Occurrence of Fungi and Fungal Toxins in Fish Feed during Storage
Abstract
1. Introduction
2. Results
2.1. Storage Conditions
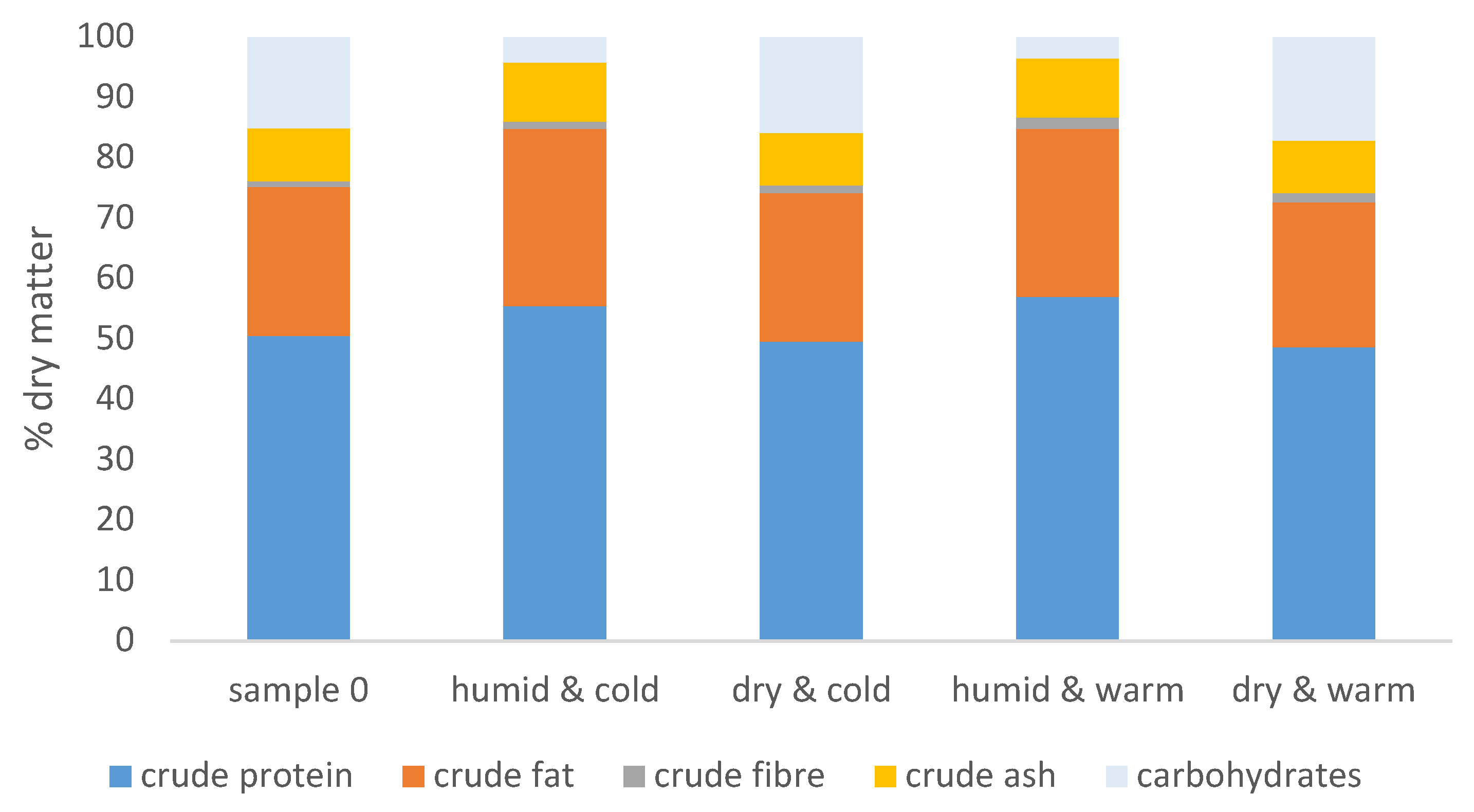
2.2. Changes in the Nutritional Composition
2.3. Detection of Microorganisms on the Feeds
2.4. Fungal Identification via PCR
2.5. Mycotoxin Analyses
3. Discussion
3.1. Influence of Storage Conditions on the Nutritional Composition of the Feeds
3.2. Microbiological Colonization of Feed Samples
3.3. Identification of Fungi
3.4. Storage Conditions and Mycotoxin Formation
3.5. Potential Effects on Fish
4. Conclusions
5. Methods
5.1. Storage Conditions
5.2. Nutritional Composition of the Feeds
5.3. Detection of Microorganisms on the Feeds
5.4. Fungal Identification via PCR
5.5. Toxin Analyses
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Binder, E.M.; Tan, L.M.; Chin, J.; Handl, J.; Richard, J. Worldwide occurrence of mycotoxins in commodities, feeds and feed ingredients. Anim. Feed Sci. Technol. 2007, 137, 265–282. [Google Scholar] [CrossRef]
- Rodrigues, I.; Naehrer, K. Prevalence of mycotoxins in feedstuffs and feed surveyed worldwide in 2009 and 2010. Phytopathol. Mediterr. 2012, 51, 175–192. [Google Scholar]
- Duarte, S.C.; Lino, C.M.; Pena, A. Ochratoxin A in feed of food-producing animals: An undesirable mycotoxin with health and performance effects. Vet. Microbiol. 2011, 154, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Duarte, S.C.; Pena, A.; Lino, C.M. A review on ochratoxin A occurrence and effects of processing of cereal and cereal derived food products. Food Microbiol. 2010, 27, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Alwatban, M.A.; Hadi, S.; Moslem, M.A. Mycotoxin production in Cladosporium species influenced by temperature regimes. J. Pure Appl. Microbiol. 2014, 8, 4061–4069. [Google Scholar]
- Magan, N.; Medina, A.; Aldred, D. Possible climate-change effects on mycotoxin contamination of food crops pre- and postharvest. Plant Pathol. 2011, 60, 150–163. [Google Scholar] [CrossRef]
- Soliman, A.K.; Jauncey, K.; Roberts, R.J. Stability of L-ascorbic acid (vitamin C) and its forms in fish feeds during processing, storage and leaching. Aquacult 1987, 60, 73–83. [Google Scholar] [CrossRef]
- Alabi, J.O.; Fafiolu, A.O.; Oso, A.O.; Jegede, A.V.; Dada, I.D.; Teniola, A.A.; Oluwatosin, O.O. Physico-chemical and compositional changes in proprietary finished feeds stored under different conditions. Archivos de Zootecnia 2017, 66, 535–541. [Google Scholar] [CrossRef]
- Laohabanjong, R.; Tantikitti, C.; Benjakul, S.; Supamattaya, K.; Boonyaratpalin, M. Lipid oxidation in fish meal stored under different conditions on growth, feed efficiency and hepatopancreatic cells of black tiger shrimp (Penaeus monodon). Aquacult 2009, 286, 283–289. [Google Scholar] [CrossRef]
- Tola, M.; Kebede, B. Occurrence, importance and control of mycotoxins: A review. Cogent Food Agricult. 2016, 2, 1191103. [Google Scholar] [CrossRef]
- Villers, P. Aflatoxins and safe storage. Front. Microbiol. 2014, 5, 158. [Google Scholar] [CrossRef] [PubMed]
- Scudamore, K.A. Prevention of ochratoxin A in commodities and likely effects of processing fractionation and animal feeds. Food Addit. Contam. 2005, 22, 17–25. [Google Scholar] [CrossRef] [PubMed]
- EFSA, Opinion of the scientific panel on contaminants in the food chain on a request from the commission related to ochratoxin A in food. EFSA J. 2004, 101, 1–36.
- Noonim, P.; Mahakarnchanakul, W.; Varga, J.; Samson, R.A. Aspergilli and ochratoxin A in coffee. In Aspergillus in the Genomic Era; Varga, J., Samson, R.A., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2008; pp. 213–231. [Google Scholar]
- Taniwaki, M.H. An update on ochratoxigenic fungi and ochratoxin A in coffee. In Advances in Experimental Medicine and Biology, 57, 1st ed.; Hocking, A.D., Pitt, J.I., Samson, R.A., Thrane, U., Eds.; Springer Science: New York, NY, USA, 2005; pp. 189–202. [Google Scholar]
- Cabañes, F.J.; Bragulat, M.R.; Castellá, G. Ochratoxin A producing species in the genus Penicillium. Toxins (Basel) 2010, 2, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.H.; Szczech, G.M.; Purmalis, B.P. Teratogenic and toxic effects of ochratoxin A in rats. Toxicol. Appl. Pharmacol. 1976, 32, 331–338. [Google Scholar] [CrossRef]
- Kőszegi, T.; Poór, M. Ochratoxin a: Molecular interactions, mechanisms of toxicity and prevention at the molecular level. Toxins (Basel) 2016, 8, 111. [Google Scholar] [CrossRef]
- Boorman, G.A. Toxicology and carcinogenic studies of ochratoxin A (CAS No. 303-47-9) in F344/n rats (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 1989, 358, 1–142. [Google Scholar]
- Arora, R.G.; Frölén, H.; Fellner-Feldegg, H. Inhibition of ochratoxin a teratogenesis by zearalenone and diethylstilboestrol. Food Chem. Toxicol. 1983, 21, 779–783. [Google Scholar] [CrossRef]
- Zahran, E.; Manning, B.; Seo, J.K.; Noga, E.J. The effect of ochratoxin A on antimicrobial polypeptide expression and resistance to water mold infection in channel catfish (Ictalurus punctatus). Fish Shellfish Immunol. 2016, 57, 60–67. [Google Scholar] [CrossRef]
- Caldwell, R.W.; Tuite, J.; Stob, M.; Baldwin, R. Zearalenone production by Fusarium species. Appl. Microbiol. 1970, 20, 31–34. [Google Scholar] [CrossRef]
- Semaskiene, R.; Mankeviciene, A.; Dabkevicius, Z.; Leistrumaite, A. Toxic fungi infection and mycotoxin level in organic grain. Bot. Lith 2005, 7, 17–25. [Google Scholar]
- Nicolaisen, M.; Suproniene, S.; Nielsen, L.K.; Lazzaro, I.; Spliid, N.H.; Justesen, A.F. Real-time PCR for quantification of eleven individual Fusarium species in cereals. J. Microbiol. Methods 2009, 76, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, J.; Font, G.; Mañes, J.; Ferrer, E. Natural occurrence of emerging Fusarium mycotoxins in feed and fish from aquaculture. J. Agric. Food Chem. 2014, 62, 12462–12470. [Google Scholar] [CrossRef] [PubMed]
- European Commission, Commission Recommendation of 17 August 2006 on the presence of deoxynivalenol, zearalenone, ochratoxin A, T-2 and HT-2 and fumonisins in products intended for animal feeding. Off. J. Eur. Union 2006, L229, 7–9. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32006H0576&from=EN (accessed on 10 January 2020).
- Van Egmond, H.P.; Jonker, M.A. Worldwide Regulations for Mycotoxins in Food and Feed in 2003; Food and Agriculture Organization of the United Nation: Rome, Italy, 2004; p. 81. Available online: http://www.fao.org/3/y5499e/y5499e00.htm (accessed on 10 January 2020).
- Pietsch, C. Risk assessment for mycotoxin contamination in fish feeds in Europe. Mycotoxin Res. 2020, 36, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Mongkon, W.; Sugita-Konishi, Y.; Chaisri, W.; Suriyasathaporn, W. Aflatoxin B1 contamination of dairy feeds after storage in farm practice in tropical environment. Biocontr. Sci. 2017, 22, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Milani, J.M. Ecological conditions affecting mycotoxin production in cereals: A review. Vet. Med. 2013, 58, 405–411. [Google Scholar] [CrossRef]
- Bhattacharya, K.; Raha, S. Deteriorative changes of maize, groundnut and soybean seeds by fungi in storage. Mycopathologia 2002, 155, 135–141. [Google Scholar] [CrossRef]
- Zmysłowska; Lewandowska, D. The effect of storage temperatures on the microbiological quality of fish feeds. Polish J. Environ. Stud. 2000, 3, 223–226. [Google Scholar]
- Piotrowska, M.; Slizewska, K.; Biernasiak, J. Mycotoxins in cereal and soybean-based food and feed. In Soybean—Pest Resistance; El-Shemy, H.A., Ed.; Intech: Rijeka, Croatia, 2013; pp. 185–230. Available online: https://www.intechopen.com/books/soybean-pest-resistance/mycotoxins-in-cereal-and-soybean-based-food-and-feed (accessed on 10 January 2020).
- Verband Deutscher Landwirtschaftlicher Untersuchungs-und Forschungsanstalten e.V. VDLUFA. Futtermitteluntersuchung: Verfahrensanweisung Identifizierung. Verfahrensanweisung zur Identifizierung von Bakterien, Hefen, Schimmel- und Schwärzepilzen als produkttypische oder verderbanzeigende Indikatorkeime. Verfahrensanweisung Identifizierung 28.1.3. In Methodenbuch III, 8. Erg; VDLUFA-Verlag: Darmstadt, Germany, 2012; p. 11. Available online: https://www.vdlufa.de/Methodenbuch/ (accessed on 10 January 2020).
- Almeida, I.F.M.; Martins, H.M.L.; Santos, S.M.O.; Freitas, M.S.; da Costa, J.M.G.N.; d’Almeida Bernardo, F.M. Mycobiota and aflatoxin B1 in feed for farmed sea bass (Dicentrarchus labrax). Toxins (Basel) 2011, 3, 163–171. [Google Scholar] [CrossRef]
- Greco, M.; Pardo, A.; Pose, G. Mycotoxigenic fungi and natural co-occurrence of mycotoxins in rainbow trout (Oncorhynchus mykiss) feeds. Toxins (Basel) 2015, 7, 4595–4609. [Google Scholar] [CrossRef] [PubMed]
- Belkacemi, L.; Barton, R.C.; Hopwood, V.; Evans, E.G.V. Determination of optimum growth conditions for gliotoxin production by Aspergillus fumigatus and development of a novel method for gliotoxin detection. Med. Mycol. 1999, 37, 227–233. [Google Scholar] [CrossRef]
- Mohamed, H.M.A.; Emeish, W.F.A.; Braeuning, A.; Hammad, S. Detection of aflatoxin-producing fungi isolated from nile tilapia and fish feed. EXCLI J. 2017, 16, 1308–1318. [Google Scholar] [PubMed]
- Marijani, E.; Wainaina, J.M.; Charo-Karisa, H.; Nzayisenga, L.; Munguti, J.; Gnonlonfin, G.J.B.; Kigadye, E.; Okoth, S. Mycoflora and mycotoxins in finished fish feed and feed from smallholder farms in East Africa. Egypt. J. Aquat. Res. 2017, 3, 169–176. [Google Scholar] [CrossRef]
- Norbäck, D.; Hashim, J.H.; Cai, G.H.; Hashim, Z.; Ali, F.; Bloom, E.; Larsson, L. Rhinitis, ocular, throat and dermal symptoms, headache and tiredness among students in schools from johor bahru, Malaysia: Associations with fungal DNA and mycotoxins in classroom dust. PLoS ONE 2016, 11, e0147996. [Google Scholar] [CrossRef] [PubMed]
- Alastruey-Izquierdo, A.; Cuenca-Estrella, M.; Monzón, A.; Mellado, E.; Rodríguez-Tudela, J.L. Antifungal susceptibility profile of clinical Fusarium spp. isolates identified by molecular methods. J. Antimicrob. Chemother 2008, 61, 805–809. [Google Scholar] [CrossRef]
- Ferrigo, D.; Raiola, A.; Causin, R. Fusarium toxins in cereals: Occurrence, legislation, factors promoting the appearance and their management. Molecules 2016, 21, 627. [Google Scholar] [CrossRef]
- Stefańczyk, E.; Sobkowiak, S.; Brylińska, M.; Śliwka, J. Diversity of Fusarium spp. associated with dry rot of potato tubers in Poland. Eur. J. Plant Pathol. 2016, 145, 871–884. [Google Scholar] [CrossRef]
- Gherbawy, Y.A.M.H.; Maghraby, T.A.; Shebany, Y.M. Seasonal variations of Fusarium species in wheat fields in Upper Egypt. Arch. Phytopathol. Plant Prot. 2006, 39, 365–377. [Google Scholar] [CrossRef]
- Silvestro, L.B.; Stenglein, S.A.; Forjan, H.; Dinolfo, M.I.; Arambarri, A.M.; Manso, L.; Moreno, M.V. Occurrence and distribution of soil Fusarium species under wheat crop in zero tillage. Spanish J. Agric. Res. 2013, 11, 72–79. [Google Scholar] [CrossRef]
- Gräfenhan, T.; Schroers, H.J.; Nirenberg, H.I.; Seifert, K.A. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud. Mycol. 2011, 68, 79–113. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.G.; Brown, D.W.; Kelly, A.C.; Kim, H.-S.; Kurtzman, C.P.; Mccormick, S.P.; O’Donnell, K.L.; Proctor, R.H.; Vaughan, M.M.; Ward, T.J.; et al. Fusarium mycotoxins: A trans-disciplinary overview. Can. J. Plant Pathol. 2018, 40, 161–171. [Google Scholar] [CrossRef]
- Stoll, D.A.; Link, S.; Kulling, S.; Geisen, R.; Schmidt-Heydt, M. Comparative proteome analysis of Penicillium verrucosum grown under light of short wavelength shows an induction of stress-related proteins associated with modified mycotoxin biosynthesis. Int. J. Food Microbiol. 2014, 175, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Nielsen, K.F.; Samson, R.A. Recommendations concerning the chronic problem of misidentification of mycotoxigenic fungi associated with foods and feeds. Adv. Food Mycol. 2006, 571, 33–46. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Wang, H.; Gu, X.; Zheng, X.; Wang, Y.; Diao, J.; Peng, Y.; Zhang, H. Screening and identification of novel ochratoxin A-producing fungi from grapes. Toxins (Basel) 2016, 8, 333. [Google Scholar] [CrossRef]
- Pietsch, C.; Kersten, S.; Burkhardt-Holm, P.; Valenta, H.; Dänicke, S. Occurrence of deoxynivalenol and zearalenone in commercial fish feed: An initial study. Toxins (Basel) 2013, 5, 184–192. [Google Scholar] [CrossRef]
- Kamei, K.; Watanabe, A. Aspergillus mycotoxins and their effect on the host. Med. Mycol. 2005, 43, S95–S99. [Google Scholar] [CrossRef]
- Paulussen, C.; Hallsworth, J.E.; Álvarez-Pérez, S.; Nierman, W.C.; Hamill, P.G.; Blain, D.; Rediers, H.; Lievens, B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2017, 10, 296–322. [Google Scholar] [CrossRef]
- Soliman, S.; Li, X.-Z.; Shao, S.; Behar, M.; Svircev, A.M.; Tsao, R.; Zhou, T. Potential mycotoxin contamination risks of apple products associated with fungal flora of apple core. Food Control 2015, 47, 585–591. [Google Scholar] [CrossRef]
- Braun, U.; Crous, P.W.; Dugan, F.; Groenewald, J.Z.; Sybren De Hoog, G. Phylogeny and taxonomy of Cladosporium-like hyphomycetes, including Davidiella gen. nov., the teleomorph of Cladosporium s. str. Mycol. Prog. 2003, 2, 3–18. [Google Scholar] [CrossRef]
- Zhang, N.; Blackwell, M. Molecular phylogeny of dogwood anthracnose fungus (Discula destructiva) and the Diaporthales. Mycologia 2001, 93, 355–365. [Google Scholar] [CrossRef]
- Smith, J.A.; Blanchette, R.A.; Ostry, M.E.; Anderson, N.A. Etiology of bronze leaf disease of Populus. Plant Dis. 2002, 86, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Embaby, E.M.; Ayaat, N.M.; Abd El-Galil, M.M.; Allah Abdel-Hameid, N.; Gouda, M.M. Mycoflora and mycotoxin contaminated chicken and fish feeds. Middle East J. Appl. Sci. 2015, 5, 1044–1054. [Google Scholar]
- McDonald, M.R.; Boland, G.J. Forecasting diseases caused by Sclerotinia spp. in eastern Canada: Fact or fiction? Can. J. Plant Pathol. 2004, 26, 480–488. [Google Scholar] [CrossRef]
- Mitchell, S.J.; Jellis, G.J.; Cox, T.W. Sclerotinia sclerotiorum on linseed. Plant Pathol. 1986, 35, 403–405. [Google Scholar] [CrossRef]
- Prova, A.; Akanda, A.M.; Islam, S.; Hossain, M.M. Characterization of Sclerotinia sclerotiorum, an emerging fungal pathogen causing blight in hyacinth bean (Lablab purpureus). Plant Pathol. J. 2018, 34, 367–380. [Google Scholar] [PubMed]
- Manning, B.B.; Ulloa, R.M.; Li, M.H.; Robinson, E.H.; Rottinghaus, G.E. Ochratoxin A fed to channel catfish (Ictalurus punctatus) causes reduced growth and lesions of hepatopancreatic tissue. Aquaculture 2003, 219, 739–750. [Google Scholar] [CrossRef]
- Tschirren, L.; Siebenmann, S.; Pietsch, C. Toxicity of ochratoxin to early life stages of zebrafish (Danio rerio). Toxins (Basel) 2018, 10, 264. [Google Scholar] [CrossRef]
- Wiger, R.; Stormer, F.C. Effects of ochratoxins A and B on prechondrogenic mesenchymal cells from chick embryo limb buds. Toxicol. Lett. 1990, 54, 129–134. [Google Scholar] [CrossRef]
- Hood, R.D.; Naughton, M.J.; Hayes, A.W. Prenatal effects of ochratoxin a in hamsters. Teratology 1976, 13, 11–14. [Google Scholar] [CrossRef]
- O’Brien, E.; Prietz, A.; Dietrich, D.R. Investigation of the teratogenic potential of ochratoxin A and B using the FETAX system. Birth Defects Res. Part B Dev. Reprod. Toxicol. 2005, 74, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Doster, R.C.; Sinnhuber, R.O.; Pawlowski, N.E. Acute intraperitoneal toxicity of ochratoxin a and B derivatives in rainbow trout (Salmo gairdneri). Food Cosmet. Toxicol. 1974, 12, 499–505. [Google Scholar] [CrossRef]
- El-Sayed, Y.S.; Khalil, R.H.; Saad, T.T. Acute toxicity of ochratoxin-A in marine water-reared sea bass (Dicentrarchus labrax L.). Chemosphere 2009, 75, 878–882. [Google Scholar] [CrossRef] [PubMed]
- Srour, T.M. Effect of ochratoxin-A with or without Biogen® on growth performance, feed utilization and carcass composition of Nile tilapia (Oreochromis niloticus) fingerlings. J. Agr. Sci. Mansoura Univ. 2004, 29, 51–61. [Google Scholar]
- Bernhoft, A.; Høgåsen, H.R.; Rosenlund, G.; Ivanova, L.; Berntssen, M.H.G.; Alexander, J.; Eriksen, G.S.; Fæste, C.K. Tissue distribution and elimination of deoxynivalenol and ochratoxin A in dietary-exposed Atlantic salmon (Salmo salar). Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2017, 34, 1211–1224. [Google Scholar] [CrossRef]
- Bernhoft, A.; Høgåsen, H.R.; Rosenlund, G.; Moldal, T.; Grove, S.; Berntssen, M.H.G.; Thoresen, S.I.; Alexander, J. Effects of dietary deoxynivalenol or ochratoxin A on performance and selected health indices in Atlantic salmon (Salmo salar). Food Chem. Toxicol. 2018, 121, 374–386. [Google Scholar] [CrossRef]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P.W. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef]
- Ogórek, R.; Lejman, A.; Pusz, W.; Miłuch, A.; Miodyńska, P. Characteristics and taxonomy of Cladosporium fungi, Charakterystyka i taksonomia grzybów z rodzaju Cladosporium. Mikol. Lek. 2012, 9, 80–85. [Google Scholar]
- Briceño, E.X.; Latorre, B.A. Characterization of Cladosporium rot in grapevines, a problem of growing importance in Chile. Plant Dis. 2008, 92, 1635–1642. [Google Scholar] [CrossRef]
- Verband Deutscher Landwirtschaftlicher Untersuchungs-und Forschungsanstalten e.V. VDLUFA. Die chemische Untersuchung von Futtermitteln. Bestimmung von Rohprotein. In Methodenbuch III, 3. Ergänzung; VDLUFA-Verlag: Darmstadt, Germany, 1993; p. 218. Available online: https://www.vdlufa.de/Methodenbuch/ (accessed on 10 January 2020).
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]




| Treatment | Sample | Temperature (°C) | Humidity (% rel H) |
|---|---|---|---|
| 1 | 8.94 ± 0.03 | 76.2 ± 0.1 | |
| humid & cold | 2 | 8.42 ± 0.02 | 79.1 ± 0.1 |
| 3 | 8.39 ± 0.04 | 79.1 ± 0.1 | |
| 4 | 8.60 ± 0.02 | 18.4 ± 0.1 | |
| dry & cold | 5 | 8.29 ± 0.04 | 26.5 ± 0.2 |
| 6 | 8.55 ± 0.03 | 21.1 ± 0.2 | |
| 7 | 27.24 ± 0.20 | 70.1 ± 0.2 | |
| humid & warm | 8 | 25.23 ± 0.14 | 73.9 ± 0.2 |
| 9 | 26.34 ± 0.12 | 69.7 ± 0.1 | |
| 10 | 28.29 ± 0.17 | 30.3 ± 0.3 | |
| dry & warm | 11 | 29.41 ± 0.22 | 20.5 ± 0.2 |
| 12 | 25.75 ± 0.13 | 24.5 ± 0.2 |
| Sample | Bengal Red | DG18 | PCA | TSA-TCC | YGA |
|---|---|---|---|---|---|
| Fungi/Bacteria | Fungi/Bacteria | Fungi/Bacteria | Fungi/Bacteria | Fungi/Bacteria | |
| 0 | -/- | -/- | +/+ | +/- | -/- |
| 1 | +/- | +/- | -/- | -/+ | +/- |
| 2 | +/+ | -/- | -/+ | -/+ | -/- |
| 3 | -/+ | +/- | -/+ | -/- | -/- |
| 4 | +/- | +/- | +/++ | -/+ | -/- |
| 5 | -/- | -/- | +/++ | -/- | +/- |
| 6 | -/- | -/- | +/- | +/- | -/- |
| 7 | ++/- | -/- | -/+ | -/+ | ++/- |
| 8 | -/- | -/- | -/+ | -/- | -/- |
| 9 | +/- | ++/- | -/+ | -/+ | -/- |
| 10 | -/- | -/- | -/+ | -/+ | -/- |
| 11 | -/- | -/- | +/- | -/- | -/- |
| 12 | -/- | -/- | +/+ | +/- | -/- |
| Sample | Agarose Type | Sequence Length | Related Fungal Species (NCBI Accession No.) |
|---|---|---|---|
| 0 | PCA | 338 bp2 | Cladosporium sp. strain CMW49962 (MH327787.1) Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) (picture of the colony on the agarose plate shown in Figure 3A) |
| TSA | 348 bp2 | Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium sp. strain CMW49962 (MH327787.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) |
| Sample | Agarose Type | Sequence Length | Related Fungal Species (NCBI Accession No.) |
|---|---|---|---|
| 1 | DG-18 | 432 bp1 | Apioplagiostoma aceriferum (AK110204.1) Cladosporium acalyphae strain SL-16 (KU574814.1) Alternaria alternata isolate CAU8331 (MH475293.1) |
| 350 bp2 | Cladosporium sp. strain CMW49962 (MH327787.1) Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium herbarum strain BEOFB1812m (MH780976.1) | ||
| (picture of the colony on the agarose plate shown in Figure 2A) | |||
| YGA | 358 bp2 | Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium sp. strain CMW49962 (MH327787.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) | |
| 2 | Bengal Red | 504 bp2 | Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) Cladosporium sp. strain CMW49962 (MH327787.1) |
| 3 | DG-18 | 395 bp2 | Sclerotinia nivalis strain KGC-S0601 (JX296007.1) Sclerotinia nivalis isolate ZJ14 (KT023311.1) Sclerotinia nivalis isolate ZJ11 (KT023308.1) |
| Sample | Agarose Type | Sequence Length | Related Fungal Species (NCBI Accession No.) |
|---|---|---|---|
| 4 | DG-18 | 425 bp1 | Apioplagiostoma aceriferum (AK110204.1) Cladosporium acalyphae strain SL-16 (KU574814.1) Alternaria alternata isolate CAU8331 (MH475293.1) |
| 355 bp2 | Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium sp. strain CMW49962 (MH327787.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) | ||
| PCA | 139 bp2 | Aspergillus fumigatus isolate 3 (MH536090.1) Aspergillus fumigatus strain m135 (MH208809.1) Aspergillus fumigatus strain m86 (MH208772.1) (picture of the colony on the agarose plate shown in Figure 3B) | |
| 340 bp2 | Cladosporium sp. strain CMW49962 (MH327787.1) Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) | ||
| 5 | PCA | 347 bp2 | Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium sp. strain CMW49962 (MH327787.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) (picture of the colony on the agarose plate shown in Figure 3C) |
| YGA | 367 bp2 | Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) Cladosporium sp. strain CMW49962 (MH327787.1) (picture of the colony on the agarose plate shown in Figure 4A) | |
| 6 | DG-18 | unknown (picture of the colony on the agarose plate shown in Figure 2B) | |
| TSA | 341 bp2 | Cladosporium sp. strain CMW49962 (MH327787.1) Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) |
| Sample | Agarose Type | Sequence Length | Related Fungal Species (NCBI Accession No.) |
|---|---|---|---|
| 7 | YGA | 497 bp2 | Aspergillus fumigatus isolate Z1201.1018 (KJ175518.1) Aspergillus fumigatus isolate M 169 (KJ175506.1) Aspergillus fumigatus (MF189897.1) (picture of the colony on the agarose plate shown in Figure 4C) |
| 353 bp2 | Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium sp. strain CMW49962 (MH327787.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) (picture of the colony on the agarose plate shown in Figure 4B) | ||
| Bengal Red | 360 bp2 | Cladosporium oxysporum isolate 015 (MF175217.1) Cladosporium oxysporum isolate 056 (MF175222.1) Cladosporium oxysporum isolate 028 (MF175220.1) | |
| 337 bp2 | Fusarium merismoides var. merismoides strain F-266,788 (EU860027.1) Fusarium merismoides isolate N271A (KP710663.1) Fusicolla septimanifiniscientiae strain CBS 144935 (MK069408.1) | ||
| 471 bp2 | Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) Cladosporium sp. strain CMW49962 (MH327787.1) | ||
| 341 bp2 | Cladosporium sp. strain CMW49962 (MH327787.1) Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) | ||
| 8 | DG-18 | 333 bp2 | Cladosporium sp. strain CMW49962 (MH327787.1) Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium allicinum culture-collection CBS:177.71(EF101451.1) (picture of the colony on the agarose plate shown in Figure 2C) |
| 9 | DG-18 | 401 bp2 | Penicillium chrysogenum strain BEOFB11120m (MH780066.1) Penicillium chrysogenum isolate 186 (MH018233.1) Penicillium chrysogenum culture MUT<ITA>:2255 (MG832189.1) (picture of the colony on the agarose plate shown in Figure 2D) |
| Sample | Agarose Type | Sequence Length | Related Fungal Species (NCBI Accession No.) |
|---|---|---|---|
| 10 | - | - | - |
| 11 | YGA | 481 bp2 | Aspergillus fumigatus isolate Z1201.1018 (KJ175518.1) Aspergillus fumigatus isolate M 169 (KJ175506.1) Aspergillus fumigatus isolate 3 (MH536090.1) (picture of the colony on the agarose plate shown in Figure 4D) |
| 12 | PCA | 501 bp2 | Davidiella tassiana strain ATCC 66670 (EF101452.1) Cladosporium allicinum culture-collection CBS:177.71 (EF101451.1) Cladosporium sp. strain CMW49962 (MH327787.1) (picture of the colony on the agarose plate shown in Figure 3D) |
| Mycotoxin (LOQ) | Aflatoxins 1 (2.5) | DON (12.5) | ZEN (12.5) | OTA (1.2) | ENN B 2 (12.7) | BEA (12.7) | FB1 + FB2 (50) | T2 + HT2 (25) |
|---|---|---|---|---|---|---|---|---|
| sample 0 | <2.5 | 34.1 | <12.5 | <1.2 | 17.7 | <13 | <50 | <25 |
| sample 1 | <2.5 | 39.8 | <12.5 | <1.2 | n.d. | n.d. | <50 | <25 |
| sample 2 | <2.5 | 27.0 | <12.5 | <1.2 | n.d. | n.d. | <50 | <25 |
| sample 3 | <2.5 | 37.0 | <12.5 | <1.2 | n.d. | n.d. | <50 | <25 |
| sample 4 | <2.5 | 34.9 | <12.5 | <1.2 | n.d. | n.d. | <50 | <25 |
| sample 5 | <2.5 | 28.4 | <12.5 | <1.2 | n.d. | n.d. | <50 | <25 |
| sample 6 | <2.5 | 33.8 | <12.5 | <1.2 | n.d. | n.d. | <50 | <25 |
| sample 7 | <2.5 | 30.0 | <12.5 | 501.7 | <12.7 | <12.7 | <50 | <25 |
| sample 8 | <2.5 | 37.4 | <12.5 | 7.1 | 18.5 | <12.7 | <50 | <25 |
| sample 9 | <2.5 | 25.1 | <12.5 | 5.7 | <12.7 | <12.7 | <50 | <25 |
| sample 10 | <2.5 | 33.4 | <12.5 | <1.2 | n.d. | n.d. | <50 | <25 |
| sample 11 | <2.5 | 43.0 | <12.5 | <1.2 | n.d. | n.d. | <50 | <25 |
| sample 12 | <2.5 | 28.1 | <12.5 | <1.2 | n.d. | n.d. | <50 | <25 |
| Sample | Treatment | Duration of Exposure (Days) |
|---|---|---|
| 0 | none | none |
| 1 | 32 | |
| 2 | humid & cold | 32 |
| 3 | 32 | |
| 4 | 32 | |
| 5 | dry & cold | 32 |
| 6 | 32 | |
| 7 | 30 | |
| 8 | humid & warm | 30 |
| 9 | 30 | |
| 10 | 30 | |
| 11 | dry & warm | 30 |
| 12 | 30 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietsch, C.; Müller, G.; Mourabit, S.; Carnal, S.; Bandara, K. Occurrence of Fungi and Fungal Toxins in Fish Feed during Storage. Toxins 2020, 12, 171. https://doi.org/10.3390/toxins12030171
Pietsch C, Müller G, Mourabit S, Carnal S, Bandara K. Occurrence of Fungi and Fungal Toxins in Fish Feed during Storage. Toxins. 2020; 12(3):171. https://doi.org/10.3390/toxins12030171
Chicago/Turabian StylePietsch, Constanze, Georg Müller, Sulayman Mourabit, Simon Carnal, and Kasun Bandara. 2020. "Occurrence of Fungi and Fungal Toxins in Fish Feed during Storage" Toxins 12, no. 3: 171. https://doi.org/10.3390/toxins12030171
APA StylePietsch, C., Müller, G., Mourabit, S., Carnal, S., & Bandara, K. (2020). Occurrence of Fungi and Fungal Toxins in Fish Feed during Storage. Toxins, 12(3), 171. https://doi.org/10.3390/toxins12030171






