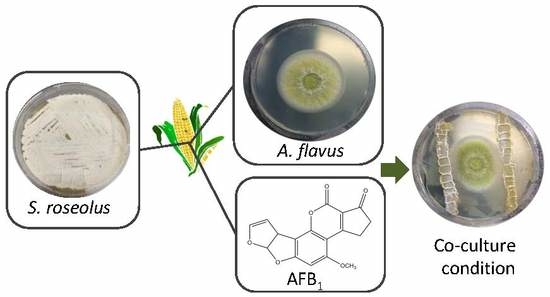
Streptomyces roseolus, A Promising Biocontrol Agent Against Aspergillus flavus, the Main Aflatoxin B1 Producer
Abstract
:1. Introduction
2. Results
2.1. Production of Secondary Metabolites in A. flavus
2.2. Morphological Changes
2.3. Expression Analysis of Genes Involved in Fungal Secondary Metabolite Pathways
2.4. Expression Analysis of Genes Involved in Fungal Development
2.5. Expression Analysis of Genes Involved in Fungal Response to External Stimuli
2.6. Expression Analysis of Genes Involved in Fungal Oxidative Stress Response
3. Discussion
3.1. Effects of S. roseolus on AFB1 Concentration and Gene Expression of AFB1 Cluster Genes
3.2. Effect of S. roseolus on the KA and CPA Concentrations
3.3. Morphological Changes in A. flavus in Co-Culture with S. roseolus
3.4. Impact of S. roseolus on Fungal Cellular Signalization and the Oxidative Stress Response
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Reagents
5.2. Strains
5.3. Culture Conditions
5.4. Secondary Metabolite Extraction and Quantification by UHPLC/FLD/DAD
5.5. Fungal RNA Extraction and Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
5.6. Quantitative Polymerase Chain Reaction (q-PCR) Analysis
5.7. Fungal Spore Quantification
5.8. Microscopic Analysis
5.9. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Hove, M.; Van Poucke, C.; Njumbe-Ediage, E.; Nyanga, L.K.; De Saeger, S. Review on the natural co-occurrence of AFB1 and FB1 in maize and the combined toxicity of AFB1 and FB1. Food Control 2016, 59, 675–682. [Google Scholar] [CrossRef]
- World Health Organization (WHO); International Agency for Research on Cancer (IARC). Aflatoxins, some naturally occurring substances: Food items and constituents, heterocyclic aromatic amines and mycotoxins. IARC Monogr. Eval. Carcinog. Risk Chem. Hum. 1993, 56, 245–395. [Google Scholar]
- Meissonnier, G.M.; Pinton, P.; Laffitte, J.; Cossalter, A.-M.; Gong, Y.Y.; Wild, C.P.; Bertin, G.; Galtier, P.; Oswald, I.P. Immunotoxicity of aflatoxin B1: Impairment of the cell-mediated response to vaccine antigen and modulation of cytokine expression. Toxicol. Appl. Pharmacol. 2008, 231, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Bbosa, G.; Lubega, A. Review of the biological and health effects of aflatoxins on body organs and body systems. In Aflatoxins—Recent Advances and Future Prospects; Intech: London, UK, 2013; Volume 12, pp. 239–265. [Google Scholar]
- Battilani, P.; Toscano, P.; Moretti, A.; Leggieri, M.C. Aflatoxin B 1 contamination in maize in Europe increases due to climate change. Sci. Rep. 2016, 6, 24328. [Google Scholar] [CrossRef] [PubMed]
- Streit, E.; Schatzmayr, G.; Tassis, P.; Tzika, E.; Marin, D.; Taranu, I.; Tabuc, C.; Nicolau, A.; Aprodu, I.; Puel, O.; et al. Current situation of mycotoxin contamination and co-occurrence in animal feed—Focus on Europe. Toxins 2012, 4, 788–809. [Google Scholar] [CrossRef] [PubMed]
- Hymery, N.; Masson, F.; Barbier, G.; Coton, E. Cytotoxicity and immunotoxicity of cyclopiazonic acid on human cells. Toxicol. In Vitro 2014, 28, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Ostry, V.; Toman, J.; Grosse, Y.; Malir, F. Cyclopiazonic acid: 50th anniversary of its discovery. World Mycotoxin J. 2018, 11, 135–148. [Google Scholar] [CrossRef]
- Bentley, R. From miso, saké and shoyu to cosmetics: A century of science for kojic acid. Nat. Prod. Rep. 2006, 23, 1046–1062. [Google Scholar] [CrossRef] [PubMed]
- Danezis, G.P.; Anagnostopoulos, C.J.; Liapis, K.; Koupparis, M.A. Multi-residue analysis of pesticides, plant hormones, veterinary drugs and mycotoxins using HILIC chromatography-MS/MS in various food matrices. Anal. Chim. Acta 2016, 942, 121–138. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, R.; Mathieu, F.; Atoui, A.; Kawtharani, H.; El Khoury, A.; Afif, C.; Maroun, R.G.; El Khoury, A. Ability of soil isolated actinobacterial strains to prevent, bind and biodegrade ochratoxin A. Toxins 2017, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, R.; Caceres, I.; Puel, O.; Bailly, S.; Atoui, A.; Oswald, I.P.; El Khoury, A.; Bailly, J.D. Identification of the anti-aflatoxinogenic activity of Micromeria graeca and elucidation of its molecular mechanism in Aspergillus flavus. Toxins 2017, 9, 87. [Google Scholar] [CrossRef] [PubMed]
- Olarte, R.A.; Horn, B.W.; Dorner, J.W.; Monacell, J.T.; Singh, R.; Stone, E.A.; Carbone, I. Effect of sexual recombination on population diversity in aflatoxin production by Aspergillus flavus and evidence for cryptic heterokaryosis. Mol. Ecol. 2012, 21, 1453–1476. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C. Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: Advantages and limitations. Front. Microbiol. 2014, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- Sakuda, S.; Yoshinari, T.; Furukawa, T.; Jermnak, U.; Takagi, K.; Iimura, K.; Yamamoto, T.; Suzuki, M. Search for aflatoxin and trichothecene production inhibitors and analysis of their modes of action. Biosci. Biotechnol. Biochem. 2016, 80, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Chi, C.; Yu, J.; Shan, S.; Li, Q.; Li, Q.; Guan, B.; Nierman, W.C.; Bennett, J.W. The inhibitory effect of Bacillus megaterium on aflatoxin and cyclopiazonic acid biosynthetic pathway gene expression in Aspergillus flavus. Appl. Microbiol. Biotechnol. 2014, 98, 5161–5172. [Google Scholar] [CrossRef] [PubMed]
- Shams-Ghahfarokhi, M.; Kalantari, S.; Razzaghi-Abyaneh, M. Terrestrial bacteria from agricultural soils: Versatile weapons against aflatoxigenic fungi. In Aflatoxins—Recent Advances And Future Prospects; InTech: London, UK, 2013; pp. 23–39. ISBN 9789535109044. [Google Scholar]
- El Khoury, R.; Choque, E.; El Khoury, A.; Snini, S.P.; Cairns, R.; Andriantsiferana, C.; Mathieu, F. OTA prevention and detoxification by actinobacterial strains and activated carbon fibers: Preliminary results. Toxins 2018, 10, 137. [Google Scholar] [CrossRef] [PubMed]
- Verheecke, C.; Liboz, T.; Anson, P.; Zhu, Y.; Mathieu, F. Streptomyces-Aspergillus flavus interactions: Impact on aflatoxin B accumulation. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2015, 32, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Verheecke, C.; Liboz, T.; Darriet, M.; Sabaou, N.; Mathieu, F. In vitro interaction of actinomycetes isolates with Aspergillus flavus: Impact on aflatoxins B1 and B2 production. Lett. Appl. Microbiol. 2014, 58, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Chang, P. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol Genet. Genom. 2003, 268, 711–719. [Google Scholar] [CrossRef]
- Ehrlich, K.C. Predicted roles of the uncharacterized clustered genes in aflatoxin biosynthesis. Toxins 2009, 1, 37–58. [Google Scholar] [CrossRef] [PubMed]
- Caceres, I.; El Khoury, R.; Medina, Á.; Lippi, Y.; Naylies, C.; Atoui, A.; El Khoury, A.; Oswald, I.P.; Bailly, J.-D.; Puel, O. Deciphering the anti-aflatoxinogenic properties of eugenol using a Large-Scale q-PCR approach. Toxins 2016, 8, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marui, J.; Yamane, N.; Ohashi-Kunihiro, S.; Ando, T.; Terabayashi, Y.; Sano, M.; Ohashi, S.; Ohshima, E.; Tachibana, K.; Higa, Y.; et al. Kojic acid biosynthesis in Aspergillus oryzae is regulated by a Zn(II)2Cys6 transcriptional activator and induced by kojic acid at the transcriptional level. J. Biosci. Bioeng. 2011, 112, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-D.; Han, L.; Yan, S.; Liu, C.-M. The non-metabolizable glucose analog D-glucal inhibits aflatoxin biosynthesis and promotes kojic acid production in Aspergillus flavus. BMC Microbiol. 2014, 14, 95. [Google Scholar] [CrossRef] [PubMed]
- Yoshinari, T.; Akiyama, T.; Nakamura, K.; Kondo, T.; Takahashi, Y.; Muraoka, Y.; Nonomura, Y.; Nagasawa, H.; Sakuda, S. Dioctatin A is a strong inhibitor of aflatoxin production by Aspergillus parasiticus. Microbiology 2007, 153, 2774–2780. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Liang, Y.; Zhang, J.; Chen, Z.; Liu, C.M. Autoxidated linolenic acid inhibits aflatoxin biosynthesis in Aspergillus flavus via oxylipin species. Fungal Genet. Biol. 2014, 81, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-K.; Ehrlich, K.C.; Fujii, I. Cyclopiazonic acid biosynthesis of Aspergillus flavus and Aspergillus oryzae. Toxins 2009, 1, 74–99. [Google Scholar] [CrossRef] [PubMed]
- Etxebeste, O.; Garzia, A.; Espeso, E.A.; Ugalde, U. Aspergillus nidulans asexual development: Making the most of cellular modules. Trends Microbiol. 2010, 18, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Adams, T.H.; Boylan, M.T.; Timberlake, W.E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 1988, 54, 353–362. [Google Scholar] [CrossRef]
- Han, S.; Adams, T.H. Complex control of the developmental regulatory locus brlA in Aspergillus nidulans. Mol. Genet. Genom. 2001, 266, 260–270. [Google Scholar] [CrossRef]
- Adams, T.H.; Wieser, J.K.; Yu, J.H. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 1998, 62, 35–54. [Google Scholar] [PubMed]
- Sewall, T.C.; Mims, C.W.; Timberlake, W.E. abaA controls phialide differentiation in Aspergillus nidulans. Plant Cell 1990, 2, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Adrianopoulos, A.; Timberlake, W.E. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 1994, 14, 2503–2515. [Google Scholar] [CrossRef]
- Lv, C.; Wang, P.; Ma, L.; Zheng, M.; Liu, Y.; Xing, F. Large-scale comparative analysis of eugenol-induced/repressed genes expression in Aspergillus flavus using RNA-seq. Front. Microbiol. 2018, 9, 1116. [Google Scholar] [CrossRef] [PubMed]
- Wieser, J.; Lee, B.N.; Fondon, J.W.; Adams, T.H. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 1994, 27, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.N.; Adams, T.H. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 1994, 8, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.N.; Adams, T.H. FluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlAβ activation. EMBO J. 1996, 15, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.K.; Mack, B.M.; Wei, Q.; Bland, J.M.; Bhatnagar, D.; Cary, J.W. RNA sequencing of an nsdC mutant reveals global regulation of secondary metabolic gene clusters in Aspergillus flavus. Microbiol. Res. 2016, 182, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, V.; Dhingra, S.; Kincaid, A.; Shantappa, S.; Feng, X.; Calvo, A.M. The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS ONE 2013, 8, e74122. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Yu, J.H. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS ONE 2007, 2, e970. [Google Scholar] [CrossRef] [PubMed]
- Bayram, Ö.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Georgianna, D.R.; Payne, G.A. Genetic regulation of aflatoxin biosynthesis: From gene to genome. Fungal Genet. Biol. 2009, 46, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhi, Q.-Q.; Li, J.-Y.; Keller, N.; He, Z.-M. The antioxidant gallic acid inhibits aflatoxin formation in Aspergillus flavus by modulating transcription factors FarB and CreA. Toxins 2018, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Affeldt, K.; Carrig, J.; Amare, M.G.; Keller, N. Global survey of canonical Aspergillus flavus G Protein-Coupled Receptors. mBio 2014, 5, 1501–1514. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-H.; Keller, N. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 2005, 43, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Tsitsigiannis, D.I.; Keller, N.P. Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans. Mol. Microbiol. 2006, 59, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Hicks, J.K.; Huang, T.P.; Keller, N.P. Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans. Genetics 2003, 165, 1095–1104. [Google Scholar] [PubMed]
- Roze, L.V.; Chanda, A.; Wee, J.; Awad, D.; Linz, J.E. Stress-related transcription factor AtfB integrates secondary metabolism with oxidative stress response in Aspergilli. J. Biol. Chem. 2011, 286, 35137–35148. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Arima, T.H.; Iwashita, K.; Yamada, O.; Gomi, K.; Akita, O. Aspergillus oryzae atfB encodes a transcription factor required for stress tolerance in conidia. Fungal Genet. Biol. 2008, 45, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Navarro, R.E.; Stringer, M.A.; Hansberg, W.; Timberlake, W.E.; Aguirre, J. catA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr. Genet. 1996, 29, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Verheecke, C.; Liboz, T.; Anson, P.; Diaz, R.; Mathieu, F. Reduction of aflatoxin production by Aspergillus flavus and Aspergillus parasiticus in interaction with Streptomyces. Microbiology 2015, 161, 967–972. [Google Scholar] [CrossRef] [PubMed]
- EL Mahgubi, A.; Bailly, S.; Tadrist, S.; Querin, A.; Ouadia, A.; Oswald, I.P.; Bailly, J.-D. Distribution and toxigenicity of Aspergillus section Flavi in spices marketed in Morocco. Food Control 2013, 32, 143–148. [Google Scholar] [CrossRef]
- Sultan, Y.; Magan, N. Impact of a Streptomyces (AS1) strain and its metabolites on control of Aspergillus flavus and aflatoxin B1 contamination in vitro and in stored peanuts. Biocontrol Sci. Technol. 2011, 21, 1437–1455. [Google Scholar] [CrossRef]
- Leite, G.M.; Magan, N.; Medina, Á. Comparison of different bead-beating RNA extraction strategies: An optimized method for filamentous fungi. J. Microbiol. Methods 2012, 88, 413–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caceres, I.; El Khoury, R.; Bailly, S.; Oswald, I.P.; Puel, O.; Bailly, J.D. Piperine inhibits aflatoxin B1 production in Aspergillus flavus by modulating fungal oxidative stress response. Fungal Genet. Biol. 2017, 107, 77–85. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caceres, I.; Snini, S.P.; Puel, O.; Mathieu, F. Streptomyces roseolus, A Promising Biocontrol Agent Against Aspergillus flavus, the Main Aflatoxin B1 Producer. Toxins 2018, 10, 442. https://doi.org/10.3390/toxins10110442
Caceres I, Snini SP, Puel O, Mathieu F. Streptomyces roseolus, A Promising Biocontrol Agent Against Aspergillus flavus, the Main Aflatoxin B1 Producer. Toxins. 2018; 10(11):442. https://doi.org/10.3390/toxins10110442
Chicago/Turabian StyleCaceres, Isaura, Selma P. Snini, Olivier Puel, and Florence Mathieu. 2018. "Streptomyces roseolus, A Promising Biocontrol Agent Against Aspergillus flavus, the Main Aflatoxin B1 Producer" Toxins 10, no. 11: 442. https://doi.org/10.3390/toxins10110442
APA StyleCaceres, I., Snini, S. P., Puel, O., & Mathieu, F. (2018). Streptomyces roseolus, A Promising Biocontrol Agent Against Aspergillus flavus, the Main Aflatoxin B1 Producer. Toxins, 10(11), 442. https://doi.org/10.3390/toxins10110442