Transcriptomic Insights into Benzenamine Effects on the Development, Aflatoxin Biosynthesis, and Virulence of Aspergillus flavus
Abstract
:1. Introduction
2. Results and Discussion
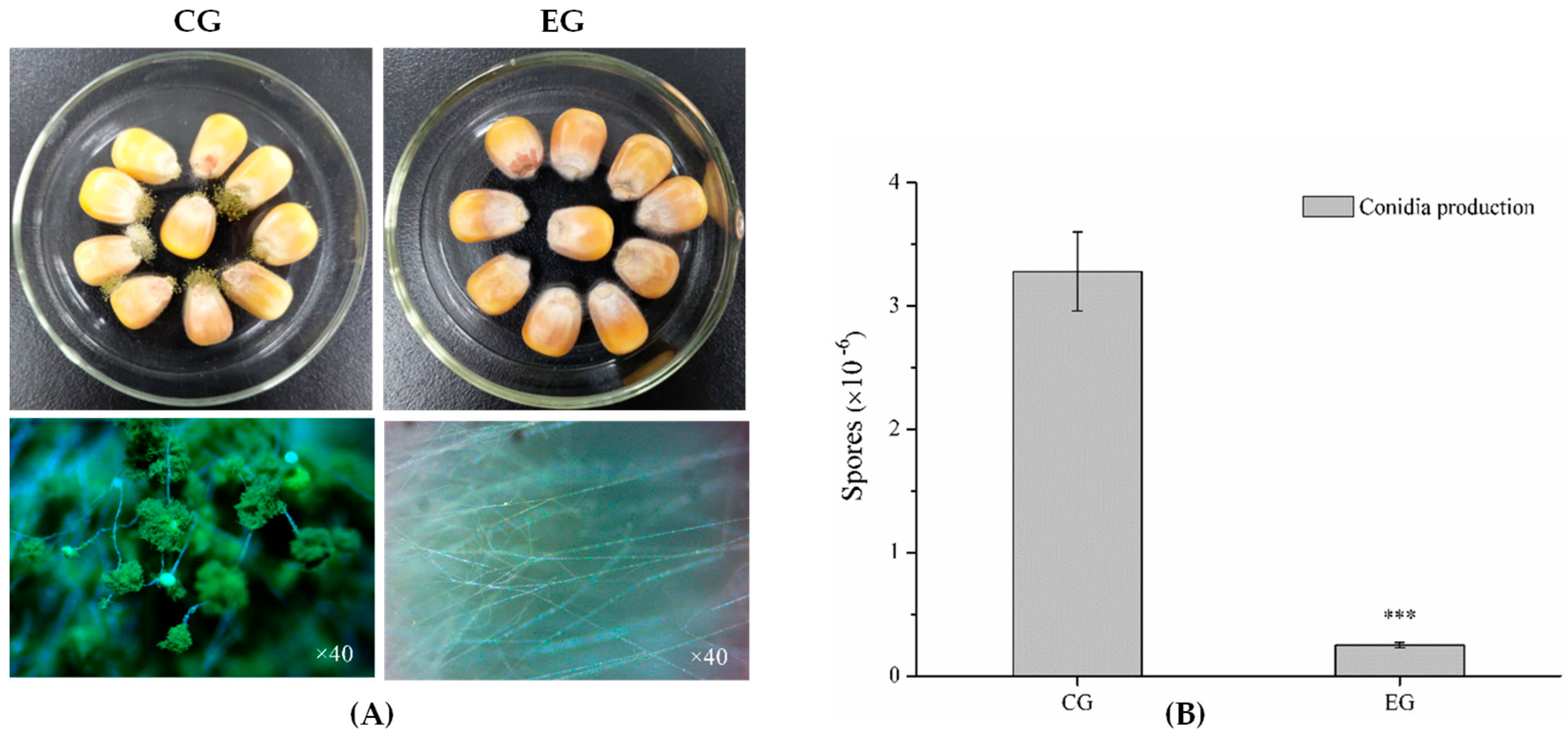
2.1. Antagonistic Activity of Benzenamine against A. flavus
2.2. Transcriptome Overview
2.3. Annotation and Analysis of All Unigenes
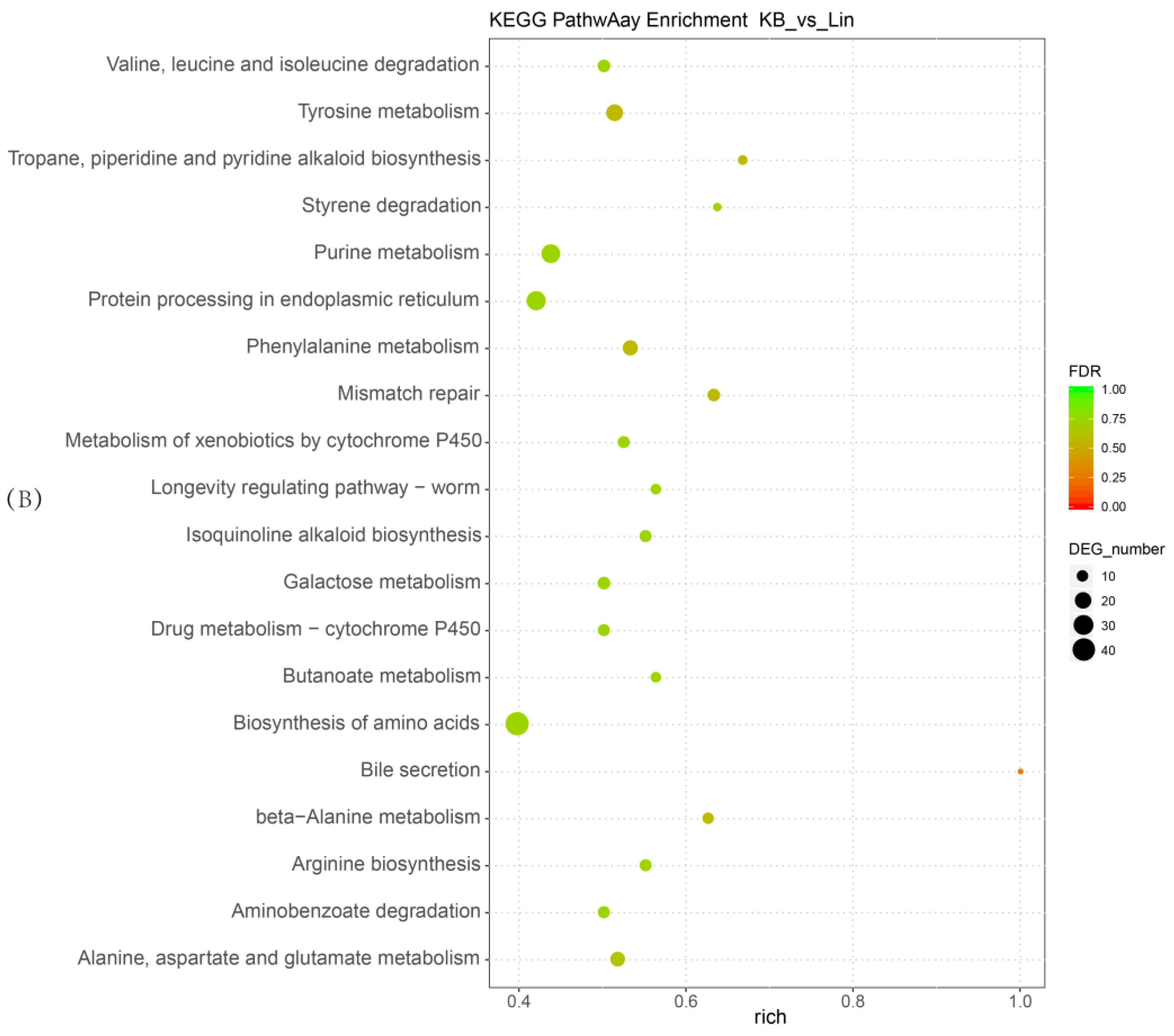
2.4. Functional and Pathway Enrichment Analysis of Differentially Expressed Genes
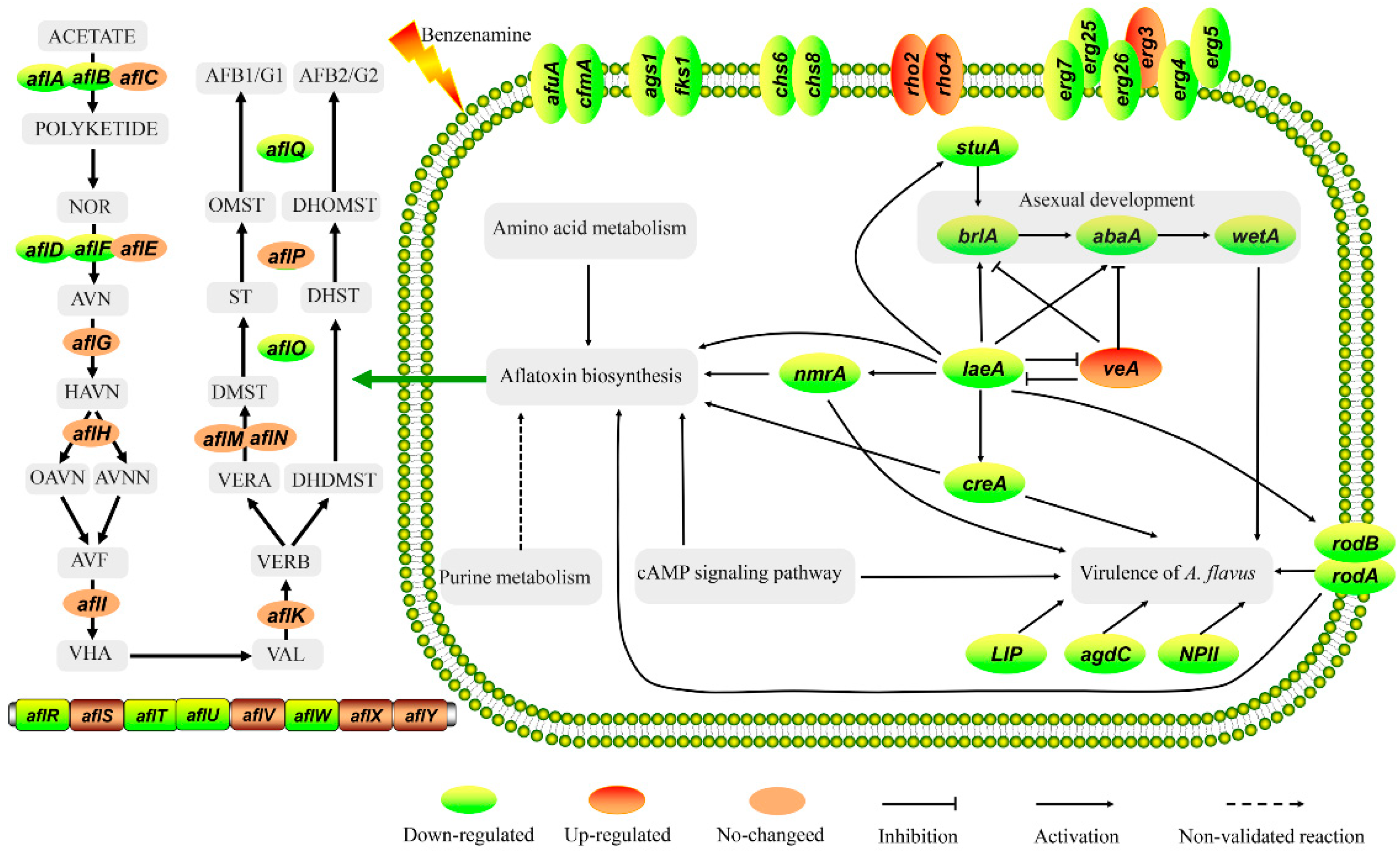
2.5. Analysis of DEGs Involved in Development
2.5.1. DEGs Involved in the Cell Wall
2.5.2. DEGs Involved in the Cell Membrane
2.5.3. DEGs Involved in Conidia
2.5.4. DEGs Involved in Transcription Factors
2.6. Analysis of DEGs Involved in Aflatoxin Biosynthesis
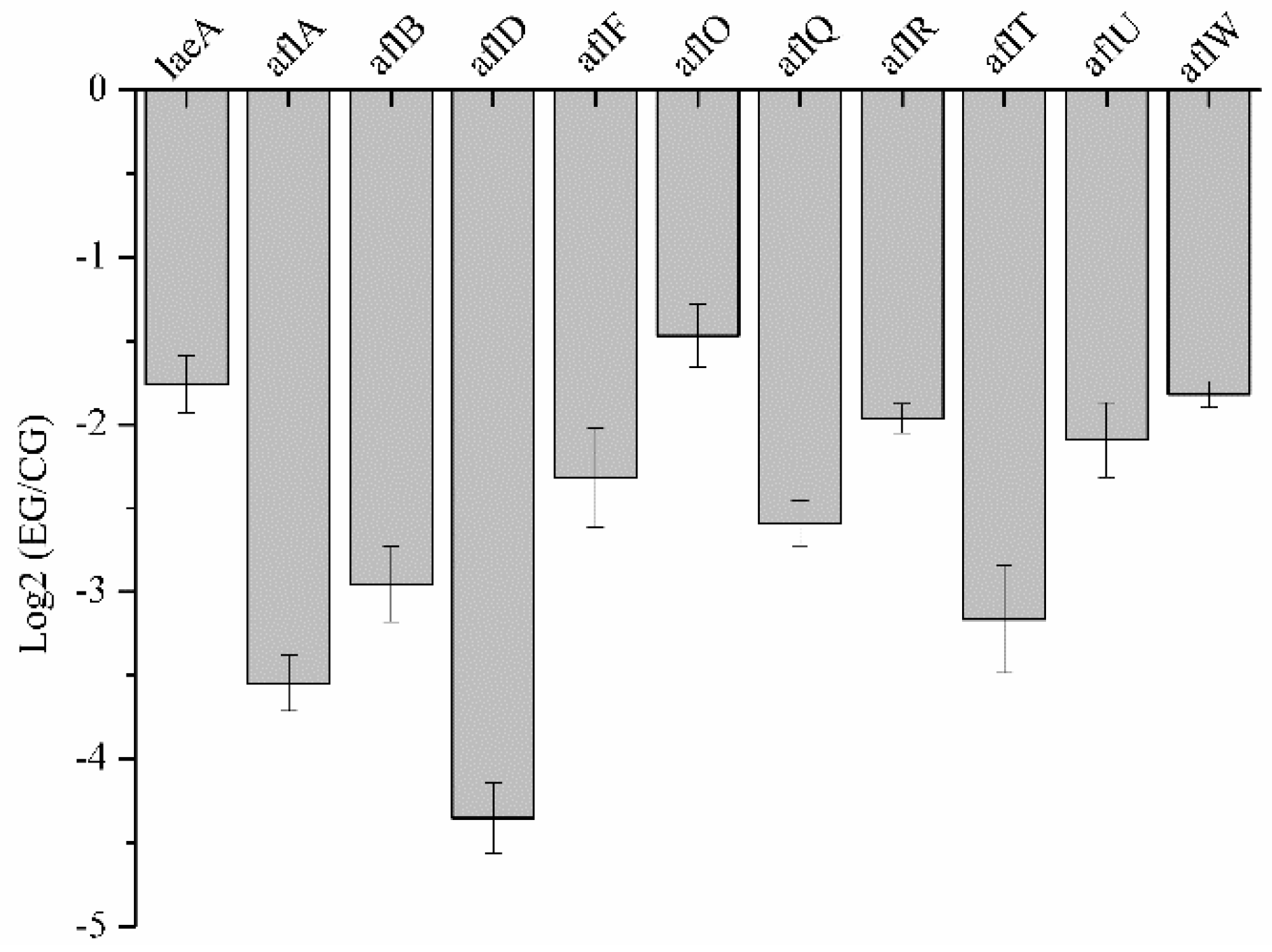
2.6.1. DEGs Involved in Aflatoxin Biosynthesis
2.6.2. DEGs Involved in Carbon/Nitrogen Metabolism
2.6.3. Other Related DEGs
2.7. Analysis of DEGs Involved in the Virulence of A. flavus
2.7.1. DEGs Involved in Hydrolases
2.7.2. DEGs Involved in the Development and Metabolism of A. flavus
2.8. Validation of RNA-Seq Data by qRT-PCR
3. Conclusions
4. Materials and Methods
4.1. Microorganisms
4.2. Antifungal Assays
4.2.1. Inhibition of Hyphal Growth of A. flavus
4.2.2. Inhibition of Conidial Germination of A. flavus
4.3. Determination of Aflatoxin B1
4.4. Virulence of A. flavus in Maize
4.5. Preparation of cDNA Libraries and Illumina Sequencing
4.6. De Novo Transcriptome Assembly and Annotation
4.7. Identification and Analysis of DEGs
4.8. qRT-PCR Analysis
4.9. Availability of Supporting Data
4.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Peng, Z.; Chen, L.; Zhu, Y.; Huang, Y.; Hu, X.; Wu, Q.; Nussler, A.K.; Liu, L.; Yang, W. Current major degradation methods for aflatoxins: A review. Trends Food Sci. Technol. 2018. [Google Scholar] [CrossRef]
- Sangare, L.; Zhao, Y.; Folly, Y.M.E.; Chang, J.; Liu, J.; Xing, F.; Zhou, L.; Wang, Y.; Liu, Y. Aflatoxin B1 degradation by a pseudomonas strain. Toxins 2015, 7, 3538–3539. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Persp. 2010, 118, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Fountain, J. Resistance to Aspergillus flavus in maize and peanut: Molecular biology, breeding, environmental stress and future perspectives. Crop. J. 2015, 3, 229–237. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. FAO Cereal Supply and Demand Brief. 2018. Available online: http://www.fao.org/worldfoodsituation/csdb/en/ (accessed on 6 December 2018).
- Food and Agriculture Organization. The State of Food Security and Nutrition in the World 2018; FAO: Rome, Italy, 2018. [Google Scholar]
- Cleveland, T.E.; Carter-Wientjes, C.H.; De Lucca, A.J.; Boue, S.M. Effect of soybean volatile compounds on Aspergillus flavus growth and aflatoxin production. J. Food Sci. 2009, 74, 83–87. [Google Scholar] [CrossRef] [PubMed]
- De Lucca, A.J.; Carter-Wientjes, C.H.; Boue, S.; Bhatnagar, D. Volatile trans-2-hexenal, a soybean aldehyde, inhibits Aspergillus flavus growth and aflatoxin production in corn. J. Food Sci. 2011, 76, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.S.; Greene-McDowelle, D.M.; Zeringue, H.J., Jr.; Bhatnagar, D.; Cleveland, T.E. Effects of volatile aldehydes from Aspergillus-resistant varieties of corn on Aspergillus parasiticus growth and aflatoxin biosynthesis. Toxicon 2000, 38, 1215–1223. [Google Scholar] [CrossRef]
- Liang, D.; Xing, F.; Selvaraj, J.N.; Liu, X.; Wang, L.; Hua, H.; Liu, Y. Inhibitory effect of cinnamaldehyde, citral, and eugenol on aflatoxin biosynthetic gene expression and aflatoxin B1 biosynthesis in Aspergillus flavus. J. Food Sci. 2015, 80, 2917–2924. [Google Scholar] [CrossRef]
- Mercier, J.; Jiménez, J.I. Control of fungal decay of apples and peaches by the biofumigant fungus Muscodor albus. Postharvest Biol. Biotechnol. 2004, 31, 1–8. [Google Scholar] [CrossRef]
- Passone, M.A.; Etcheverry, M. Antifungal impact of volatile fractions of Peumus boldus and Lippia turbinata on Aspergillus section Flavi and residual levels of these oils in irradiated peanut. Int. J. Food Microbiol. 2014, 168, 17–23. [Google Scholar] [CrossRef]
- Li, Q.; Ning, P.; Zheng, L.; Huang, J.; Li, G.; Hsiang, T. Fumigant activity of volatiles of Streptomyces globisporus JK-1 against Penicillium italicum on Citrus microcarpa. Postharvest Biol. Technol. 2010, 58, 157–165. [Google Scholar] [CrossRef]
- Wu, Y.; Yuan, J.; E, Y.; Raza, W.; Shen, Q.; Huang, Q. Effects of volatile organic compounds from Streptomyces albulus NJZJSA2 on growth of two fungal pathogens. J. Basic Microbiol. 2015, 55, 1104–1117. [Google Scholar] [CrossRef]
- Yang, M.; Lu, L.; Pang, J.; Hu, Y.; Guo, Q.; Li, Z.; Wu, S.; Liu, H.; Wang, C. Biocontrol activity of volatile organic compounds from Streptomyces alboflavus TD-1 against Aspergillus flavus growth and aflatoxin production. J. Microbiol. Accept.
- Payne, G.A.; Nierman, W.C.; Wortman, J.R.; Pritchard, B.L.; Brown, D.; Dean, R.A.; Bhatnagar, D.; Cleveland, T.E.; Machida, M.; Yu, J. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 2006, 44, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Yue, Y.; Fu, Y.; Fang, Z.; Xu, Z.; Ma, G.; Wang, S. Exploration of the regulatory mechanism of secondary metabolism by comparative transcriptomics in Aspergillus flavus. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Chang, P.K.; Scharfenstein, L.L.; Ehrlich, K.C.; Wei, Q.; Bhatnagar, D.; Ingber, B.F. Effects of laeA deletion on Aspergillus flavus conidial development and hydrophobicity may contribute to loss of aflatoxin production. Fungal. Bio. 2012, 116, 298–307. [Google Scholar] [CrossRef]
- Amaike, S.; Keller, N.P. Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell 2009, 8, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Ma, L.; Hu, Z.; Li, H.; Ai, M.; Long, C.; Zeng, B. Deep sequencing analysis of transcriptomes in Aspergillus oryzae in response to salinity stress. Appl. Microbiol. Biotechnol. 2018, 102, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Scharfenstein, L.L.; Mack, B.; Yu, J.; Ehrlich, K.C. Transcriptomic profiles of Aspergillus flavus CA42, a strain that produces small sclerotia, by decanal treatment and after recovery. Fungal Genet. Biol. 2014, 68, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Lara-Pezzi, E.; Desco, M.; Gatto, A.; Gómez-Gaviro, M.V. Neurogenesis: Regulation by alternative splicing and related posttranscriptional processes. Neuroscientist 2017, 23, 466–477. [Google Scholar] [CrossRef]
- Lee, M.J.; Sheppard, D.C. Recent advances in the understanding of the Aspergillus fumigatus cell wall. J. Microbiol. 2016, 54, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, K.; Helmschrott, C.; Dirr, F.; Wagener, J. Deciphering cell wall integrity signalling in Aspergillus fumigatus: Identification and functional characterization of cell wall stress sensors and relevant Rho GTPases. Mol. Microbiol. 2012, 83, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Latgé, J.P.; Beauvais, A. α1, 3 glucans are dispensable in Aspergillus fumigatus. Eukaryot. Cell. 2012, 11, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Meetei, P.A.; Rathore, R.S.; Prabhu, N.P.; Vindal, V. In silico screening for identification of novel β-1, 3-glucan synthase inhibitors using pharmacophore and 3D-QSAR methodologies. Springerplus 2016, 5, 965. [Google Scholar] [CrossRef] [PubMed]
- Henar, M.V.; Durán, A.; Roncero, C. Chitin synthases in yeast and fungi. Exs 1999, 87, 55–69. [Google Scholar]
- Wang, Y.; Feng, K.; Yang, H.; Zhang, Z.; Yuan, Y.; Yue, T. Effect of cinnamaldehyde and citral combination on transcriptional profile, growth, oxidative damage and patulin biosynthesis of Penicillium expansum. Front. Microbiol. 2018, 9, 597. [Google Scholar] [CrossRef]
- Kitagaki, H.; Wu, H.; Shimoi, H.; Ito, K. Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 2002, 46, 1011–1022. [Google Scholar] [CrossRef]
- Parveen, M.; Hasan, M.K.; Takahashi, J.; Murata, Y.; Kitagawa, E.; Kodama, O.; Iwahashi, H. Response of Saccharomyces cerevisiae to a monoterpene: Evaluation of antifungal potential by DNA microarray analysis. J. Antimicrob. Chemother. 2004, 54, 46–55. [Google Scholar] [CrossRef]
- Henriques, S.T.; Craik, D.J. Importance of the cell membrane on the mechanism of action of cyclotides. ACS Chem. Biol. 2012, 7, 626–636. [Google Scholar] [CrossRef]
- Hu, Z.; He, B.; Ma, L.; Sun, Y.; Niu, Y.; Zeng, B. Recent advances in ergosterol biosynthesis and regulation mechanisms in Saccharomyces cerevisiae. Indian J. Microbiol. 2017, 57, 270–277. [Google Scholar] [CrossRef] [PubMed]
- OuYang, Q.; Tao, N.; Jing, G. Transcriptional profiling analysis of Penicillium digitatum, the causal agent of citrus green mold, unravels an inhibited ergosterol biosynthesis pathway in response to citral. BMC Genom. 2016, 17, 599. [Google Scholar] [CrossRef] [PubMed]
- Malik, P.; Chaudhry, N.; S. Kitawat, B.; Kumar, R.; K. Mukherjee, T. Relationship of azole resistance with the structural alteration of the target sites: Novel synthetic compounds for better antifungal activities. Nat. Prod. J. 2014, 4, 131–139. [Google Scholar] [CrossRef]
- Klug, L.; Daum, G. Yeast lipid metabolism at a glance. FEMS Yeast Res. 2014, 14, 369–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.Y.; Mead, M.E.; Kim, S.C.; Rokas, A.; Yu, J.H. WetA bridges cellular and chemical development in Aspergillus flavus. PLoS ONE 2017, 12, e0179571. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Mead, M.E.; Lee, M.K.; Loss, E.M.O.; Kim, S.C.; Rokas, A.; Yu, J.H. Systematic dissection of the evolutionarily conserved WetA developmental regulator across a genus of Filamentous Fungi. mBio 2018, 9, e01130-18. [Google Scholar] [CrossRef]
- Valsecchi, I.; Dupres, V.; Stephen-Victor, E.; Guijarro, J.I.; Gibbons, J.; Beau, R.; Bayry, J.; Coppee, J.Y.; Lafont, F.; Latge, J.P.; et al. Role of Hydrophobins in Aspergillus fumigatus. J. Fungi 2017, 4, 2. [Google Scholar] [CrossRef]
- Lv, Y.; Lv, A.; Zhai, H.; Zhang, S.; Li, L.; Cai, J.; Hu, Y. Insight into the global regulation of laeA in Aspergillus flavus based on proteomic profiling. Int. J. Food Microbiol. 2018, 284, 11–21. [Google Scholar] [CrossRef]
- 43 Cary, J.W.; Han, Z.; Yin, Y.; Lohmar, J.M.; Shantappa, S.; Harris-Coward, P.Y.; Mack, B.; Ehrlich, K.C.; Wei, Q.; Arroyo-Manzanares, N.; et al. Transcriptome analysis of Aspergillus flavus reveals veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryot. Cell 2015, 14, 983–997. [Google Scholar] [CrossRef]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef]
- Kale, S.P.; Milde, L.; Trapp, M.K.; Frisvad, J.C.; Keller, N.P.; Bok, J.W. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 2008, 45, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Gacem, M.A.; El Hadj-Khelil, A.O. Toxicology, biosynthesis, bio-control of aflatoxin and new methods of detection. Asian Pac. J. Trop Biomed. 2016, 6, 808–814. [Google Scholar] [CrossRef] [Green Version]
- Theumer, M.G.; Henneb, Y.; Khoury, L.; Snini, S.P.; Tadrist, S.; Canlet, C.; Puel, O.; Oswald, I.P.; Audebert, M. Genotoxicity of aflatoxins and their precursors in human cells. Toxicol. Lett. 2018, 287, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.W.; Lee, L.S.; Vinnett, C. The correlation of aflatoxin and norsolorinic acid production. J. Am. Oil Chem. Soc. 1971, 48, 368. [Google Scholar] [CrossRef]
- Yu, J. Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 2012, 4, 1024–1057. [Google Scholar] [CrossRef] [PubMed]
- Trail, F.; Chang, P.K.; Cary, J.; Linz, J.E. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl. Environ. Microb. 1994, 60, 4078–4085. [Google Scholar] [CrossRef]
- Zhang, F.; Zhong, H.; Han, X.; Guo, Z.; Yang, W.; Liu, Y.; Yang, K.; Zhuang, Z.; Wang, S. Proteomic profile of Aspergillus flavus in response to water activity. Fungal Biol. 2015, 119, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Guo, Z.; Zhong, H.; Wang, S.; Yang, W.; Liu, Y.; Wang, S. RNA-Seq-based transcriptome analysis of aflatoxigenic Aspergillus flavus in response to water activity. Toxins 2014, 6, 3187–3207. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Yu, J.; Yu, J.H. aflT, a MFS transporter-encoding gene located in the aflatoxin gene cluster, does not have a significant role in aflatoxin secretion. Fungal Genet. Biol. 2004, 41, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Montalbano, B.G.; Cary, J.W. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 1999, 230, 249–257. [Google Scholar] [CrossRef]
- Wang, B.; Han, X.; Bai, Y.; Lin, Z.; Qiu, M.; Nie, X.; Wang, S.; Zhang, F.; Zhuang, Z.; Yuan, J.; et al. Effects of nitrogen metabolism on growth and aflatoxin biosynthesis in Aspergillus flavus. J. Hazard. Mater. 2017, 324, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Fasoyin, O.E.; Wang, B.; Qiu, M.; Han, X.; Chung, K.R.; Wang, S. Carbon catabolite repression gene creA regulates morphology, aflatoxin biosynthesis and virulence in Aspergillus flavus. Fungal Genet. Biol. 2018, 115, 41–51. [Google Scholar] [CrossRef] [PubMed]
- OBrian, G.R.; Fakhoury, A.M.; Payne, G.A. Identification of genes differentially expressed during aflatoxin biosynthesis in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet. Biol. 2003, 39, 118–127. [Google Scholar] [CrossRef]
- Adnan, M.; Zheng, W.; Islam, W.; Arif, M.; Abubakar, Y.; Wang, Z.; Lu, G. Carbon catabolite repression in filamentous fungi. Int. J. Mol. Sci. 2017, 19, 48. [Google Scholar] [CrossRef] [PubMed]
- Ries, L.N.; Beattie, S.R.; Espeso, E.A.; Cramer, R.A.; Goldman, G.H. Diverse regulation of the CreA carbon catabolite repressor in Aspergillus nidulans. Genetics 2016, 203, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.A.; Kelly, J.M. Proteins interacting with CreA and CreB in the carbon catabolite repression network in Aspergillus nidulans. Curr. Genet. 2017, 63, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Beattie, S.R.; Mark, K.M.; Thammahong, A.; Ries, L.N.A.; Dhingra, S.; Caffrey-Carr, A.K.; Cheng, C.; Black, C.C.; Bowyer, P.; Bromley, M.J.; et al. Filamentous fungal carbon catabolite repression supports metabolic plasticity and stress responses essential for disease progression. PLoS Pathog. 2017, 13, e1006340. [Google Scholar] [CrossRef]
- Cubero, B.; Scazzocchio, C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994, 13, 407–415. [Google Scholar] [CrossRef]
- Zhao, X.; Zhi, Q.Q.; Li, J.Y.; Keller, N.; He, Z.M. The antioxidant gallic acid inhibits aflatoxin formation in Aspergillus flavus by modulating transcription factors FarB and CreA. Toxins 2018, 10, 270. [Google Scholar] [CrossRef]
- Georgianna, D.R.; Payne, G.A. Genetic regulation of aflatoxin biosynthesis: From gene to genome. Fungal Genet. Biol. 2009, 46, 113–125. [Google Scholar] [CrossRef]
- Tudzynski, B. Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 2014, 5, 656. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.R.; Yu, J.; Bland, J.M.; Nierman, W.C.; Bhatnagar, D.; Cleveland, T.E. Amino acid supplementation reveals differential regulation of aflatoxin biosynthesis in Aspergillus flavus NRRL 3357 and Aspergillus parasiticus SRRC 143. Appl. Microbiol. Biotechnol. 2007, 74, 1308–1319. [Google Scholar] [CrossRef]
- Şenyuva, H.Z.; Gilbert, J.; Öztürkoǧlu, S.; Özcan, S.; Gürel, N. Changes in free amino acid and sugar levels of dried figs during aflatoxin B1 production by Aspergillus flavus and Aspergillus parasiticus. J. Agric. Food Chem. 2008, 56, 9661–9666. [Google Scholar] [CrossRef] [PubMed]
- Narendja, F.; Goller, S.P.; Wolschek, M.; Strauss, J. Nitrate and the GATA factor AreA are necessary for in vivo binding of NirA, the pathway-specific transcriptional activator of Aspergillus nidulans. Mol. Microbiol. 2002, 44, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Qiu, M.; Wang, B.; Yin, W.B.; Nie, X.; Qin, Q.; Ren, S.; Yang, K.; Zhang, F.; Zhuang, Z.; et al. Functional analysis of the nitrogen metabolite repression regulator gene nmrA in Aspergillus flavus. Front. Microbiol. 2016, 7, 1794. [Google Scholar] [CrossRef] [PubMed]
- Chanda, A.; Roze, L.V.; Kang, S.; Artymovich, K.A.; Hicks, G.R.; Raikhel, N.V.; Calvo, A.M.; Linz, J.E. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 19533–19538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fillinger, S.; Chaveroche, M.K.; Shimizu, K.; Keller, N.; Enfert, C. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2002, 44, 1001–1016. [Google Scholar] [CrossRef]
- Xue, C.; Hsueh, Y.P.; Chen, L.; Heitman, J. The RGS protein Crg2 regulates both pheromone and cAMP signalling in Cryptococcus neoformans. Mol. Microbiol. 2008, 70, 379–395. [Google Scholar] [CrossRef]
- Choi, Y.E.; Xu, J.R. The cAMP signaling pathway in Fusarium verticillioides is important for conidiation, plant infection, and stress responses but not fumonisin production. Mol. Plant-Microbe Interact. 2010, 23, 522–533. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, K.; Zhang, X.; Tang, W.; Wang, J.; Guo, M.; Zhao, Q.; Zheng, X.; Wang, P.; Zhang, Z. Two phosphodiesterase genes, PDEL and PDEH, regulate development and pathogenicity by modulating intracellular cyclic AMP levels in Magnaporthe oryzae. PLoS ONE 2011, 6, e17241. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, C.; Wu, C.; Sun, P.; Hou, R.; Liu, H.; Wang, C.; Xu, J.R. TRI6 and TRI10 play different roles in the regulation of deoxynivalenol (DON) production by cAMP signalling in Fusarium graminearum. Environ. Microbiol. 2016, 18, 3689–3701. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Zhou, X.; Gu, X.; Cao, S.; Wang, C.; Xu, J.R. The cAMP-PKA pathway regulates growth, sexual and asexual differentiation, and pathogenesis in Fusarium graminearum. Mol. Plant-Microbe Interact. 2014, 27, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, K.; Qin, Q.; Lin, G.; Hu, T.; Xu, Z.; Wang, S. G protein α subunit GpaB is required for asexual development, aflatoxin biosynthesis and pathogenicity by regulating cAMP signaling in Aspergillus flavus. Toxins 2018, 10, 117. [Google Scholar] [CrossRef]
- Yang, K.; Qin, Q.; Liu, Y.; Zhang, L.; Liang, L.; Lan, H.; Chen, C.; You, Y.; Zhang, F.; Wang, S. Adenylate cyclase AcyA regulates development, aflatoxin biosynthesis and fungal virulence in Aspergillus flavus. Front. Cell Infect. Microbiol. 2016, 6, 190. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Brown, R.L.; Damann, K.E.; Cleveland, T.E. Characterization of an alkaline protease excreted by Aspergillus flavus and its function in fungal infection of corn kernels. Phytopathology 1999, 89, S15. [Google Scholar]
- Luo, M.; Brown, R.L.; Chen, Z.Y.; Cleveland, T.E. Host genes involved in the interaction between Aspergillus flavus and maize. Toxin Rev. 2009, 28, 118–128. [Google Scholar] [CrossRef]
- Mellon, J.E.; Cotty, P.J.; Dowd, M.K. Aspergillus flavus hydrolases: Their roles in pathogenesis and substrate utilization. Appl. Microbiol. Biotechnol. 2007, 77, 497–504. [Google Scholar] [CrossRef]
- Amaike, S.; Keller, N.P. Aspergillus flavus. Annu. Rev. Phytopathol. 2011, 49, 107–133. [Google Scholar] [CrossRef]
- Rementeria, A.; Lopez-Molina, N.; Ludwig, A.; Vivanco, A.B.; Bikandi, J.; Pontpon, J.; Garaizar, J. Genes and molecules involved in Aspergillus fumigatus virulence. Rev. Iberoam. Micol. 2005, 22, 1–23. [Google Scholar] [CrossRef]
- Valiante, V.; Macheleidt, J.; Föge, M.; Brakhage, A.A. The Aspergillus fumigatus cell wall integrity signaling pathway: Drug target, compensatory pathways, and virulence. Front. Microbiol. 2015, 6, 325. [Google Scholar] [CrossRef]
- Mah, J.H.; Yu, J.H. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 1585–1595. [Google Scholar] [CrossRef]
- Lamarre, C.; Sokol, S.; Debeaupuis, J.P.; Henry, C.; Lacroix, C.; Glaser, P.; Coppee, J.Y.; Francois, J.M.; Latge, J.P. Transcriptomic analysis of the exit from dormancy of Aspergillus fumigatus conidia. BMC Genom. 2008, 9, 417. [Google Scholar] [CrossRef] [PubMed]
- Novodvorska, M.; Stratford, M.; Blythe, M.J.; Wilson, R.; Beniston, R.G.; Archer, D.B. Metabolic activity in dormant conidia of Aspergillus niger and developmental changes during conidial outgrowth. Fungal Genet. Biol. 2016, 94, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Shankar, J.; Tiwari, S.; Shishodia, S.K.; Gangwar, M.; Hoda, S.; Thakur, R.; Vijayaraghavan, P. Molecular insights into development and virulence determinants of Aspergilli: A proteomic perspective. Front. Cell. Infect. Microbiol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Aimanianda, V.; Bayry, J.; Bozza, S.; Kniemeyer, O.; Perruccio, K.; Elluru, S.R.; Clavaud, C.; Paris, S.; Brakhage, A.A.; Kaveri, S.V.; et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 2009, 460, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Dagenais, T.R.; Giles, S.S.; Aimanianda, V.; Latgé, J.P.; Hull, C.M.; Keller, N.P. Aspergillus fumigatus LaeA-mediated phagocytosis is associated with a decreased hydrophobin layer. Infect. Immun. 2010, 78, 823–829. [Google Scholar] [CrossRef]
- D’Souza, C.A.; Heitman, J. Conserved cAMP signaling cascades regulate fungal development and virulence. Fems Microbiol. Rev. 2001, 25, 349–364. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; D’Souza, C.A.; Kronstad, J.W. Of smuts, blasts, mildews, and blights: cAMP signaling in phytopathogenic fungi. Annu. Rev. Phytopathol. 2003, 41, 399–427. [Google Scholar] [CrossRef]
- Krijgsheld, P.; Bleichrodt, R.V.; Van Veluw, G.J.; Wang, F.; Müller, W.H.; Dijksterhuis, J.; Wösten, H.A.B. Development in Aspergillus. Stud. Mycol. 2013, 74, 1–29. [Google Scholar] [CrossRef]
- Li, H.X.; Xiao, C.L. Characterization of fludioxonil-resistant and pyrimethanil-resistant phenotypes of Penicillium expansum from apple. Phytopathology 2008, 98, 427–435. [Google Scholar] [CrossRef]
- Bavaro, S.L.; D’Antuono, I.; Cozzi, G.; Haidukowski, M.; Cardinali, A.; Logrieco, A.F. Inhibition of aflatoxin B1 production by verbascoside and other olive polyphenols. World Mycotoxin. J. 2016, 9, 545–553. [Google Scholar] [CrossRef]
- Diao, J.; Liu, H.; Hu, F.; Li, L.; Wang, X.; Gai, C.; Yu, X.; Fan, Y.; Xu, L.; Ye, H. Transcriptome analysis of immune response in fat greenling (Hexagrammos otakii) against Vibrio harveyi infection. Fish Shellfish Immunol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Zhu, Y.; Xu, J.; Sun, C.; Zhou, S.; Xu, H.; Nelson, D.R.; Qian, J.; Song, J.; Luo, H.; Xiang, L.; et al. Chromosome-level genome map provides insights into diverse defense mechanisms in the medicinal fungus Ganoderma sinense. Sci. Rep. 2015, 5, 11087. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Gene ID | Gene Name | Log2 (EG/CG) * | p-Value | Function |
|---|---|---|---|---|
| Cell wall | ||||
| TRINITY_DN6715_c0_g1 | crh11 | −3.98 | 8.95 × 10−8 | cell wall glucanase |
| TRINITY_DN10780_c1_g3 | ags1 | −2.24 | 1.53 × 10−5 | Alpha-1,3-glucan synthase |
| TRINITY_DN17727_c0_g1 | fks1 | −1.38 | 8.42 × 10−3 | beta-1,3 -glucan synthase |
| TRINITY_DN9747_c0_g1 | rho2 | 1.42 | 2.71 × 10−4 | GTP-binding protein |
| TRINITY_DN9087_c0_g1 | rho4 | 1.31 | 5.24 × 10−3 | GTP-binding protein |
| TRINITY_DN11443_c2_g2 | cfmA | −1.51 | 3.20 × 10−4 | GPI-anchored CFEM domain protein A |
| TRINITY_DN11909_c4_g1 | afuA | −1.75 | 2.47 × 10−3 | GPI-anchored membrane protein |
| TRINITY_DN9375_c0_g1 | chs6 | −1.02 | 1.65 × 10−3 | Chitin synthase |
| TRINITY_DN10195_c0_g1 | chs8 | −1.31 | 1.13 × 10−3 | Chitin synthase |
| Cell membrane | ||||
| TRINITY_DN8232_c0_g1 | erg3 | 3.58 | 1.91 × 10−2 | C-5 sterol desaturase |
| TRINITY_DN10457_c0_g1 | erg4 | −2.87 | 8.85 × 10−6 | C6 transcription factor |
| TRINITY_DN7343_c0_g1 | erg5 | −1.80 | 5.19 × 10−3 | Cytochrome P450 |
| TRINITY_DN6009_c0_g1 | erg6 | −1.23 | 4.12 × 10−2 | 24-C-methyltransferase |
| TRINITY_DN9690_c0_g1 | erg7 | −6.36 | 6.64 × 10−21 | Lanosterol synthase |
| TRINITY_DN11958_c0_g2 | erg13 | −1.34 | 2.89 × 10−2 | Hydroxymethylglutaryl-CoA synthase |
| TRINITY_DN10380_c0_g1 | erg25 | −1.64 | 1.37 × 10−2 | Methylsterol monooxygenase |
| TRINITY_DN15242_c0_g1 | erg26 | −1.59 | 1.48 × 10−3 | Sterol-4-alpha-carboxylate 3-dehydrogenase |
| Conidia | ||||
| TRINITY_DN11281_c0_g1 | brlA | −1.27 | 8.69 × 10−4 | C2H2 type master regulator of conidiophore development |
| TRINITY_DN11770_c0_g1 | wetA | −3.99 | 1.85 × 10−21 | developmental regulatory protein |
| TRINITY_DN9659_c0_g1 | abaA | −3.17 | 1.23 × 10−12 | Conidiophore development regulator |
| TRINITY_DN11518_c0_g4 | rodA | −9.10 | 6.30 × 10−19 | Conidial hydrophobin |
| TRINITY_DN15172_c0_g1 | rodB | −6.94 | 4.97 × 10−23 | Conidial hydrophobin |
| TRINITY_DN11879_c4_g1 | stuA | −1.54 | 7.43 × 10−6 | Cell pattern formation-associated protein |
| Transcription regulator | ||||
| TRINITY_DN9345_c0_g1 | laeA | −1.82 | 5.63 × 10−5 | Secondary metabolism regulator |
| TRINITY_DN11910_c4_g1 | veA | 1.42 | 2.68 × 10−3 | Developmental and secondary metabolism regulator |
| TRINITY_DN8021_c0_g1 | fig1 | −2.59 | 4.64 × 10−3 | Ca2+ regulator and membrane fusion protein |
| TRINITY_DN10388_c0_g1 | sebA | −2.44 | 2.36 × 10−4 | C2H2 finger domain transcription factor |
| TRINITY_DN12068_c7_g5 | mtfA | 1.06 | 2.65 × 10−2 | C2H2 finger domain transcription factor |
| TRINITY_DN12136_c0_g2 | hir3 | −1.57 | 1.24 × 10−3 | Histone transcription regulator |
| Developmental signal | ||||
| TRINITY_DN7900_c0_g1 | fluG | −2.25 | 3.28 × 10−5 | Extracellular developmental signal biosynthesis protein |
| Apoptosis | ||||
| TRINITY_DN12180_c5_g1 | cycA | 2.33 | 5.30 × 10−5 | Cytochrome c |
| G-protein | ||||
| TRINITY_DN10123_c0_g1 | gblP | −1.90 | 1.37 × 10−6 | Nucleotide-binding protein subunit beta-like protein |
| TRINITY_DN9739_c0_g1 | gna12 | −2.16 | 2.48 × 10−6 | G-protein alpha subunit |
| Gene ID | Gene Name | Log2 (EG/CG) * | p-Value | Function |
|---|---|---|---|---|
| Aflatoxin biosynthesis | ||||
| TRINITY_DN9945_c0_g1 | aflA | −3.42 | 5.80 × 10−10 | Fatty acid synthase alpha subunit |
| TRINITY_DN6952_c0_g1 | aflB | −3.32 | 4.61 × 10−2 | Fatty acid synthase subunit beta |
| TRINITY_DN61_c0_g1 | aflD | −4.96 | 2.64 × 10−8 | Norsolorinic acid ketoreductase |
| TRINITY_DN12173_c2_g2 | aflF | −1.65 | 4.36 × 10−2 | Norsolorinic acid ketoreductase |
| TRINITY_DN10698_c0_g2 | aflO | −1.83 | 5.63 × 10−3 | O-methyltransferase |
| TRINITY_DN10938_c0_g1 | aflQ | −1.49 | 2.16 × 10−4 | Oxidoreductase |
| TRINITY_DN10167_c0_g1 | aflR | −1.86 | 1.68 × 10−2 | Aflatoxin biosynthesis regulatory protein |
| TRINITY_DN9418_c0_g1 | aflT | −2.72 | 4.27 × 10−2 | Transmembrane protein |
| TRINITY_DN399_c0_g1 | aflU | −3.24 | 2.31 × 10−8 | P450 monooxygenase |
| TRINITY_DN10373_c0_g1 | aflW | −1.60 | 1.37 × 10−2 | FAD-binding monooxygenase |
| Carbon metabolism | ||||
| TRINITY_DN7925_c1_g1 | creA | −1.14 | 9.62 × 10−5 | DNA-binding protein |
| TRINITY_DN10031_c0_g1 | mexAM | 1.55 | 1.84 × 10−2 | Oxidoreductase |
| TRINITY_DN10394_c0_g1 | pot1 | 1.90 | 1.45 × 10−6 | Acetyl-CoA C-acyltransferase |
| TRINITY_DN11110_c0_g1 | rntA | 1.88 | 5.21 × 10−6 | Guanyl-specific ribonuclease |
| TRINITY_DN11497_c0_g1 | ppoA | −1.83 | 7.67 × 10−6 | Linoleate 8R-dioxygenase like protein |
| TRINITY_DN12116_c3_g3 | ppoC | −2.79 | 9.25 × 10−4 | Fatty acid oxygenase |
| Nitrogen metabolism | ||||
| TRINITY_DN5952_c0_g1 | nmrAL1 | −1.60 | 2.14 × 10−4 | NmrA-like family domain-containing protein 1 |
| TRINITY_DN11972_c8_g2 | gdh-1 | −2.42 | 1.52 × 10−9 | Specific glutamate dehydrogenase |
| TRINITY_DN11667_c1_g3 | glt1 | −2.40 | 4.57 × 10−5 | Putative glutamate synthase |
| TRINITY_DN11707_c0_g1 | niiA | −1.73 | 4.23 × 10−8 | Nitrite reductase |
| TRINITY_DN10046_c0_g1 | nirA | −1.93 | 2.28 × 10−4 | Nitrogen assimilation transcription factor |
| TRINITY_DN13321_c0_g1 | ddc | 1.36 | 1.02 × 10−2 | Aromatic-L-amino-acid decarboxylase |
| TRINITY_DN9125_c0_g1 | aat2 | −3.66 | 1.66 × 10−2 | Aspartate aminotransferase |
| TRINITY_DN19445_c0_g1 | melO | −3.40 | 8.11 × 10−8 | Tyrosinase |
| TRINITY_DN10664_c0_g1 | orsC | −1.06 | 4.78 × 10−2 | Tyrosinase-like protein |
| TRINITY_DN11859_c0_g1 | gad1 | −4.38 | 1.27 × 10−27 | Glutamate decarboxylase |
| TRINITY_DN11916_c1_g2 | gfa1 | −1.46 | 2.29 × 10−4 | Glutamine--fructose-6-phosphate aminotransferase |
| TRINITY_DN12169_c4_g1 | sch9 | 1.86 | 2.72 × 10−4 | Serine/threonine-protein kinase |
| Purine metabolism | ||||
| TRINITY_DN12187_c1_g2 | ade17 | −1.58 | 6.35 × 10−5 | Bifunctional purine biosynthesis protein |
| TRINITY_DN8936_c0_g1 | uaY | −3.71 | 2.76 × 10−4 | Positive regulator of purine utilization |
| TRINITY_DN9209_c0_g1 | uapC | −3.11 | 1.58 × 10−9 | Purine permease |
| TRINITY_DN9614_c0_g1 | fcy2 | −2.36 | 3.47 × 10−4 | Purine-cytosine permease |
| TRINITY_DN10376_c0_g2 | pol12 | −2.28 | 3.34 × 10−2 | DNA polymerase alpha subunit B |
| TRINITY_DN8807_c0_g1 | hxA | −1.16 | 2.02 × 10−3 | Xanthine dehydrogenase |
| cAMP signaling pathway | ||||
| TRINITY_DN10852_c0_g2 | ATP12A | −2.10 | 8.03 × 10−3 | ATPase alpha 1 subunit |
| TRINITY_DN11913_c3_g2 | pld1 | −1.55 | 8.45 × 10−5 | Phospholipase D1 |
| TRINITY_DN11065_c0_g2 | pka-C3 | −1.02 | 1.11 × 10−2 | cAMP-dependent protein kinase |
| Gene ID | Gene Name | Log2 (EG/CG) * | p-Value | Function |
|---|---|---|---|---|
| Hydrolase | ||||
| TRINITY_DN11687_c2_g1 | mal1 | −3.39 | 6.39 × 10−10 | Alpha-glucosidase |
| TRINITY_DN18434_c0_g1 | mal2 | −2.86 | 2.02 × 10−3 | Alpha-glucosidase |
| TRINITY_DN9284_c0_g1 | agdC | −2.16 | 5.88 × 10−6 | Alpha -glucosidase |
| TRINITY_DN4002_c0_g1 | lip | −3.41 | 2.15 × 10−3 | Lipase |
| TRINITY_DN11096_c0_g3 | NPII | −7.81 | 2.84 × 10−61 | Neutral protease |
| TRINITY_DN10289_c0_g1 | ctf1B | −2.77 | 2.89 × 10−2 | Cutinase transcription factor 1 beta |
| TRINITY_DN11791_c0_g4 | ctf1A | −1.59 | 4.29 × 10−2 | Cutinase transcription factor 1 alpha |
| TRINITY_DN8062_c0_g1 | abfA | −2.71 | 2.69 × 10−3 | Alpha-L-arabinofuranosidase A |
| TRINITY_DN12177_c2_g1 | dvrA | 1.03 | 1.97 × 10−2 | C2H2 finger domain transcription factor |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Lu, L.; Li, S.; Zhang, J.; Li, Z.; Wu, S.; Guo, Q.; Liu, H.; Wang, C. Transcriptomic Insights into Benzenamine Effects on the Development, Aflatoxin Biosynthesis, and Virulence of Aspergillus flavus. Toxins 2019, 11, 70. https://doi.org/10.3390/toxins11020070
Yang M, Lu L, Li S, Zhang J, Li Z, Wu S, Guo Q, Liu H, Wang C. Transcriptomic Insights into Benzenamine Effects on the Development, Aflatoxin Biosynthesis, and Virulence of Aspergillus flavus. Toxins. 2019; 11(2):70. https://doi.org/10.3390/toxins11020070
Chicago/Turabian StyleYang, Mingguan, Laifeng Lu, Shuhua Li, Jing Zhang, Zhenjing Li, Shufen Wu, Qingbin Guo, Huanhuan Liu, and Changlu Wang. 2019. "Transcriptomic Insights into Benzenamine Effects on the Development, Aflatoxin Biosynthesis, and Virulence of Aspergillus flavus" Toxins 11, no. 2: 70. https://doi.org/10.3390/toxins11020070
APA StyleYang, M., Lu, L., Li, S., Zhang, J., Li, Z., Wu, S., Guo, Q., Liu, H., & Wang, C. (2019). Transcriptomic Insights into Benzenamine Effects on the Development, Aflatoxin Biosynthesis, and Virulence of Aspergillus flavus. Toxins, 11(2), 70. https://doi.org/10.3390/toxins11020070






