Synergistic Phytotoxic Effects of Culmorin and Trichothecene Mycotoxins
Abstract
1. Introduction
2. Results
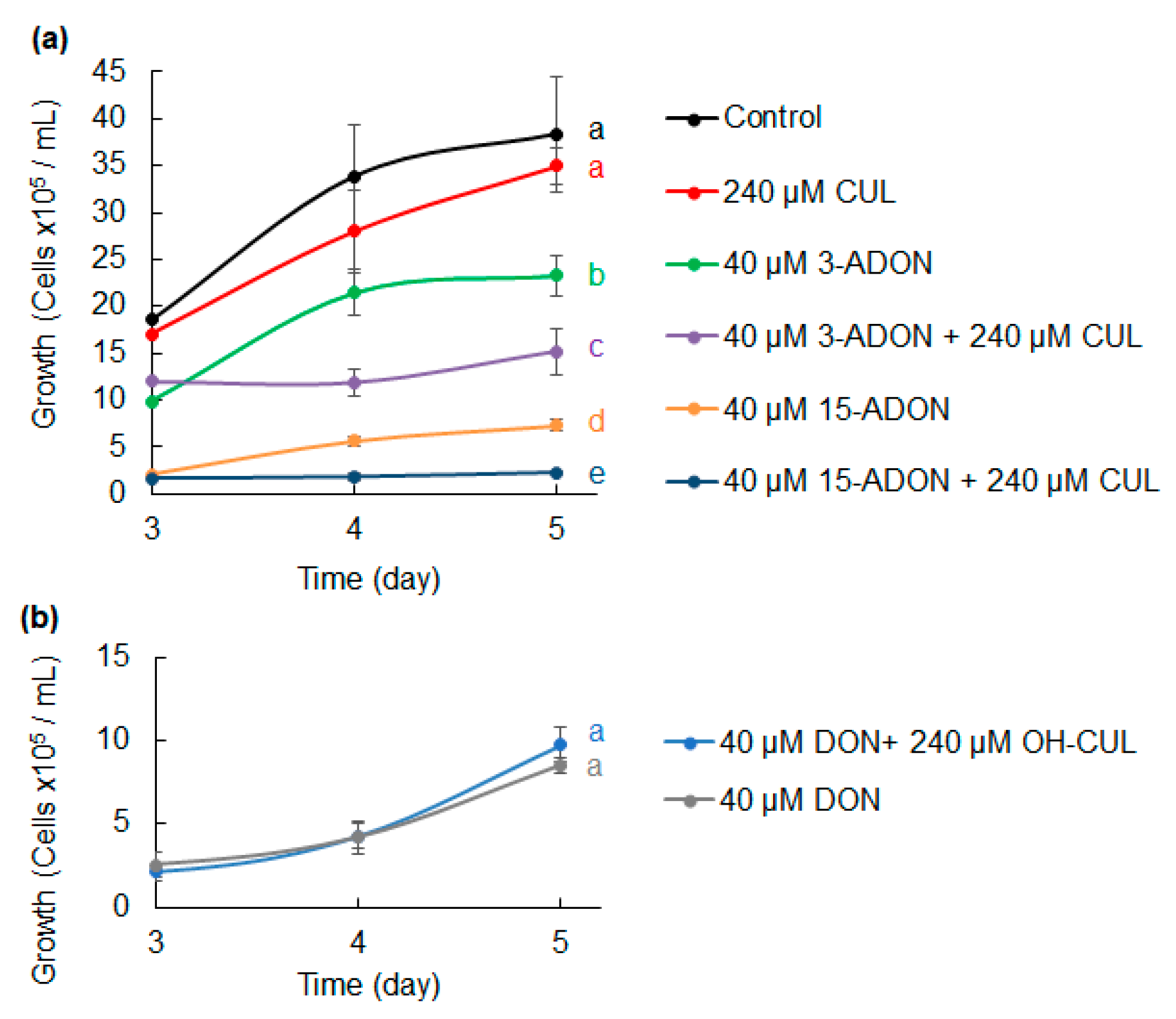
2.1. Effects of CUL and Trichothecenes on C. reinhardtii Growth
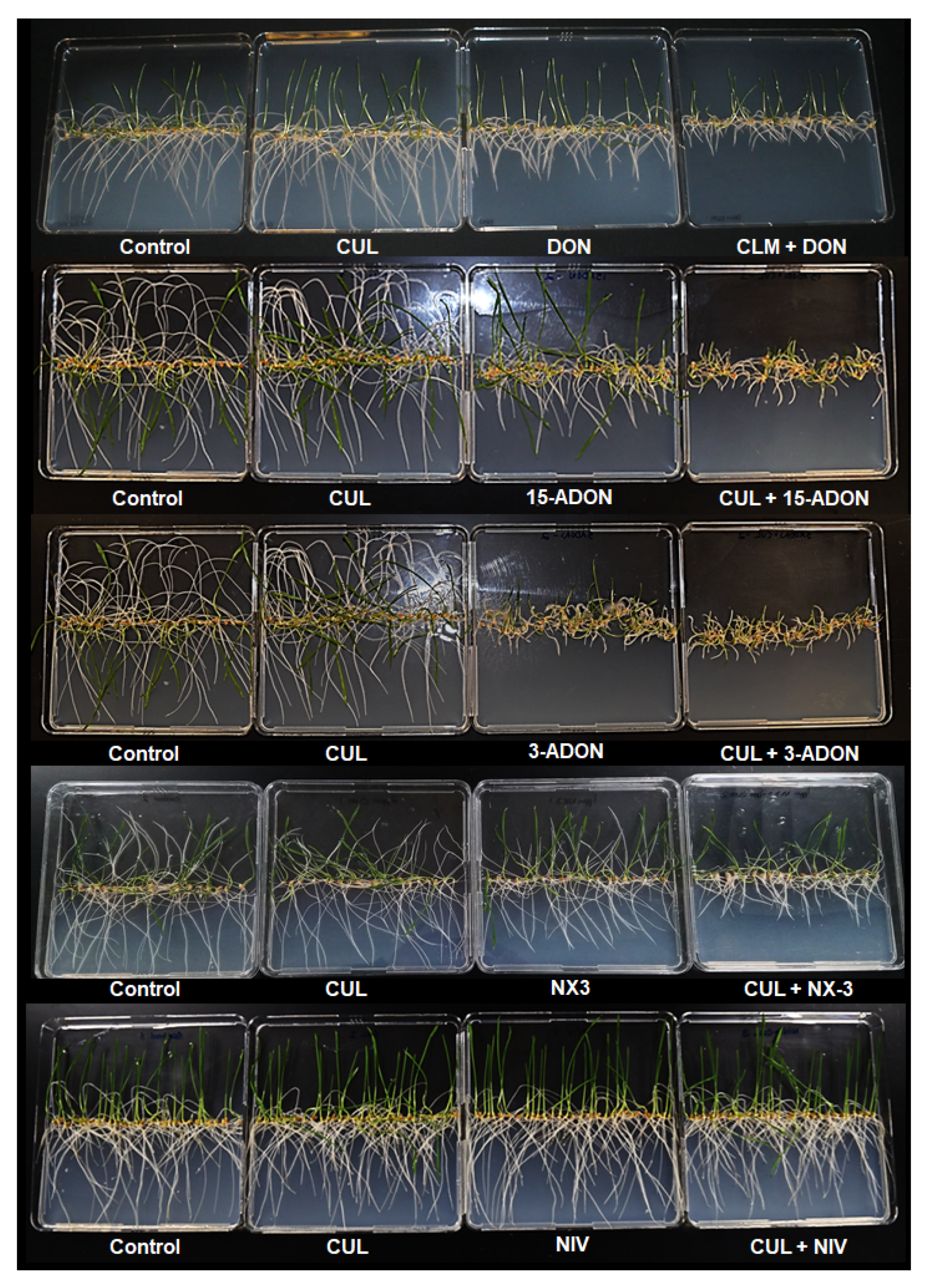
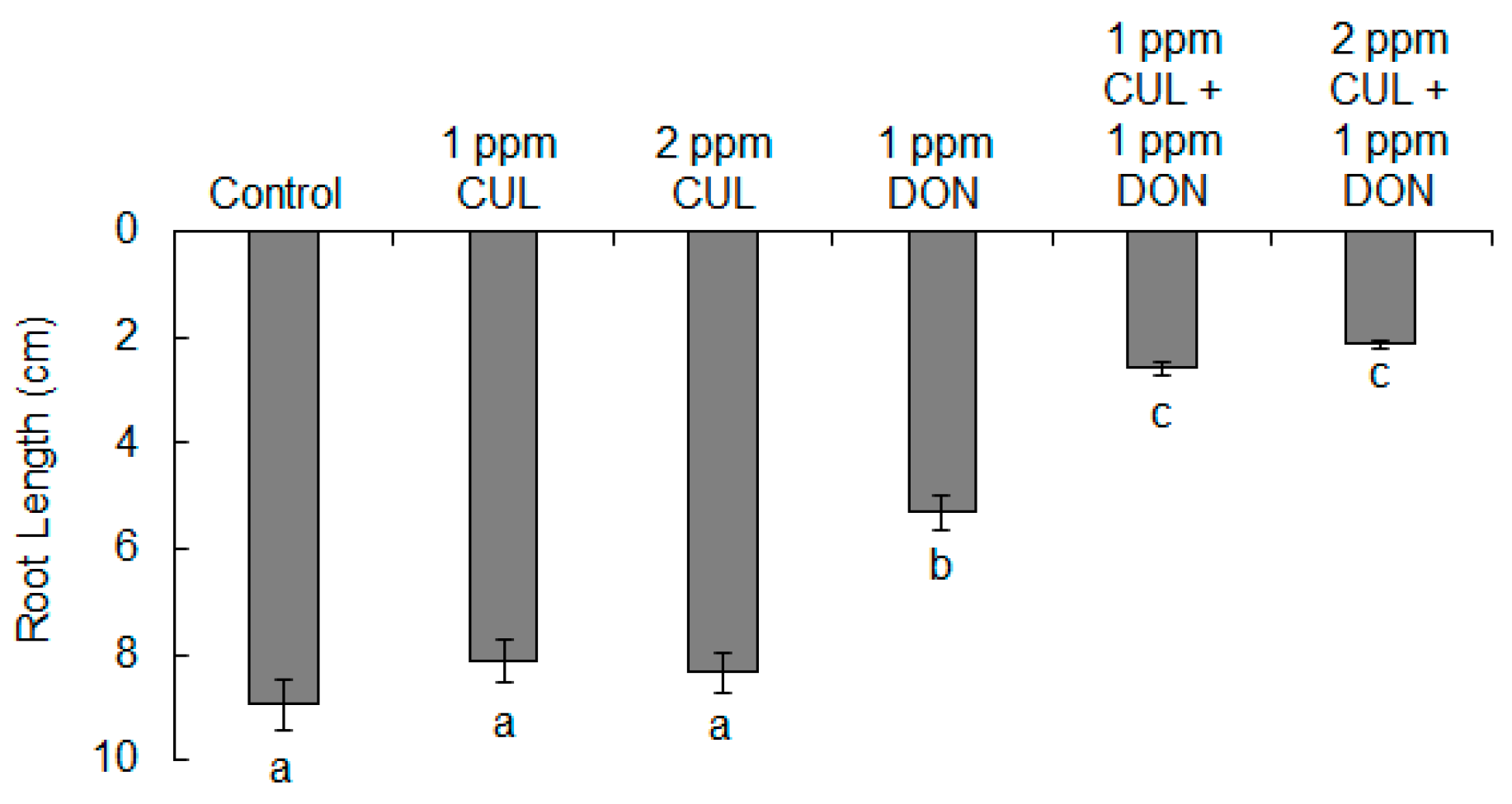
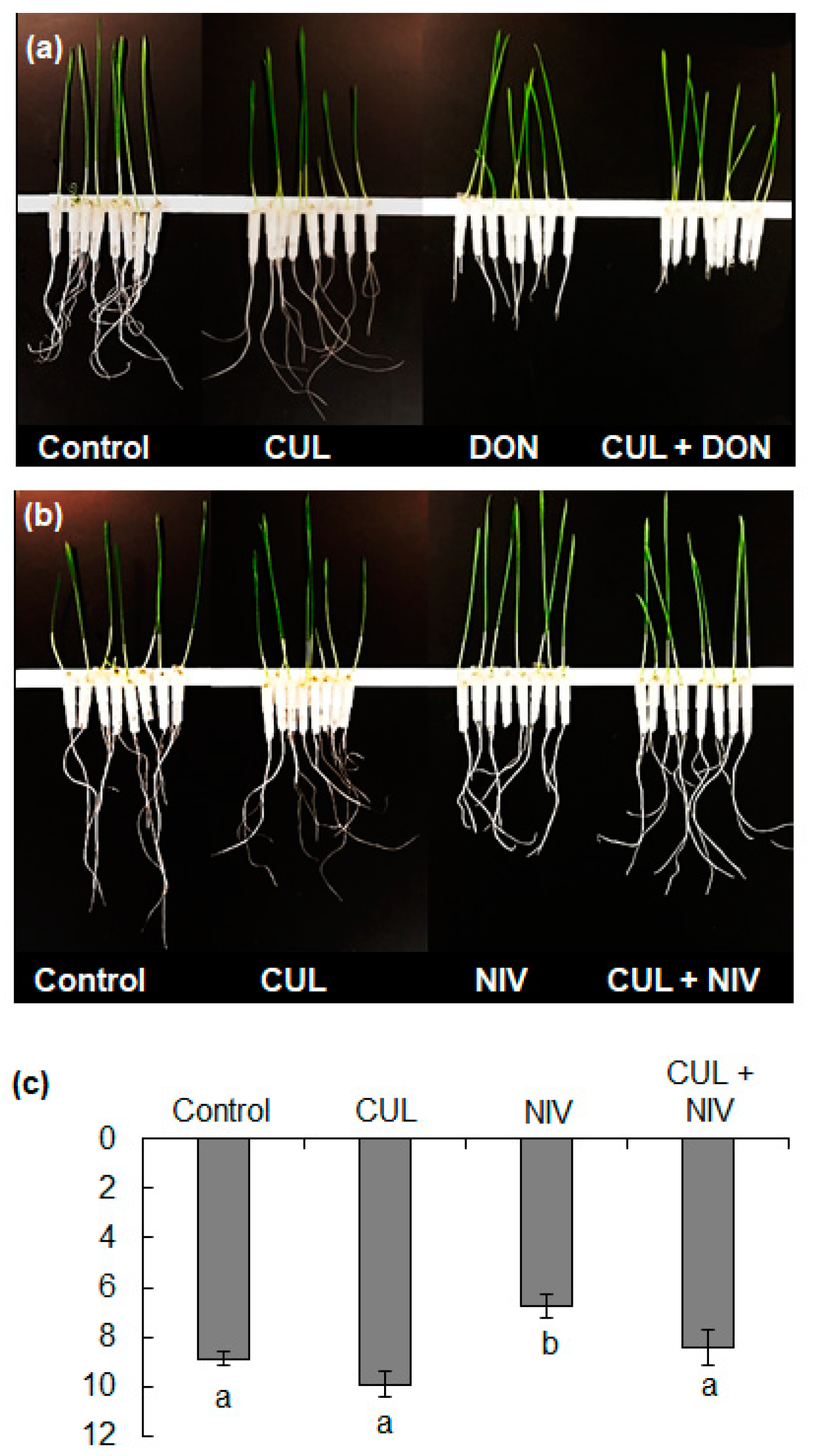
2.2. Effects of CUL and Trichothecenes on the Growth of Wheat Roots
2.3. Synergistic Phytotoxicity of CUL and DON on Multiple Varieties of Wheat, Barley and Corn
2.4. Relationship between FHB and the Ratio of CUL/DON
2.5. Transcriptional Analysis of CUL and Trichothecene Biosynthetic Genes
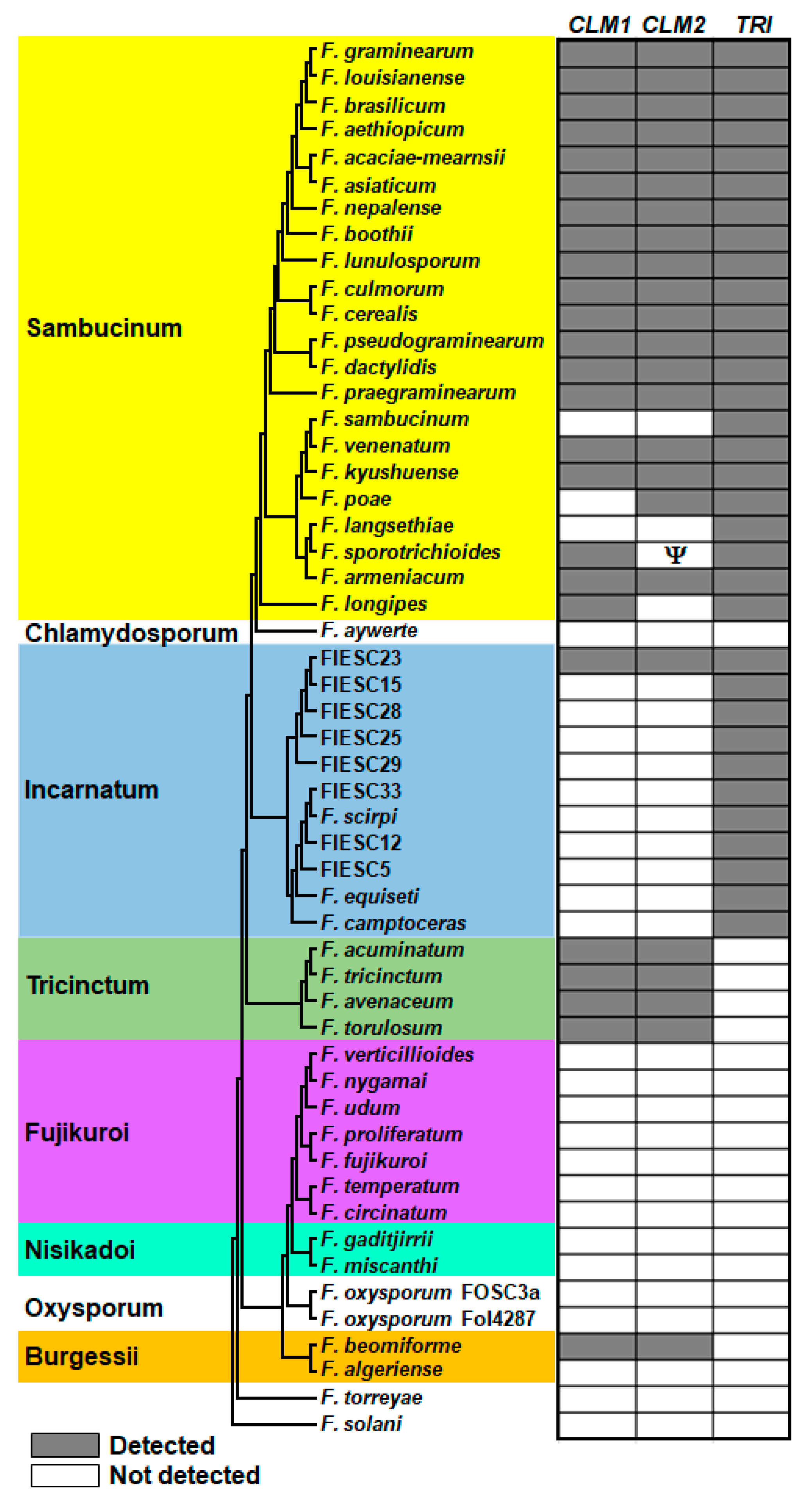
2.6. Distribution of CUL and Trichothecene Biosynthetic Genes in Selected Species of Fusarium
3. Discussion
4. Materials and Methods
4.1. Mycotoxins
4.2. Phytotoxicity Assays Using Chlamydomonas reinhardtii
4.3. Root Phytotoxicity Assays
4.4. Wheat Fusarium Head Blight Assays
4.5. Mycotoxin Extraction and Quantification
4.6. Transcriptional Analysis of CUL and DON Biosynthetic Genes
4.7. Statistical Analyses
4.8. Genomic Analysis of the Distribution of CUL and Trichothecene Biosynthetic Genes
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McCormick, S.P.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From simple to complex. mycotoxins. Toxins 2011, 3, 802. [Google Scholar] [PubMed]
- Khaneghah, A.M.; Kamani, M.H.; Fakhri, Y.; Coppa, C.F.S.C.; de Oliveira, C.A.F.; Sant’Ana, A.S. Changes in masked forms of deoxynivalenol and their co-occurrence with culmorin in cereal-based products: A systematic review and meta-analysis. Food Chem. 2019, 294, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Kovalsky, P.; Kos, G.; Nährer, K.; Schwab, C.; Jenkins, T.; Schatzmayr, G.; Krska, R.; Sulyok, M. Co-occurrence of regulated, masked and emerging mycotoxins and secondary metabolites in finished feed and maize—An extensive survey. Toxins 2016, 8, 363. [Google Scholar] [CrossRef] [PubMed]
- Varga, E.; Wiesenberger, G.; Hametner, C.; Ward, T.J.; Dong, Y.; Schöfbeck, D.; Krska, R.; McCormick, S.; Broz, K.; Adam, G.; et al. New tricks of an old enemy: Isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ. Microbiol. 2015, 17, 2588–2600. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Z.; Miller, J.D. Effects of Fusarium graminearum metabolites on wheat tissue in relation to Fusarium head blight resistance. J. Phytopathol. 1988, 122, 118–125. [Google Scholar] [CrossRef]
- Bruins, M.B.M.; Karsai, I.; Schepers, J.; Snijders, C.H.A. Phytotoxicity of deoxynivalenol to wheat tissue with regard to in vitro selection for Fusarium head blight resistance. Plant Sci. 1993, 94, 195–206. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Ziegenhorn, S.L. Phytotoxicity of selected trichothecenes using Chlamydomonas reinhardtii as a model system. Nat. Toxins 1999, 7, 265–269. [Google Scholar] [CrossRef]
- Pestka, J.J. Deoxynivalenol: Toxicity, mechanisms and animal health risks. Anim. Feed Sci. Technol. 2007, 137, 283–298. [Google Scholar] [CrossRef]
- Bai, G.-H.; Desjardins, A.E.; Plattner, R.D. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 2002, 153, 91–98. [Google Scholar] [CrossRef]
- Mesterházy, Á. Role of deoxynivalenol in aggressiveness of Fusarium graminearum and F. culmorum and in resistance to Fusarium head blight. In Mycotoxins in Plant Disease: Under the Aegis of COST Action 835 ‘Agriculturally Important Toxigenic Fungi 1998–2003′, EU project (QLK 1-CT-1998-01380), and ISPP ‘Fusarium Committee’; Logrieco, A., Corazza, L., Bailey, J.A., Cooke, B.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 675–684. [Google Scholar]
- Maier, F.J.; Miedaner, T.; Hadeler, B.; Felk, A.; Salomon, S.; Lemmens, M.; Schaefer, W.; Kassner, H. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 2006, 7, 449–461. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M.; Manners, J.M. On the trail of a cereal killer: Recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 2012, 13, 399–413. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.P.; Alexander, N.J.; Harris, L.J. CLM1 of Fusarium graminearum encodes a longiborneol synthase required for culmorin production. Appl. Environ. Microbiol. 2010, 76, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Bahadoor, A.; Schneiderman, D.; Gemmill, L.; Bosnich, W.; Blackwell, B.; Melanson, J.E.; Harris, L.J.; McRae, G. Hydroxylation of longiborneol by a Clm2-encoded CYP450 monooxygenase to produce culmorin in Fusarium graminearum. J. Nat. Prod. 2016, 79, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Uhlig, S.; Eriksen, G.; Hofgaard, I.; Krska, R.; Beltrán, E.; Sulyok, M. Faces of a changing climate: Semi-quantitative multi-mycotoxin analysis of grain grown in exceptional climatic conditions in Norway. Toxins 2013, 5, 1682–1697. [Google Scholar] [CrossRef]
- Vaclavikova, M.; Malachova, A.; Veprikova, Z.; Dzuman, Z.; Zachariasova, M.; Hajslova, J. ‘Emerging’ mycotoxins in cereals processing chains: Changes of enniatins during beer and bread making. Food Chem. 2013, 136, 750–757. [Google Scholar] [CrossRef]
- Dowd, P.F.; Miller, J.D.; Greenhalgh, R. Toxicity and interactions of some Fusarium graminearum metabolites to caterpillars. Mycologia 1989, 81, 646–650. [Google Scholar] [CrossRef]
- Rotter, R.G.; Trenholm, H.L.; Prelusky, D.B.; Hartin, K.E.; Thompson, B.K.; Miller, J.D. A preliminary examination of potential interactions between deoxynivalenol (DON) and other selected Fusarium metabolites in growing pigs. Can. J. Anim. Sci. 1992, 72, 107–116. [Google Scholar] [CrossRef]
- Woelflingseder, L.; Warth, B.; Vierheilig, I.; Schwartz-Zimmermann, H.; Hametner, C.; Nagl, V.; Marko, D.; Novak, B.; Berthiller, F.; Adam, G.; et al. The Fusarium metabolite culmorin suppresses the in vitro glucuronidation of deoxynivalenol. Arch. Toxicol. 2019, 93, 1729–1743. [Google Scholar] [CrossRef]
- Bryła, M.; Waśkiewicz, A.; Ksieniewicz-Woźniak, E.; Szymczyk, K.; Jędrzejczak, R. Modified Fusarium mycotoxins in cereals and their products-metabolism, occurrence, and toxicity: An. Updated Review. Molecules 2018, 23, 963. [Google Scholar] [CrossRef]
- Ovando-Martínez, M.; Ozsisli, B.; Anderson, J.; Whitney, K.; Ohm, J.B.; Simsek, S. Analysis of deoxynivalenol and deoxynivalenol-3-glucoside in hard red spring wheat inoculated with Fusarium graminearum. Toxins 2013, 5, 2522–2532. [Google Scholar] [CrossRef]
- Xing, L.P.; He, L.Q.; Xiao, J.; Chen, Q.G.; Li, M.H.; Shang, Y.; Zhu, Y.-F.; Chen, P.-D.; Cao, A.-Z.; Wang, X.E.; et al. An udp-glucosyltransferase gene from barley confers disease resistance to Fusarium head blight. Plant Mol. Biol. Report. 2017, 35, 224–236. [Google Scholar] [CrossRef]
- Lemmens, M.; Scholz, U.; Berthiller, F.; Dall’Asta, C.; Koutnik, A.; Schuhmacher, R.; Ruckenbauer, P.; Adam, G.; Buerstmayr, H.; Krska, R.; et al. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol. Plant Microbe Interact. 2005, 18, 1318–1324. [Google Scholar] [CrossRef] [PubMed]
- Arruda, M.P.; Lipka, A.E.; Brown, P.J.; Krill, A.M.; Thurber, C.; Brown-Guedira, G.; Kolb, F.L.; Dong, Y.; Foresman, B.J. Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.). Mol. Breed. 2016, 36, 84. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Gatti, M.; Choulet, F.; Macadré, C.; Guérard, F.; Seng, J.M.; Langin, T.; Dufresne, M. Identification, molecular cloning, and functional characterization of a wheat UDP-glucosyltransferase involved in resistance to Fusarium head blight and to mycotoxin accumulation. Front. Plant Sci. 2018, 9, 1853. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ma, X.; Su, P.; Ge, W.; Wu, H.; Guo, X.; Kong, L.; Li, A.; Wang, H. Cloning and characterization of a specific UDP-glycosyltransferase gene induced by DON and Fusarium graminearum. Plant Cell Rep. 2018, 37, 641–652. [Google Scholar] [CrossRef]
- Michlmayr, H.; Varga, E.; Malachová, A.; Fruhmann, P.; Piątkowska, M.; Hametner, C.; Berthiller, F.; Jaunecker, G.; Lemmens, M.; Adam, G.; et al. UDP-Glucosyltransferases from rice, Brachypodium, and barley: Substrate specificities and synthesis of Type A and B trichothecene-3-O-β-d-glucosides. Toxins 2018, 10, 111. [Google Scholar] [CrossRef]
- Wetterhorn, K.M.; Newmister, S.A.; Caniza, R.K.; Busman, M.; McCormick, S.P.; Berthiller, F.; Rayment, I.; Adam, G. Crystal structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase involved in the detoxification of deoxynivalenol. Biochemistry 2016, 55, 6175–6186. [Google Scholar] [CrossRef]
- Li, X.; Shin, S.; Heinen, S.; Dill-Macky, R.; Berthiller, F.; Nersesian, N.; Muehlbauer, G.J.; Clemente, N.; McCormick, S. Transgenic wheat expressing a barley UDP-glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum. Mol. Plant Microbe Interact. 2015, 28, 1237–1246. [Google Scholar] [CrossRef]
- Gardiner, D.M.; Kazan, K.; Manners, J.M. Novel genes of Fusarium graminearum that negatively regulate deoxynivalenol production and virulence. Mol. Plant Microbe Interact. 2009, 22, 1588–1600. [Google Scholar] [CrossRef]
- Ikram, N.K.B.K.; Zhan, X.; Pan, X.W.; King, B.C.; Simonsen, H.T. Stable heterologous expression of biologically active terpenoids in green plant cells. Front. Plant Sci. 2015, 6, 129. [Google Scholar] [CrossRef] [PubMed]
- Masuda, D.; Ishida, M.; Yamaguchi, K.; Yamaguchi, I.; Kimura, M.; Nishiuchi, T. Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, C.C.; Fernando, W.G.D. Comparative analysis of deoxynivalenol biosynthesis related gene expression among different chemotypes of Fusarium graminearum in spring wheat. Front. Microbiol. 2016, 7, 1229. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Rooney, A.P.; Proctor, R.H.; Brown, D.W.; McCormick, S.P.; Ward, T.J.; Robert, V.A.; Frandsen, R.J.N.; Rehner, S.A.; Aoki, T.; et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 2013, 52, 20–31. [Google Scholar] [CrossRef]
- Villani, A.; Moretti, A.; De Saeger, S.; Han, Z.; Di Mavungu, J.D.; Soares, C.M.; Paciolla, C.; Proctor, R.H.; Lima, N.; Stea, G.; et al. A polyphasic approach for characterization of a collection of cereal isolates of the Fusarium incarnatum-equiseti species complex. Int. J. Food Microbiol. 2016, 234, 24–35. [Google Scholar] [CrossRef]
- Villani, A.; Proctor, R.H.; Kim, H.S.; Brown, D.W.; Logrieco, A.F.; Amatulli, M.T.; Susca, A.; Moretti, A. Variation in secondary metabolite production potential in the Fusarium incarnatum-equiseti species complex revealed by comparative analysis of 13 genomes. BMC Genom. 2019, 20, 314. [Google Scholar] [CrossRef]
- Wetterhorn, K.M.; Gabardi, K.; Michlmayr, H.; Malachova, A.; Busman, M.; McCormick, S.P.; Rayment, I.; Berthiller, F.; Adam, G. Determinants and expansion of specificity in a trichothecene UDP-glucosyltransferase from Oryza sativa. Biochemistry 2017, 56, 6585–6596. [Google Scholar] [CrossRef]
- Bryła, M.; Ksieniewicz-Woźniak, E.; Yoshinari, T.; Waśkiewicz, A.; Szymczyk, K. Contamination of wheat cultivated in various regions of Poland during 2017 and 2018 agricultural seasons with selected trichothecenes and their modified forms. Toxins 2019, 11, 88. [Google Scholar] [CrossRef]
- Li, X.; Michlmayr, H.; Schweiger, W.; Malachova, A.; Shin, S.; Huang, Y.; Fruhmann, P.; Dong, Y.; Lemmens, M.; Hametner, C.; et al. A barley UDP-glucosyltransferase inactivates nivalenol and provides Fusarium head blight resistance in transgenic wheat. J. Exp. Bot. 2017, 68, 2187–2197. [Google Scholar] [CrossRef]
- Ghebremeskel, M.; Langseth, W. The occurrence of culmorin and hydroxy-culmorins in cereals. Mycopathologia 2001, 152, 103–108. [Google Scholar] [CrossRef]
- Aoki, T.; Vaughan, M.M.; McCormick, S.P.; Busman, M.; Ward, T.J.; Kelly, A.; Geiser, D.M.; O’Donnell, K.; Johnston, P.R. Fusarium dactylidis sp. nov., a novel nivalenol toxin-producing species sister to F. pseudograminearum isolated from orchard grass (Dactylis glomerata) in Oregon and New Zealand. Mycologia 2015, 107, 409–418. [Google Scholar] [CrossRef]
- Cerón-Bustamante, M.; Ward, T.J.; Kelly, A.; Vaughan, M.M.; McCormick, S.P.; Cowger, C.; Nava-Díaz, C.; Leyva-Mir, S.G.; Villaseñor-Mir, H.E.; Ayala-Escobar, V.; et al. Regional differences in the composition of Fusarium head blight pathogens and mycotoxins associated with wheat in Mexico. Int. J. Food Microbiol. 2018, 273, 11–19. [Google Scholar] [CrossRef]
- McCormick, S. Personal Communication; National Center for Agricultural Utilization Research, USDA, ARS: Peoria, IL, USA, 2019.
- Chen, L.; McCormick, S.P.; Hohn, T.M. Altered regulation of 15-acetyldeoxynivalenol production in Fusarium graminearum. Appl. Environ. Microbiol. 2000, 66, 2062–2065. [Google Scholar] [CrossRef]
- McCormick, S.P.; Alexander, N.J.; Trapp, S.E.; Hohn, T.M. Disruption of TRI101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides. Appl. Environ. Microbiol. 1999, 65, 5252–5256. [Google Scholar]
- Kelly, A.C.; Ward, T.J. Population genomics of Fusarium graminearum reveals signatures of divergent evolution within a major cereal pathogen. PLoS ONE 2018, 13, e0194616. [Google Scholar] [CrossRef]
- Hao, G.; McCormick, S.; Vaughan, M.M.; Naumann, T.A.; Kim, H.S.; Proctor, R.; Ward, T.J.; Kelly, A. Fusarium graminearum arabinanase (Arb93B) enhances wheat head blight susceptibility by suppressing plant immunity. Mol. Plant Microbe Interact. 2019, 32, 888–898. [Google Scholar] [CrossRef]
- Geiser, D.M.; Aoki, T.; Bacon, C.W.; Baker, S.E.; Bhattacharyya, M.K.; Brandt, M.E.; Correll, J.C.; Brown, D.W.; Burgess, L.W.; Chulze, S.; et al. One fungus, one name: Defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 2013, 103, 400–408. [Google Scholar] [CrossRef]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- Ma, L.J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.J.; Di Pietro, A.; Houterman, P.M.; Dufresne, M.; Freitag, M.; Baker, S.E.; et al. Comparative genomics reveal mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Brown, D.W.; Proctor, R.H. Insights into natural products biosynthesis from analysis of 490 polyketide synthases from Fusarium. Fungal Genet. Biol. 2016, 89, 37–51. [Google Scholar] [CrossRef]
- Starkey, D.E.; Ward, T.J.; Aoki, T.; Gale, L.R.; Kistler, H.C.; Geiser, D.M.; O’Donnell, K.; Suga, H.; Tóth, B.; Varga, J. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 2007, 44, 1191–1204. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Kobert, K.; Salichos, L.; Rokas, A.; Stamatakis, A. Computing the internode certainty and related measures from partial gene trees. Mol. Biol. Evol. 2016, 33, 1606–1617. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wipfler, R.; McCormick, S.P.; Proctor, R.; Teresi, J.; Hao, G.; Ward, T.; Alexander, N.; Vaughan, M.M. Synergistic Phytotoxic Effects of Culmorin and Trichothecene Mycotoxins. Toxins 2019, 11, 555. https://doi.org/10.3390/toxins11100555
Wipfler R, McCormick SP, Proctor R, Teresi J, Hao G, Ward T, Alexander N, Vaughan MM. Synergistic Phytotoxic Effects of Culmorin and Trichothecene Mycotoxins. Toxins. 2019; 11(10):555. https://doi.org/10.3390/toxins11100555
Chicago/Turabian StyleWipfler, Rebecca, Susan P. McCormick, Robert Proctor, Jennifer Teresi, Guixia Hao, Todd Ward, Nancy Alexander, and Martha M. Vaughan. 2019. "Synergistic Phytotoxic Effects of Culmorin and Trichothecene Mycotoxins" Toxins 11, no. 10: 555. https://doi.org/10.3390/toxins11100555
APA StyleWipfler, R., McCormick, S. P., Proctor, R., Teresi, J., Hao, G., Ward, T., Alexander, N., & Vaughan, M. M. (2019). Synergistic Phytotoxic Effects of Culmorin and Trichothecene Mycotoxins. Toxins, 11(10), 555. https://doi.org/10.3390/toxins11100555




