Investigation into Cellular Glycolysis for the Mechanism Study of Energy Metabolism Disorder Triggered by Lipopolysaccharide
Abstract
:1. Introduction
2. Results
2.1. The Effects of LPS on Intracellular ATP Levels
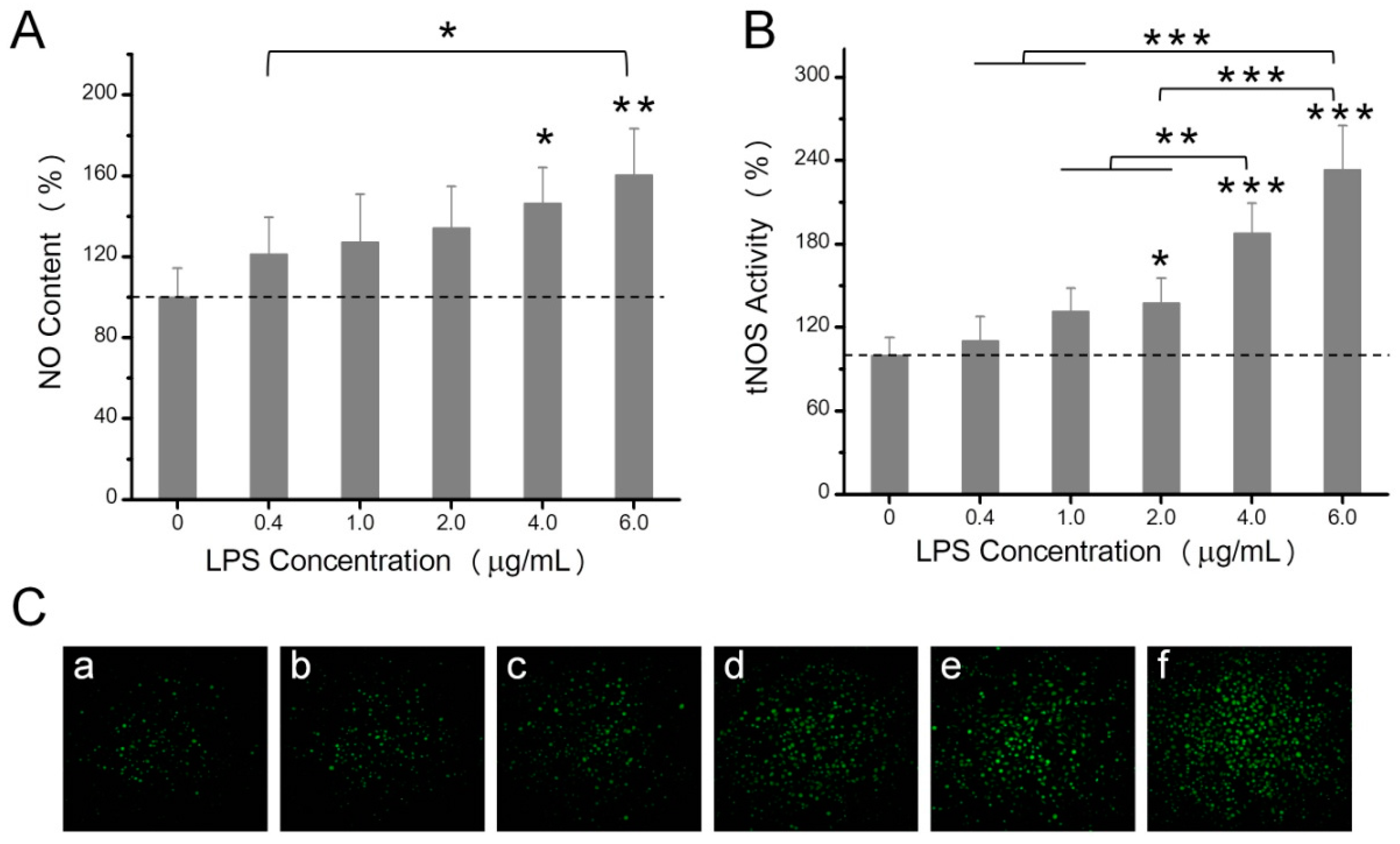
2.2. The Effects of LPS on NO Production, Mitochondrial ROS, and Mitochondrial Membrane Potential
2.3. The Effects of LPS on Enzyme Activities Related to Energy Metabolism
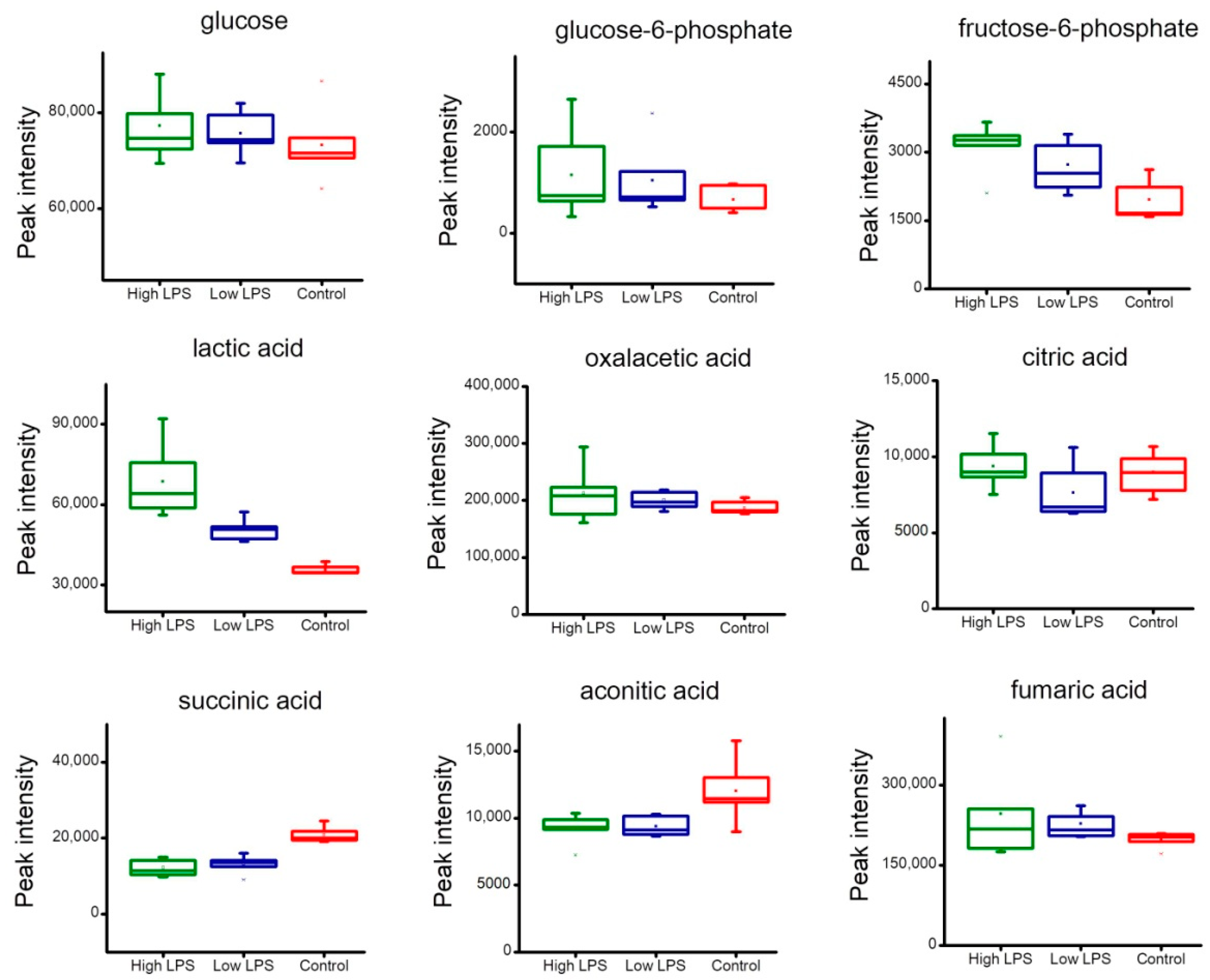
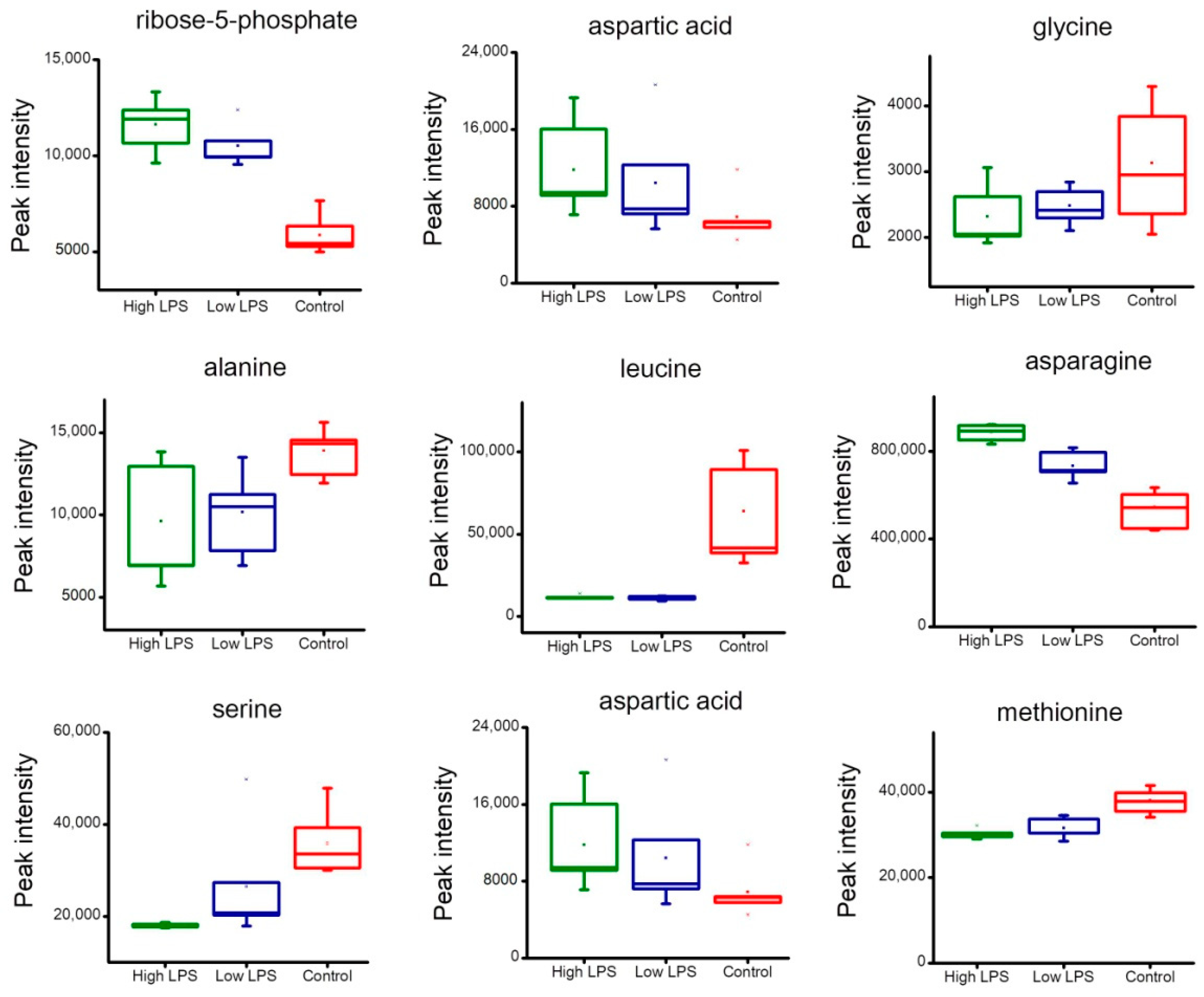
2.4. Analysis of Metabolites Related to Energy Metabolism
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Reagents
5.2. Cell Culture and Treatment
5.3. ATP Measurements
5.4. Nitrite Oxide Measurements
5.5. Mitochondrial ROS Detection
5.6. Mitochondrial Membrane Potential Assessment
5.7. Determination of Enzyme Activities Related to Energy Metabolisms
5.8. Metabolite Extraction
5.9. Derivatization
5.10. GC-TOF/MS Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Favoriti, P.; Carbone, G.; Greco, M.; Pirozzi, F.; Pirozzi, R.E.; Corcione, F. Worldwide burden of colorectal cancer: A review. Updat. Surg. 2016, 68, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Andersen, A.P.; Moreira, J.M.; Pedersen, S.F. Interactions of ion transporters and channels with cancer cell metabolism and the tumour microenvironment. Philso. Trans. R. Soc. Lond. 2014, 369, 20130098. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, A.K.; Melo, J.M.L.D.; Mørup, N.; Tritsaris, K.; Pedersen, S.F. Tumor microenvironment conditions alter Akt and Na+/H+ exchanger NHE1 expression in endothelial cells more than hypoxia alone: Implications for endothelial cell function in cancer. BMC Cancer 2017, 17, 542. [Google Scholar] [CrossRef] [PubMed]
- Airley, R.E.; Mobasheri, A. Hypoxic regulation of glucose transport, anaerobic metabolism and angiogenesis in cancer: Novel pathways and targets for anticancer therapeutics. Chemotherapy 2007, 53, 233–256. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Birsoy, K.; Possemato, R.; Lorbeer, F.K.; Bayraktar, E.C.; Thiru, P.; Yucel, B.; Wang, T.; Chen, W.W.; Clish, C.B.; Sabatini, D.M. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 2014, 508, 108–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbet, C.; Pinto, A.; Martherus, R.; Jp, S.D.J.; Polet, F.; Feron, O. Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation. Cell Metab. 2016, 24, 311–323. [Google Scholar] [CrossRef] [PubMed]
- DeNicola, G.M.; Cantley, L.C. Cancer’s Fuel Choice: New Flavors for a Picky Eater. Mol. Cell 2015, 60, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Rietschel, E.T.; Brade, H.; Holst, O.; Brade, L.; Müller-Loennies, S.; Mamat, U.; Zähringer, U.; Beckmann, F.; Seydel, U.; Brandenburg, K. Bacterial Endotoxin: Chemical Constitution, Biological Recognition, Host Response, and Immunological Detoxification; Springer: Heidelberg, Germany, 1996; pp. 39–81. [Google Scholar]
- Caroff, M.; Karibian, D. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 2003, 338, 2431–2447. [Google Scholar] [CrossRef] [PubMed]
- Needham, B.D.; Trent, M.S. Fortifying the barrier: The impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 2013, 11, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Badshah, H.; Ali, T.; Rehman, S.U.; Amin, F.U.; Ullah, F.; Kim, T.H.; Kim, M.O. Protective Effect of Lupeol Against Lipopolysaccharide-Induced Neuroinflammation via the p38/c-Jun N-Terminal Kinase Pathway in the Adult Mouse Brain. J. Neuroimmune Pharmacol. 2016, 11, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, B.; Sonobe, Y.; Kawanokuchi, J.; Doi, Y.; Noda, M.; Takeuchi, H.; Mizuno, T.; Suzumura, A. GM-CSF increases LPS-induced production of proinflammatory mediators via upregulation of TLR4 and CD14 in murine microglia. J. Neuroinflammation 2012, 9, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, A.C.; Massagué, J. Molecular basis of metastasis. N. Engl. J. Med. 2005, 159, 2814–2823. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabol. Off. J. Metabol. Soc. 2016, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y. Cancer Energy Metabolism: Shutting Power off Cancer Factory. Biomol. Ther. 2018, 26, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Snezhkina, A.V.; Krasnov, G.S.; Zaretsky, A.R.; Zhavoronkov, A.; Nyushko, K.M.; Moskalev, A.A.; Karpova, I.Y.; Afremova, A.I.; Lipatova, A.V.; Kochetkov, D.V. Differential expression of alternatively spliced transcripts related to energy metabolism in colorectal cancer. BMC Genom. 2016, 17, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Korniluk, A.; Koper, O.; Kemona, H.; Dymickapiekarska, V. From inflammation to cancer. Ir. J. Med. Sci. 2017, 186, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Pesic, M.; Greten, F.R. Inflammation and cancer: Tissue regeneration gone awry. Curr. Opin. Cell Biol. 2016, 43, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Yan, G.T.; Wang, L.H.; Hao, X.H.; Zhang, K.; Xue, H. The mediating role of cPLA2 in IL-1 beta and IL-6 release in LPS-induced HeLa cells. Cell Biochem. Funct. 2004, 22, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, D.; Peng, H.; Shao, J.; Gu, J. Cancer-derived immunoglobulin G promotes LPS-induced proinflammatory cytokine production via binding to TLR4 in cervical cancer cells. Oncotarget 2014, 5, 9727–9743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mineshiba, J.; Myokai, F.F.; Matsuura, K.; Nishimura, F.; Takashiba, S. Transcriptional regulation of beta-defensin-2 by lipopolysaccharide in cultured human cervical carcinoma (HeLa) cells. Pathog. Dis. 2005, 45, 37–44. [Google Scholar]
- Lin, M.C.; Pan, C.Y.; Hui, C.F.; Chen, J.Y.; Wu, J.L. Shrimp anti-lipopolysaccharide factor (SALF), an antimicrobial peptide, inhibits proinflammatory cytokine expressions through the MAPK and NF-κB pathways in LPS-induced HeLa cells. Peptides 2013, 40, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.X.; Qi, X.Y.; Huang, J.L.; Hu, M.; Zhou, L.M.; Li, B.S.; Xu, X.X. Toll-like receptor 4 signaling promotes the immunosuppressive cytokine production of human cervical cancer. Eur. J. Gynaecol. Oncol. 2012, 33, 291–294. [Google Scholar] [PubMed]
- Shimura, T.; Sasatani, M.; Kamiya, K.; Kawai, H.; Inaba, Y.; Kunugita, N. Mitochondrial reactive oxygen species perturb AKT/cyclin D1 cell cycle signaling via oxidative inactivation of PP2A in lowdose irradiated human fibroblasts. Oncotarget 2016, 7, 3559–3570. [Google Scholar] [CrossRef] [PubMed]
- Martínezreyes, I.; Diebold, L.P.; Kong, H.; Schieber, M.; Huang, H.; Hensley, C.T.; Mehta, M.M.; Wang, T.; Santos, J.H.; Woychik, R. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Mol. Cell 2016, 61, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fetterman, J.L.; Holbrook, M.; Flint, N.; Feng, B.; Bretónromero, R.; Linder, E.A.; Berk, B.D.; Duess, M.A.; Farb, M.G.; Gokce, N. Restoration of autophagy in endothelial cells from patients with diabetes mellitus improves nitric oxide signaling. Atherosclerosis 2016, 247, 207–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraehling, J.R.; Sessa, W.C. Contemporary Approaches to Modulating the Nitric Oxide-cGMP Pathway in Cardiovascular Disease. Circ. Res. 2017, 120, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Kulaksızoglu, S.; Karalezli, A. Aqueous Humour and Serum Levels of Nitric Oxide, Malondialdehyde and Total Antioxidant Status in Patients with Type 2 Diabetes with Proliferative Diabetic Retinopathy and Nondiabetic Senile Cataracts. Can. J. Diabetes 2016, 40, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Brix, B.; Mesters, J.R.; Pellerin, L.; Jöhren, O. Endothelial cell-derived nitric oxide enhances aerobic glycolysis in astrocytes via HIF-1α-mediated target gene activation. J. Neurosci. Off. J. Soc. Neurosci. 2012, 32, 9727–9735. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhu, L.; Hao, B.; Gao, W.; Wang, Q.; Li, K.; Wang, M.; Huang, M.; Liu, Z.; Yang, Q. iNOS-derived nitric oxide promotes glycolysis by inducing pyruvate kinase M2 nuclear translocation in ovarian cancer. Oncotarget 2017, 8, 33047–33063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sena, L.; Chandel, N. Physiological Roles of Mitochondrial Reactive Oxygen Species. Mol. Cell 2012, 48, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.Q.; Li, Q.; Wang, G.H.; Sun, F.F.; Huang, G.J.; Bian, X.W.; Yu, S.C.; Qian, G.S. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int. J. Cancer 2011, 129, 820–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietilä, M.; Lehtonen, S.; Närhi, M.; Hassinen, I.E.; Leskelä, H.V.; Aranko, K.; Nordström, K.; Vepsäläinen, A.; Lehenkari, P. Mitochondrial function determines the viability and osteogenic potency of human mesenchymal stem cells. Tissue Eng. Part C Methods 2010, 16, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Altenberg, B.; Greulich, K.O. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 2004, 84, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Neary, C.L.; Pastorino, J.G. Nucleocytoplasmic shuttling of hexokinase II in a cancer cell. Biochem. Biophys. Res. Commun. 2010, 394, 1075–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmieri, D.; Fitzgerald, D.; Shreeve, S.M.; Hua, E.; Bronder, J.L.; Weil, R.J.; Davis, S.; Stark, A.M.; Merino, M.J.; Kurek, R. Analyses of Resected Human Brain Metastases of Breast Cancer Reveal the Association between Up-regulation of Hexokinase 2 and Poor Prognosis. Mol. Cancer Res. 2009, 7, 1438–1445. [Google Scholar] [CrossRef] [PubMed]
- Benesch, C.; Schneider, C.; Voelker, H.U.; Kapp, M.; Caffier, H.; Krockenberger, M.; Dietl, J.; Kammerer, U.; Schmidt, M. The clinicopathological and prognostic relevance of pyruvate kinase M2 and pAkt expression in breast cancer. Anticancer Res. 2010, 30, 1689–1694. [Google Scholar] [PubMed]
- Shuch, B.; Linehan, W.M.; Srinivasan, R. Aerobic glycolysis: A novel target in kidney cancer. Expert Rev. Anticancer Ther. 2013, 13, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Scheffler, I.E. Mitochondria, 2nd ed.; John Wiley & Sons, Inc.: San Diego, CA, USA, 2007. [Google Scholar]
- Nelson, D.L.; Cox, M.M. Lehninger Principles of Biochemistry, 5th ed.; W.H. Freeman: New York, NY, USA, 2008. [Google Scholar]
- King, A.; Selak, M.A.; Gottlieb, E. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 2006, 25, 4675–4682. [Google Scholar] [CrossRef] [PubMed]
- Pollard, P.J.; Briã¨Re, J.J.; Alam, N.A.; Barwell, J.; Barclay, E.; Wortham, N.C.; Hunt, T.; Mitchell, M.; Olpin, S.; Moat, S.J. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum. Mol. Gen. 2005, 14, 2231–2239. [Google Scholar] [CrossRef] [PubMed]
- Tseng, P.L.; Wu, W.H.; Hu, T.H.; Chen, C.W.; Cheng, H.C.; Li, C.F.; Tsai, W.H.; Tsai, H.J.; Hsieh, M.C.; Chuang, J.H. Decreased succinate dehydrogenase B in human hepatocellular carcinoma accelerates tumor malignancy by inducing the Warburg effect. Sci. Rep. 2018, 8, 3081. [Google Scholar] [CrossRef] [PubMed]
- Han, S.J.; Jang, H.S.; Noh, M.R.; Kim, J.; Kong, M.J.; Kim, J.I.; Park, J.W.; Park, K.M. Mitochondrial NADP+-Dependent Isocitrate Dehydrogenase Deficiency Exacerbates Mitochondrial and Cell Damage after Kidney Ischemia-Reperfusion Injury. J. Am. Soc. Nephrol. Jasn 2016, 28, 1200–1215. [Google Scholar] [CrossRef] [PubMed]
- Reitman, Z.J.; Yan, H. Isocitrate Dehydrogenase 1 and 2 Mutations in Cancer: Alterations at a Crossroads of Cellular Metabolism. J. Natl. Cancer Inst. 2010, 102, 932–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tawakol, A.; Singh, P.; Mojena, M.; Pimentel-Santillana, M.; Emami, H.; Macnabb, M.; Rudd, J.H.; Narula, J.; Enriquez, J.A.; Través, P.G. HIF-1α and PFKFB3 Mediate a Tight Relationship Between Proinflammatory Activation and Anerobic Metabolism in Atherosclerotic Macrophages. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- Haschemi, A.; Kosma, P.; Gille, L.; Evans, C.R.; Burant, C.F.; Starkl, P.; Knapp, B.; Haas, R.; Schmid, J.A.; Jandl, C. The Sedoheptulose Kinase CARKL Directs Macrophage Polarization through Control of Glucose Metabolism. Cell Metab. 2012, 15, 813–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blagih, J.; Jones, R. Polarizing Macrophages through Reprogramming of Glucose Metabolism. Cell Metab. 2012, 15, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Van den Bossche, J.; Baardman, J.; Otto, N.A.; Van der Velden, S.; Neele, A.E.; Sm, V.D.B.; Luque-Martin, R.; Chen, H.J.; Boshuizen, M.C.; Ahmed, M. Mitochondrial Dysfunction Prevents Repolarization of Inflammatory Macrophages. Cell Rep. 2016, 17, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Xie, N.; Cui, H.; Moellering, D.R.; Abraham, E.; Thannickal, V.J.; Liu, G. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J. Immunol. 2015, 194, 6082–6089. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.; O’Neill, L.A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015, 25, 771–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassaganyariera, J.; Guri, A.J.; Lu, P.; Climent, M.; Carbo, A.; Sobral, B.W.; Horne, W.T.; Lewis, S.N.; Bevan, D.R.; Hontecillas, R. Abscisic Acid Regulates Inflammation via Ligand-binding Domain-independent Activation of Peroxisome Proliferator-activated Receptor γ. J. Biol. Chem. 2011, 286, 2504–2516. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Shen, L.; Liu, S.; Li, H.; Ma, Y.; Zhang, R.; Wu, K.; Yao, L.; Li, J.; Zhang, J. Chronic inflammation confers to the metabolic reprogramming associated with tumorigenesis of colorectal cancer. Cancer Biol. Ther. 2017, 18, 237–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Ji, J.; Blaženović, I.; Pi, F.; Wang, T.; Zhang, Y.; Sun, X. Investigation into Cellular Glycolysis for the Mechanism Study of Energy Metabolism Disorder Triggered by Lipopolysaccharide. Toxins 2018, 10, 441. https://doi.org/10.3390/toxins10110441
Zhang R, Ji J, Blaženović I, Pi F, Wang T, Zhang Y, Sun X. Investigation into Cellular Glycolysis for the Mechanism Study of Energy Metabolism Disorder Triggered by Lipopolysaccharide. Toxins. 2018; 10(11):441. https://doi.org/10.3390/toxins10110441
Chicago/Turabian StyleZhang, Ruyuan, Jian Ji, Ivana Blaženović, Fuwei Pi, Tingwei Wang, Yinzhi Zhang, and Xiulan Sun. 2018. "Investigation into Cellular Glycolysis for the Mechanism Study of Energy Metabolism Disorder Triggered by Lipopolysaccharide" Toxins 10, no. 11: 441. https://doi.org/10.3390/toxins10110441
APA StyleZhang, R., Ji, J., Blaženović, I., Pi, F., Wang, T., Zhang, Y., & Sun, X. (2018). Investigation into Cellular Glycolysis for the Mechanism Study of Energy Metabolism Disorder Triggered by Lipopolysaccharide. Toxins, 10(11), 441. https://doi.org/10.3390/toxins10110441




