.C. Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol. Microbiol. 2003, 49, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Schwab, C.; Ganzle, M. Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiol. Lett. 2011, 315, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Garrido, D.; Mills, D.A.; Barile, D. Hydrolysis of milk gangliosides by infant-gut associated bifidobacteria determined by microfluidic chips and high-resolution mass spectrometry. Electrophoresis 2014, 35, 1742–1750. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Young, W.; Egert, M.; Bassett, S.A.; Bibiloni, R. Detection of Sialic Acid-Utilising Bacteria in a Caecal Community Batch Culture Using RNA-Based Stable Isotope Probing. Nutrients 2015, 7, 2109-2124. https://doi.org/10.3390/nu7042109
Young W, Egert M, Bassett SA, Bibiloni R. Detection of Sialic Acid-Utilising Bacteria in a Caecal Community Batch Culture Using RNA-Based Stable Isotope Probing. Nutrients. 2015; 7(4):2109-2124. https://doi.org/10.3390/nu7042109
Chicago/Turabian StyleYoung, Wayne, Markus Egert, Shalome A. Bassett, and Rodrigo Bibiloni. 2015. "Detection of Sialic Acid-Utilising Bacteria in a Caecal Community Batch Culture Using RNA-Based Stable Isotope Probing" Nutrients 7, no. 4: 2109-2124. https://doi.org/10.3390/nu7042109