Systemic Effects of Hemorrhagic Snake Venom Metalloproteinases: Untargeted Peptidomics to Explore the Pathodegradome of Plasma Proteins
Abstract
:1. Introduction
2. Results
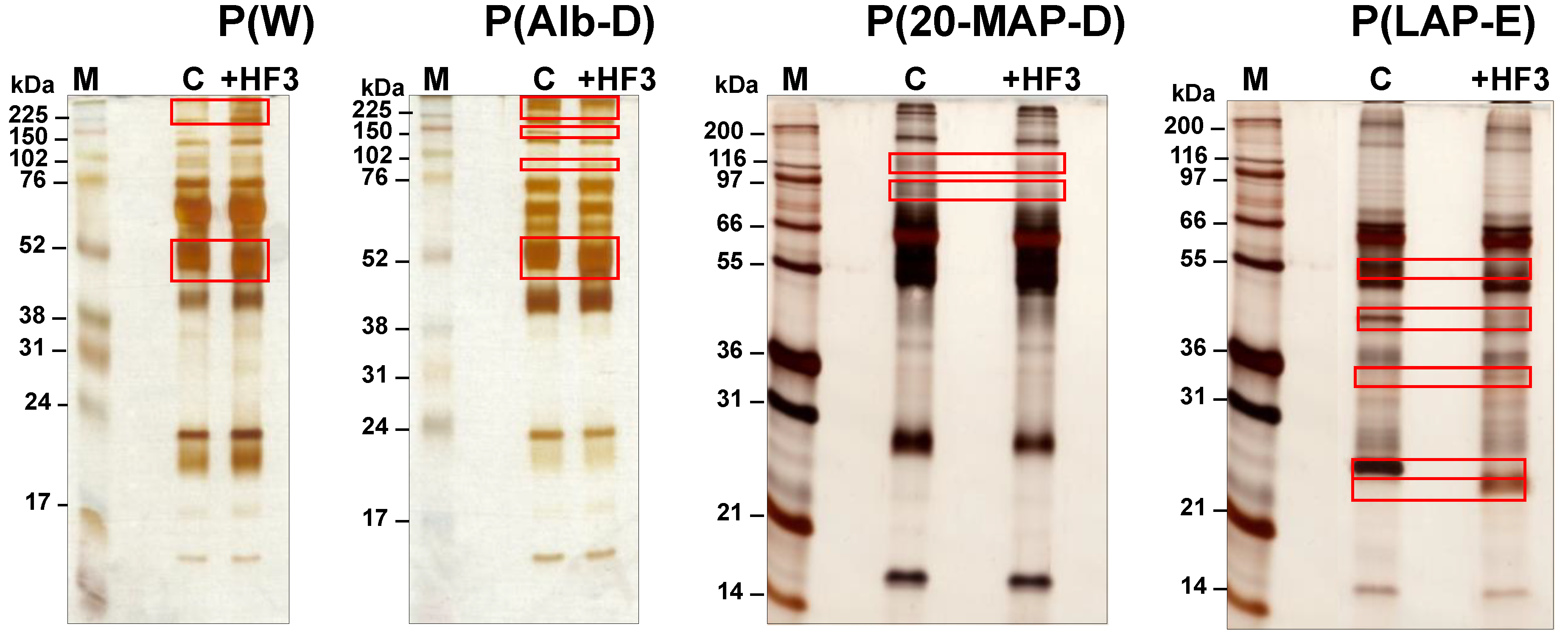
2.1. Electrophoretic Profiles of Plasma Samples Incubated with HF3
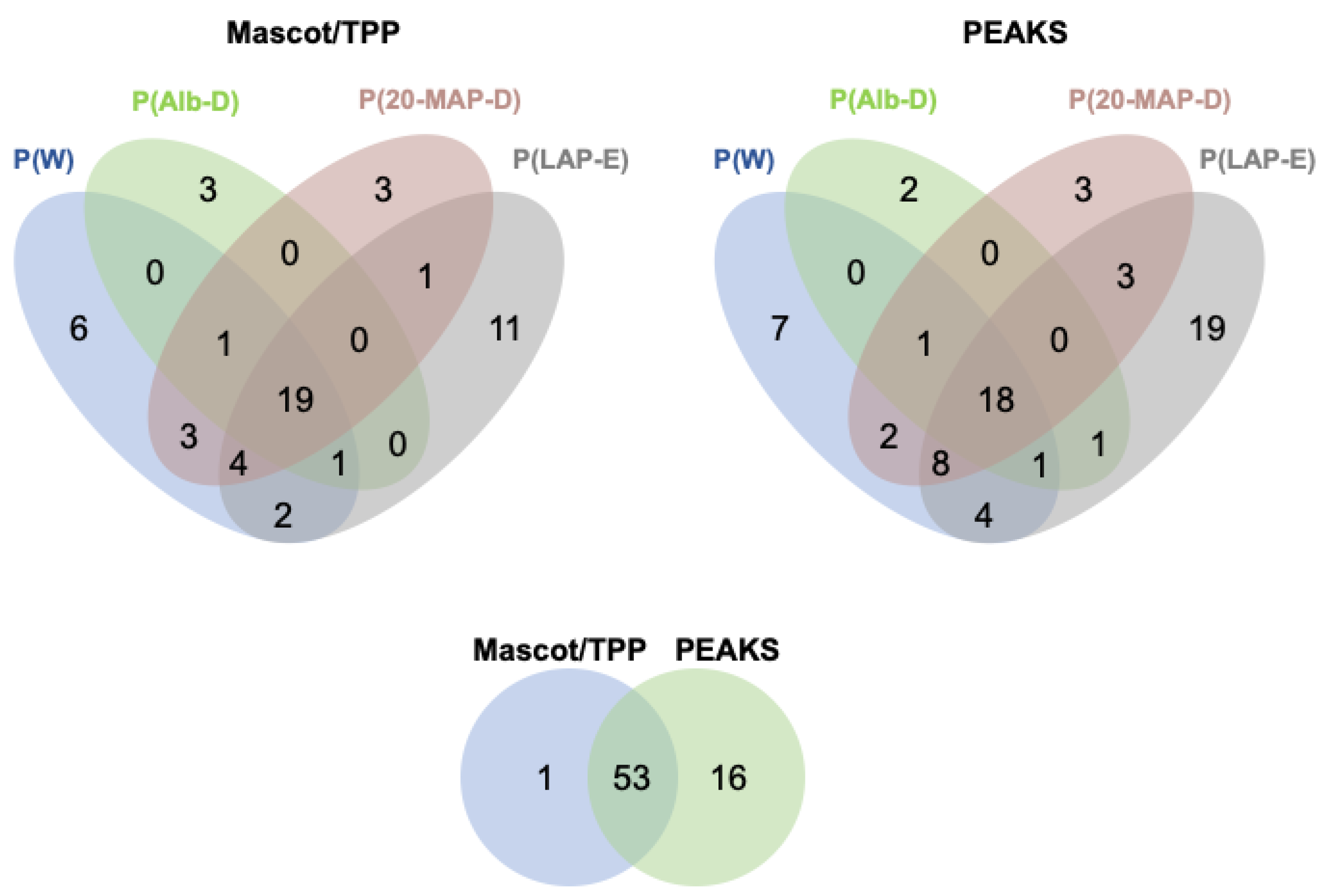
2.2. Peptidomic Profiling of Plasma Samples Incubated with HF3
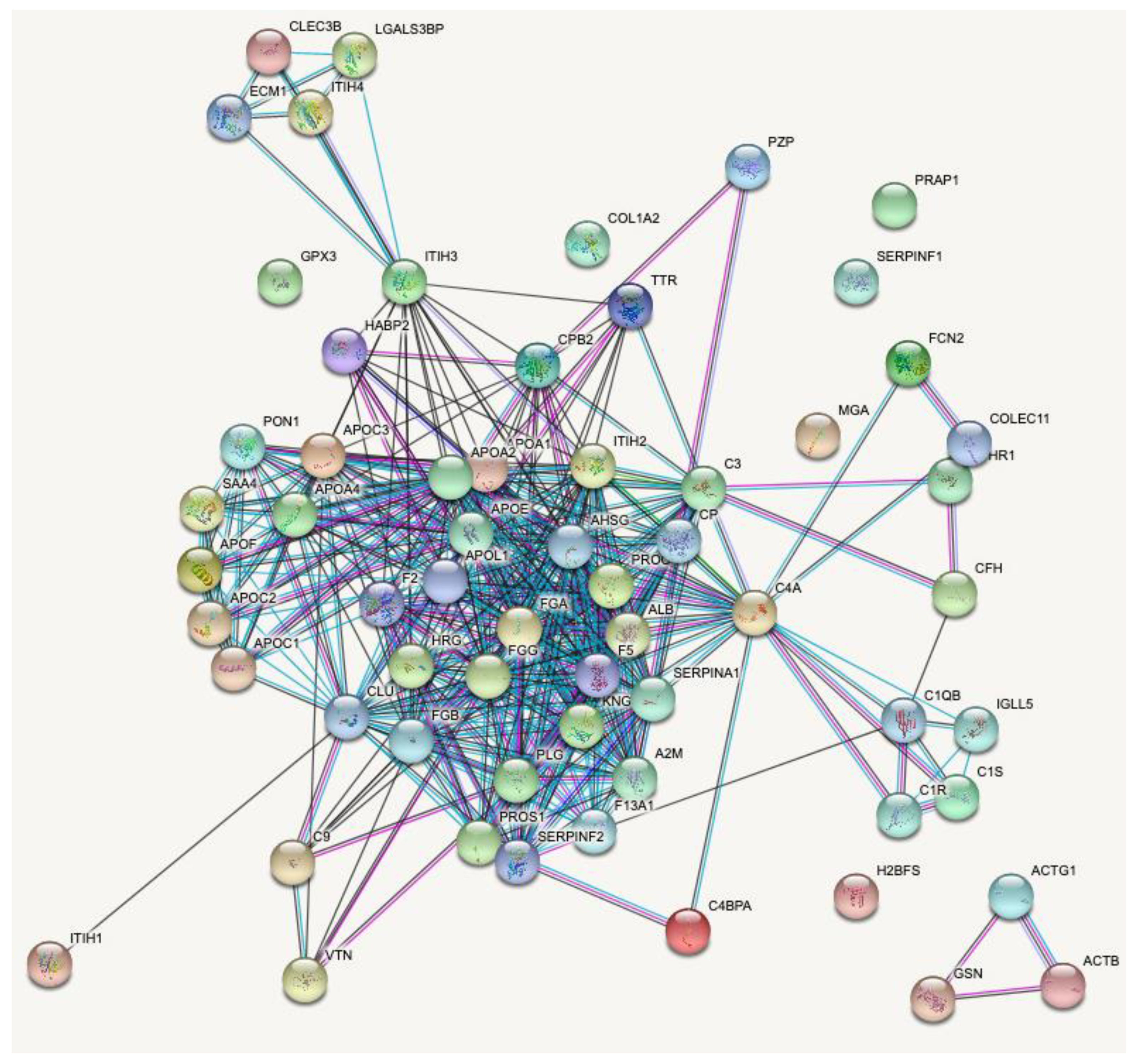
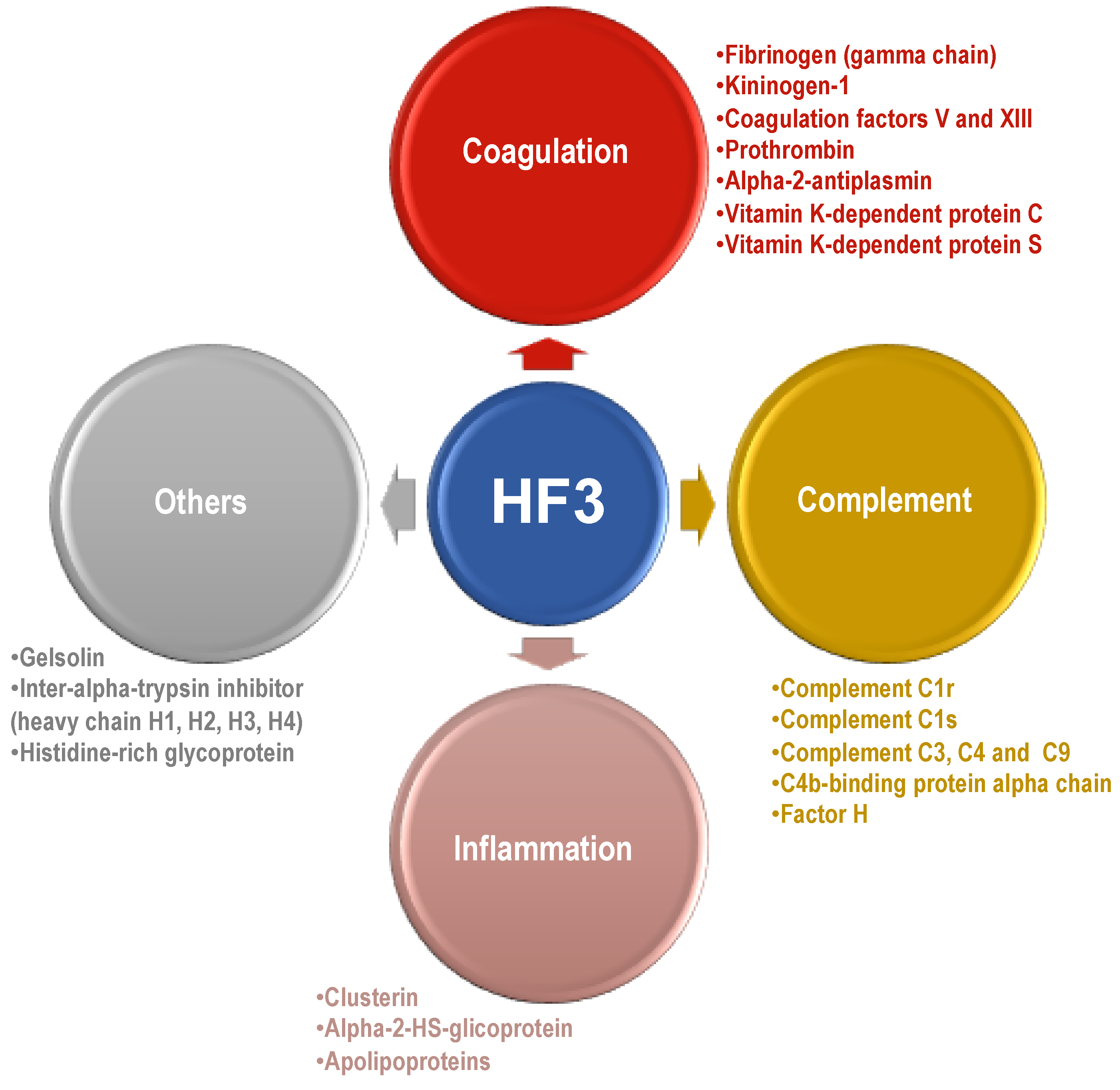
2.3. Proteins Degraded by HF3 in the Human Plasma
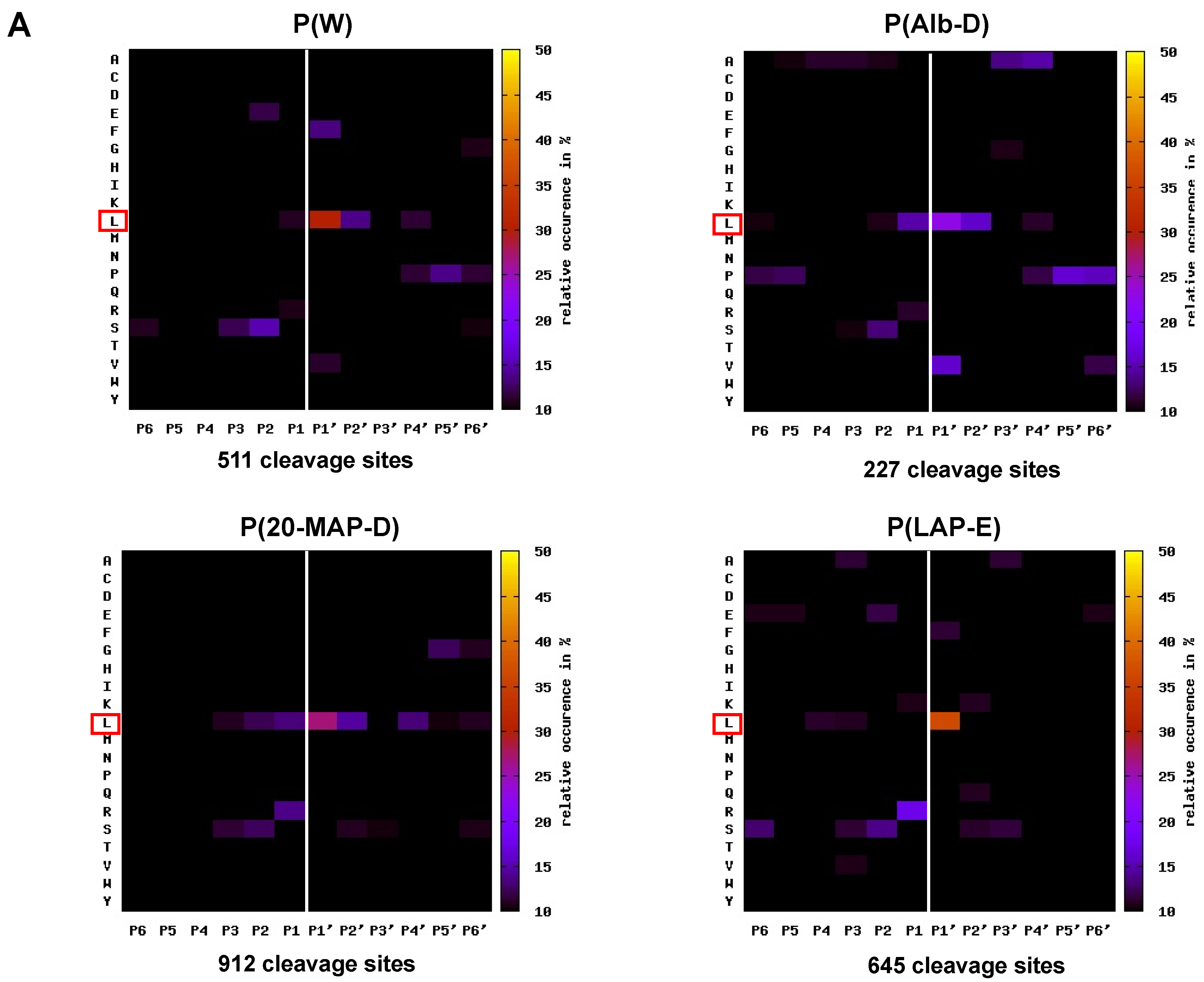
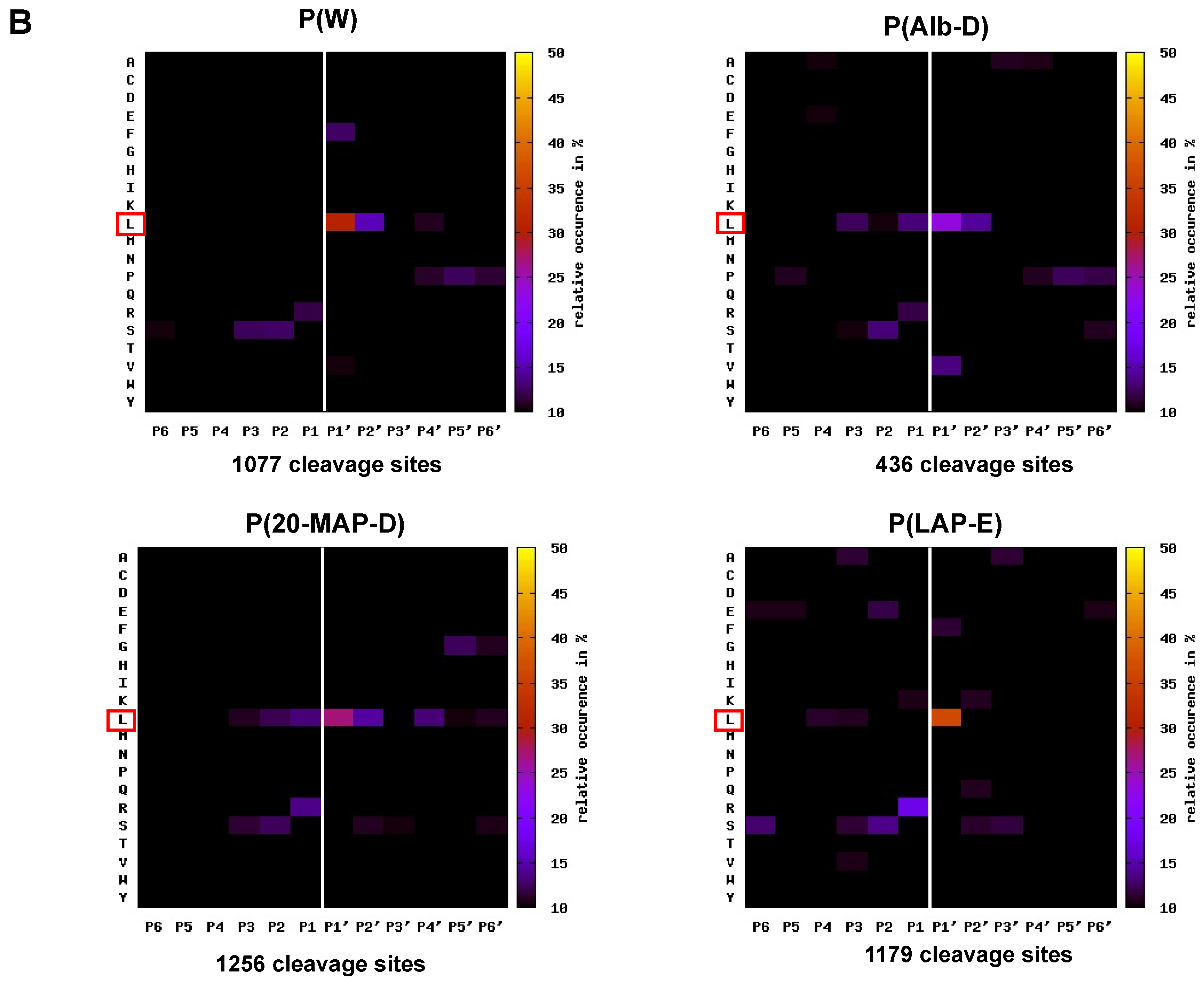
2.4. Mapping the Primary Specificity of HF3 on Plasma Proteins
3. Discussion
3.1. Human Plasma Preparations for Degradomic Analysis
3.2. Analysis of the Plasma Peptide Fraction
3.3. Analysis of the Primary Specificity of HF3 on Plasma Proteins
3.4. Plasma Proteins Cleaved by HF3
4. Conclusions
5. Materials and Methods
5.1. HF3
5.2. Analytical Procedures
5.3. Human Plasma
5.4. Albumin-Depleted Human Plasma
5.5. Human Plasma Depleted of 20 Most Abundant Proteins
5.6. Human Plasma Enriched in Low-Abundance Proteins
5.7. Proteolytic Activity of HF3 on Human Plasma
5.8. LC-MS/MS Analysis of the Plasma Peptide Fraction
5.9. Mass Spectrometry Data Analysis
5.10. Identification of Peptides Generated the Incubation of Fibrinogen with HF3
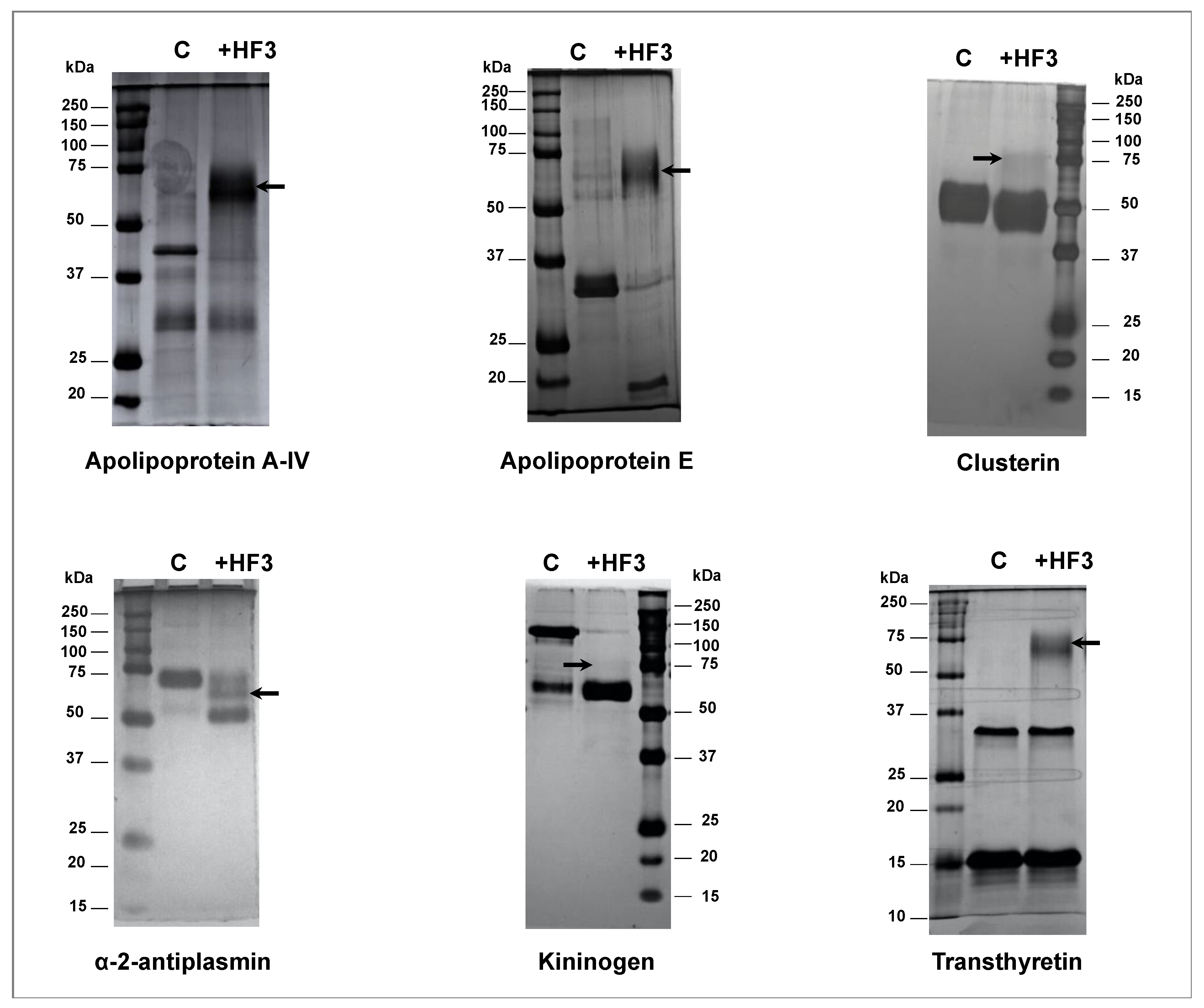
5.11. Validation of HF3 Substrates in Human Plasma by Incubation with HF3
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gutiérrez, J.M.; Escalante, T.; Rucavado, A.; Herrera, C. Hemorrhage caused by snake venom metalloproteinases: A journey of discovery and understanding. Toxins 2016, 8, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasturiratne, A.; Wickremasinghe, A.R.; De Silva, N.; Gunawardena, N.K.; Pathmeswaran, A.; Premaratna, R.; Savioli, L.; Lalloo, D.G.; De Silva, H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008, 5, e218. [Google Scholar] [CrossRef] [Green Version]
- Ownby, C.L.; Bjarnason, J.; Tu, A.T. Hemorrhagic toxins from rattlesnake (Crotalus atrox) venom. Pathogenesis of hemorrhage induced by three purified toxins. Am. J. Pathol. 1978, 93, 201–218. [Google Scholar]
- Ohsaka, A.; Ikezawa, H.; Kondo, H.; Kondo, S.; Uchida, N. Haemorrhagic activities of habu snake venom, and their relations to lethal toxicity, proteolytic activities and other pathological activities. Br. J. Exp. Pathol. 1960, 41, 478–486. [Google Scholar]
- Bjarnason, J.B.; Fox, J.W. Hemorrhagic metalloproteinases from snake venoms. Pharmacol. Ther. 1994, 62, 325–372. [Google Scholar] [CrossRef]
- White, J. Snake venoms and coagulopathy. Toxicon 2005, 45, 951–967. [Google Scholar] [CrossRef]
- Isbister, G. Procoagulant snake toxins: Laboratory studies, diagnosis, and understanding snakebite coagulopathy. Semin. Thromb. Hemost. 2009, 35, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M.T. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef]
- Oliveira, A.K.; Leme, A.P.; Asega, A.F.; Camargo, A.C.M.; Fox, J.W.; Serrano, S.M.T. New insights into the structural elements involved in the skin haemorrhage induced by snake venom metalloproteinases. Thromb. Haemost. 2010, 104, 485–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escalante, T.; Rucavado, A.; Fox, J.W.; Gutiérrez, J.M. Key events in microvascular damage induced by snake venom hemorrhagic metalloproteinases. J. Proteom. 2011, 74, 1781–1794. [Google Scholar] [CrossRef]
- Kini, R.M.; Koh, C.Y. Metalloproteases Affecting blood coagulation, fibrinolysis and platelet aggregation from snake venoms: Definition and nomenclature of interaction sites. Toxins 2016, 8, 284. [Google Scholar] [CrossRef] [Green Version]
- Assakura, M.T.; Reichl, A.P.; Mandelbaum, F.R. Comparison of immunological, biochemical and biophysical properties of three hemorrhagic factors isolated from the venom of Bothrops jararaca (jararaca). Toxicon 1986, 24, 943–946. [Google Scholar] [CrossRef]
- Oliveira, A.K.; Leme, A.F.P.; Assakura, M.T.; Menezes, M.C.; Zelanis, A.; Tashima, A.K.; Lopes-Ferreira, M.; Lima, C.; Camargo, A.C.; Fox, J.W.; et al. Simplified procedures for the isolation of HF3, bothropasin, disintegrin-like/cysteine-rich protein and a novel P-I metalloproteinase from Bothrops jararaca venom. Toxicon 2009, 53, 797–801. [Google Scholar] [CrossRef]
- Silva, C.A.; Zuliani, J.P.; Assakura, M.T.; Mentele, R.; Camargo, A.C.; Teixeira, C.F.; Serrano, S.M. Activation of αMβ2-mediated phagocytosis by HF3, a P-III class metalloproteinase isolated from the venom of Bothrops jararaca. Biochem. Biophys. Res. Commun. 2004, 322, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Asega, A.F.; Menezes, M.C.; Trevisan-Silva, D.; Carvalho, D.C.; Bertholim, L.; Oliveira, A.K.; Zelanis, A.; Serrano, S.M.T. Cleavage of proteoglycans, plasma proteins and the platelet-derived growth factor receptor in the hemorrhagic process induced by snake venom metalloproteinases. Sci. Rep. 2020, 10, 12912. [Google Scholar] [CrossRef] [PubMed]
- Asega, A.F.; Oliveira, A.K.; Menezes, M.C.; Neves-Ferreira, A.G.C.; Serrano, S.M. Interaction of Bothrops jararaca venom metalloproteinases with protein inhibitors. Toxicon 2014, 80, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menezes, M.C.; de Oliveira, A.K.; Melo, R.L.; Lopes-Ferreira, M.; Rioli, V.; Balan, A.; Leme, A.F.P.; Serrano, S.M. Disintegrin-like/cysteine-rich domains of the reprolysin HF3: Site-directed mutagenesis reveals essential role of specific residues. Biochimie 2011, 93, 345–351. [Google Scholar] [CrossRef]
- Menezes, M.C.; Imbert, L.; Kitano, E.S.; Vernet, T.; Serrano, S.M.T. Recombinant expression of the precursor of the hemorrhagic metalloproteinase HF3 and its non-catalytic domains using a cell-free synthesis system. Amino Acids 2016, 48, 2205–2214. [Google Scholar] [CrossRef] [PubMed]
- Leme, A.P.; Sherman, N.; Smalley, D.M.; Sizukusa, L.O.; Oliveira, A.K.; Menezes, M.C.; Fox, J.W.; Serrano, S.M.T. Hemorrhagic activity of HF3, a snake venom metalloproteinase: Insights from the proteomic analysis of mouse skin and blood plasma. J. Proteome Res. 2012, 11, 279–291. [Google Scholar] [CrossRef]
- Zelanis, A.; Oliveira, A.K.; Prudova, A.; Huesgen, P.F.; Tashima, A.K.; Kizhakkedathu, J.; Overall, C.M.; Serrano, S.M.T. Deep profiling of the cleavage specificity and human substrates of snake venom metalloprotease HF3 by proteomic identification of cleavage site specificity (PICS) using proteome derived peptide libraries and terminal amine isotopic labeling of substrates. J. Proteome Res. 2019, 18, 3419–3428. [Google Scholar] [CrossRef]
- Bertholim, L.; Zelanis, A.; Oliveira, A.K.; Serrano, S.M.T. Proteome-derived peptide library for the elucidation of the cleavage specificity of HF3, a snake venom metalloproteinase. Amino Acids 2016, 48, 1331–1335. [Google Scholar] [CrossRef]
- Leme, A.P.; Escalante, T.; Pereira, J.G.; Oliveira, A.K.; Sanchez, E.O.F.; Gutiérrez, J.M.; Serrano, S.M.T.; Fox, J.W. High resolution analysis of snake venom metalloproteinase (SVMP) peptide bond cleavage specificity using proteome based peptide libraries and mass spectrometry. J. Proteom. 2011, 74, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.L.; Anderson, N.G. The human plasma proteome. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef] [Green Version]
- Geyer, P.E.; Kulak, N.A.; Pichler, G.; Holdt, L.M.; Teupser, D.; Mann, M. Plasma Proteome profiling to assess human health and disease. Cell Syst. 2016, 2, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Schilling, O.; Huesgen, P.; Barré, O.; Keller, U.A.D.; Overall, C.M. Characterization of the prime and non-prime active site specificities of proteases by proteome-derived peptide libraries and tandem mass spectrometry. Nat. Protoc. 2011, 6, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kloczewiak, M.; Timmons, S.; Lukas, T.J.; Hawiger, J. Platelet receptor recognition site on human fibrinogen. Synthesis and structure-function relationship of peptides corresponding to the carboxy-terminal segment of the gamma chain. Biochemistry 1984, 23, 1767–1774. [Google Scholar] [CrossRef]
- Chen, R.; Doolittle, R.F. Isolation, Characterization, and location of a donor-acceptor unit from cross-linked fibrin. Proc. Natl. Acad. Sci. USA 1970, 66, 472–479. [Google Scholar] [CrossRef] [Green Version]
- Packham, M.A.; Taylor, D.M.; Yeo, E.L.; Gemmell, C.H.; Patil, S.; Lam, S.C.-T.; Rand, M.L. The fibrinogen γ chain dodecapeptide inhibits agonist-induced aggregation of rabbit platelets and fibrinogen binding to rabbit glycoprotein IIb-IIIa. Thromb. Haemost. 1999, 82, 1680–1686. [Google Scholar] [CrossRef]
- Millioni, R.; Tolin, S.; Puricelli, L.; Sbrignadello, S.; Fadini, G.P.; Tessari, P.; Arrigoni, G. High Abundance proteins depletion vs low abundance proteins enrichment: Comparison of methods to reduce the plasma proteome complexity. PLoS ONE 2011, 6, e19603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakimi, A.; Auluck, J.; Jones, G.D.D.; Ng, L.L.; Jones, D.J.L. Assessment of reproducibility in depletion and enrichment workflows for plasma proteomics using label-free quantitative data-independent LC-MS. Proteomics 2014, 14, 4–13. [Google Scholar] [CrossRef]
- Polaskova, V.; Kapur, A.; Khan, A.; Molloy, M.; Baker, M.S. High-abundance protein depletion: Comparison of methods for human plasma biomarker discovery. Electrophoresis 2010, 31, 471–482. [Google Scholar] [CrossRef]
- Govorukhina, N.; Keizer-Gunnink, A.; van der Zee, A.; de Jong, S.; de Bruijn, H.; Bischoff, R. Sample preparation of human serum for the analysis of tumor markers. J. Chromatogr. A 2003, 1009, 171–178. [Google Scholar] [CrossRef]
- Gong, Y.; Li, X.; Yang, B.; Ying, W.; Li, D.; Zhang, Y.; Dai, S.; Cai, Y.; Wang, J.; He, A.F.; et al. Different immunoaffinity fractionation strategies to characterize the human plasma proteome. J. Proteome Res. 2006, 5, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Quero, C.; Colomé, N.; Prieto, M.R.; Carrascal, M.; Posada, M.; Gelpí, E.; Abian, J. Determination of protein markers in human serum: Analysis of protein expression in toxic oil syndrome studies. Proteomics 2004, 4, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Boschetti, E.; Righetti, P.G. The ProteoMiner in the proteomic arena: A non-depleting tool for discovering low-abundance species. J. Proteom. 2008, 71, 255–264. [Google Scholar] [CrossRef]
- Guerranti, R.; Cortelazzo, A.; Hope-Onyekwere, N.S.; Furlani, E.; Cerutti, H.; Puglia, M.; Bini, L.; Leoncini, R. In vitro effects of Echis carinatus venom on the human plasma proteome. Proteomics 2010, 10, 3712–3722. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.K.; Park, S.M.; Kim, S.Y.; Lee, S.-T. A proteomic approach to identify substrates of matrix metalloproteinase-14 in human plasma. Biochim. Biophys. Acta Proteins Proteom. 2004, 1702, 79–87. [Google Scholar] [CrossRef]
- Escalante, T.; Rucavado, A.; Pinto, A.F.M.; Terra, R.M.S.; Gutiérrez, J.M.; Fox, J.W. Wound exudate as a proteomic window to reveal different mechanisms of tissue damage by snake venom toxins. J. Proteome Res. 2009, 8, 5120–5131. [Google Scholar] [CrossRef]
- Menezes, M.C.; Kitano, E.S.; Bauer, V.C.; Oliveira, A.K.; Cararo-Lopes, E.; Nishiyama, M.Y.; Zelanis, A.; Serrano, S.M. Early response of C2C12 myotubes to a sub-cytotoxic dose of hemorrhagic metalloproteinase HF3 from Bothrops jararaca venom. J. Proteom. 2019, 198, 163–176. [Google Scholar] [CrossRef]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 2012, 11, M111.010587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckhard, U.; Huesgen, P.F.; Schilling, O.; Bellac, C.L.; Butler, G.S.; Cox, J.H.; Dufour, A.; Goebeler, V.; Kappelhoff, R.; Keller, U.A.D.; et al. Active site specificity profiling of the matrix metalloproteinase family: Proteomic identification of 4300 cleavage sites by nine MMPs explored with structural and synthetic peptide cleavage analyses. Matrix Biol. 2016, 49, 37–60. [Google Scholar] [CrossRef]
- Kleifeld, O.; Doucet, A.; Keller, U.A.D.; Prudova, A.; Schilling, O.; Kainthan, R.K.; Starr, A.E.; Foster, L.J.; Kizhakkedathu, J.N.; Overall, C.M. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 2010, 28, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Prudova, A.; Keller, U.A.D.; Butler, G.S.; Overall, C.M. Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell. Proteom. 2010, 9, 894–911. [Google Scholar] [CrossRef] [Green Version]
- Schlage, P.; Egli, F.E.; Nanni, P.; Wang, L.W.; Kizhakkedathu, J.N.; Apte, S.; Keller, U.A.D. Time-resolved analysis of the matrix metalloproteinase 10 substrate degradome. Mol. Cell. Proteom. 2014, 13, 580–593. [Google Scholar] [CrossRef] [Green Version]
- Kloczewiak, M.; Timmons, S.; Bednarek, M.A.; Sakon, M.; Hawiger, J. Platelet receptor recognition domain on the .gamma. chain of human fibrinogen and its synthetic peptide analogues. Biochemistry 1989, 28, 2915–2919. [Google Scholar] [CrossRef]
- Springer, T.A.; Zhu, J.; Xiao, T. Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ. J. Cell Biol. 2008, 182, 791–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrell, D.H.; Thiagarajan, P.; Chung, D.W.; Davie, E.W. Role of fibrinogen alpha and gamma chain sites in platelet aggregation. Proc. Natl. Acad. Sci. USA 1992, 89, 10729–10732. [Google Scholar] [CrossRef] [Green Version]
- Stockley, R.A. The multiple facets of alpha-1-antitrypsin. Ann. Transl. Med. 2015, 3, 130–137. [Google Scholar] [CrossRef]
- Robbins, K.C.; Summaria, L.; Hsieh, B.; Shah, R.J. The peptide chains of human plasmin. Mechanism of activation of human plasminogen to plasmin. J. Biol. Chem. 1967, 242, 2333–2342. [Google Scholar] [CrossRef]
- O’Reilly, M.S. Angiostatin: A novel angiogenesis inhibitor that mediates the suppression of metastases by a lewis lung carcinoma. Cell 1994, 79, 315–328. [Google Scholar] [CrossRef]
- Cao, Y.; Xue, L. Angiostatin. Semin. Thromb. Hemost. 2004, 30, 83–93. [Google Scholar] [CrossRef]
- Cornelius, A.L.; Nehring, L.C.; Harding, E.; Bolanowski, M.; Welgus, H.G.; Kobayashi, D.K.; Pierce, R.A.; Shapiro, S.D. Matrix metalloproteinases generate angiostatin: Effects on neovascularization. J. Immunol. 1998, 161, 6845–6852. [Google Scholar]
- Ho, P.L.; Serrano, S.M.T.; Chudzinski-Tavassi, A.M.; Moura-Da-Silva, A.M.; Mentele, R.; Caldas, C.; Oliva, M.L.V.; Batista, I.D.F.C.; de Oliveira, M.L.S. Angiostatin-like molecules are generated by snake venom metalloproteinases. Biochem. Biophys. Res. Commun. 2002, 294, 879–885. [Google Scholar] [CrossRef]
- Moroi, M.; Aoki, N. Isolation and characterization of alpha2-plasmin inhibitor from human plasma. A novel proteinase inhibitor which inhibits activator-induced clot lysis. J. Biol. Chem. 1976, 251, 5956–5965. [Google Scholar] [CrossRef]
- Lee, K.N.; Lee, C.S.; Tae, W.-C.; Jackson, K.W.; Christiansen, V.J.; Mckee, P.A. Crosslinking of α2-Antiplasmin to Fibrin. Ann. N. Y. Acad. Sci. 2006, 936, 335–339. [Google Scholar] [CrossRef]
- Koyama, T.; Koike, Y.; Toyota, S.; Miyagi, F.; Suzuki, N.; Aoki, N. Different NH2-terminal form with 12 additional residues of α2-plasmin inhibitor from human plasma and culture media of HEP G2 cells. Biochem. Biophys. Res. Commun. 1994, 200, 417–422. [Google Scholar] [CrossRef]
- Sumi, Y.; Ichikawa, Y.; Nakamura, Y.; Miura, O.; Aoki, N. Expression and characterization of Pro α2-Plasmin Inhibitor. J. Biochem. 1989, 106, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Muszbek, L.; Bereczky, Z.; Bagoly, Z.; Komáromi, I.; Katona, É. Factor XIII: A coagulation factor with multiple plasmatic and cellular functions. Physiol. Rev. 2011, 91, 931–972. [Google Scholar] [CrossRef] [Green Version]
- Pisano, J.J.; Finlayson, J.S.; Peyton, M.P. Cross-link in fibrin polymerized by Factor XIII: Egr-(ggr-Glutamyl) lysine. Science 1968, 160, 892–893. [Google Scholar] [CrossRef] [PubMed]
- Komáromi, I.; Bagoly, Z.; Muszbek, L. Factor XIII: Novel structural and functional aspects. J. Thromb. Haemost. 2011, 9, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, N.; Kitagawa, H.; Fukushima, D.; Takagaki, Y.; Miyata, T.; Nakanishi, S. Structural organization of the human kininogen gene and a model for its evolution. J. Biol. Chem. 1985, 260, 8610–8617. [Google Scholar] [CrossRef]
- Salvesen, G.; Parkes, C.; Abrahamson, M.; Grubb, A.; Barrett, A.J. Human low-Mr kininogen contains three copies of a cystatin sequence that are divergent in structure and in inhibitory activity for cysteine proteinases. Biochem. J. 1986, 234, 429–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, A.P.; Silverberg, M. The coagulation-kinin pathway of human plasma. Blood 1987, 70, 1–15. [Google Scholar] [CrossRef]
- Thompson, E.R.; Mandle, R.; Kaplan, A.P. Characterization of human high molecular weight kininogen. Procoagulant activity associated with the light chain of kinin-free high molecular weight kininogen. J. Exp. Med. 1978, 147, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Sugo, T.; Ikari, N.; Hashimoto, N.; Maruyama, I.; Han, Y.N.; Iwanaga, S.; Fujii, S. Role of bovine high-molecular-weight (HMW) kininogen in contact-mediated activation of bovine Factor XII. Adv. Exp. Med. Biol. 1979, 120, 19–37. [Google Scholar]
- Zaitsev, V.N.; Zaitseva, I.; Papiz, M.; Lindley, P.F. An X-ray crystallographic study of the binding sites of the azide inhibitor and organic substrates to ceruloplasmin, a multi-copper oxidase in the plasma. JBIC J. Biol. Inorg. Chem. 1999, 4, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Ehrenwald, E.; Fox, P. Isolation of nonlabile human ceruloplasmin by chromatographic removal of a plasma metalloproteinase. Arch. Biochem. Biophys. 1994, 309, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Gitlin, J.D. Transcriptional regulation of ceruloplasmin gene expression during inflammation. J. Biol. Chem. 1988, 263, 6281–6287. [Google Scholar] [CrossRef]
- Fleming, R.E.; Whitman, I.P.; Gitlin, J.D. Induction of ceruloplasmin gene expression in rat lung during inflammation and hyperoxia. Am. J. Physiol. Cell. Mol. Physiol. 1991, 260, L68–L74. [Google Scholar] [CrossRef]
- Walker, F.J.; Fay, P.J. Characterization of an interaction between protein C and ceruloplasmin. J. Biol. Chem. 1990, 265, 1834–1836. [Google Scholar] [CrossRef]
- Reid, K.B.M.; Porter, R.R. The proteolytic activation systems of complement. Annu. Rev. Biochem. 1981, 50, 433–464. [Google Scholar] [CrossRef]
- Fearon, D.T.; Carroll, M.C. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu. Rev. Immunol. 2000, 18, 393–422. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.C. The complement system in regulation of adaptive immunity. Nat. Immunol. 2004, 5, 981–986. [Google Scholar] [CrossRef]
- Arlaud, G.J.; Volanakis, J.E.; Thielens, N.M.; Narayana, S.V.; Rossi, V.; Xu, Y. The Atypical serine proteases of the complement system. Adv. Immunol. 1998, 69, 249–307. [Google Scholar] [CrossRef]
- Vogel, C.-W.; Müller-Eberhard, H.J. The cobra complement system: I. The alternative pathway of activation. Dev. Comp. Immunol. 1985, 9, 311–325. [Google Scholar] [CrossRef]
- Tambourgi, D.; van den Berg, C.W. Animal venoms/toxins and the complement system. Mol. Immunol. 2014, 61, 153–162. [Google Scholar] [CrossRef]
- Jeong, S.; Ledee, D.; Gordon, G.M.; Itakura, T.; Patel, N.; Martin, A.; Fini, M.E. Interaction of Clusterin and matrix metalloproteinase-9 and its implication for epithelial homeostasis and inflammation. Am. J. Pathol. 2012, 180, 2028–2039. [Google Scholar] [CrossRef] [Green Version]
- Mori, K.; Emoto, M.; Inaba, M. Fetuin-A: A multifunctional protein. Recent Pat. Endocr. Metab. Immune Drug Discov. 2011, 5, 124–146. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, M.; Soda, K.; Sama, A.; Tracey, K.J. Fetuin protects the fetus from TNF. Lancet 1997, 350, 861–862. [Google Scholar] [CrossRef]
- Ombrellino, M.; Wang, H.; Yang, H.; Zhang, M.; Vishnubhakat, J.; Frazier, A.; Scher, L.A.; Friedman, S.G.; Tracey, K.J. Fetuin, a negative acute phase protein, attenuates TNF synthesis and the innate inflammatory response to carrageenan. Shock 2001, 15, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, J.; Lottaz, D.; Meyer, K.; Yiallouros, I.; Jahnen-Dechent, W.; Stöcker, W.; Becker-Pauly, C. Fetuin-A and cystatin C are endogenous inhibitors of human meprin metalloproteases. Biochemistry 2010, 49, 8599–8607. [Google Scholar] [CrossRef] [PubMed]
- Dholey, Y.; Chaudhuri, A.; Chakraborty, S.S. An integrated in silico approach to understand protein–protein interactions: Human meprin-β with fetuin-A. J. Biomol. Struct. Dyn. 2020, 38, 2080–2092. [Google Scholar] [CrossRef] [PubMed]
- Gisterå, A.; Hansson, G.K. The immunology of atherosclerosis. Nat. Rev. Nephrol. 2017, 13, 2368–2380. [Google Scholar] [CrossRef]
- Kim, M.-S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Park, S.M.; Lee, S.-T. Apolipoprotein C-II is a novel substrate for matrix metalloproteinases. Biochem. Biophys. Res. Commun. 2006, 339, 47–54. [Google Scholar] [CrossRef]
- Park, J.Y.; Park, J.H.; Jang, W.; Hwang, I.-K.; Kim, I.J.; Kim, H.-J.; Cho, K.-H.; Lee, S.-T. Apolipoprotein A-IV is a novel substrate for matrix metalloproteinases. J. Biochem. 2012, 151, 291–298. [Google Scholar] [CrossRef]
- El-Asmar, M.; Swaney, J. Proteolysis in vitro of low and high density lipoproteins in human plasma by Cerastes cerastes (Egyptian sand viper) venom. Toxicon 1988, 26, 809–816. [Google Scholar] [CrossRef]
- Lind, S.E.; Smith, D.B.; Janmey, P.A.; Stossel, T.P. Role of plasma gelsolin and the vitamin D-binding protein in clearing actin from the circulation. J. Clin. Investig. 1986, 78, 736–742. [Google Scholar] [CrossRef] [Green Version]
- Haddad, J.G.; Harper, K.D.; Guoth, M.; Pietra, G.G.; Sanger, J.W. Angiopathic consequences of saturating the plasma scavenger system for actin. Proc. Natl. Acad. Sci. USA 1990, 87, 1381–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epstein, F.H.; Lee, W.M.; Galbraith, R.M. The extracellular actin-scavenger system and actin toxicity. N. Engl. J. Med. 1992, 326, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-M.; Hwang, I.K.; Kim, S.Y.; Lee, S.-J.; Park, K.-S.; Lee, S.-T. Characterization of plasma gelsolin as a substrate for matrix metalloproteinases. Proteomics 2006, 6, 1192–1199. [Google Scholar] [CrossRef]
- Zhuo, L.; Hascall, V.C.; Kimata, K. Inter-α-trypsin Inhibitor, a covalent protein-glycosaminoglycan-protein complex. J. Biol. Chem. 2004, 279, 38079–38082. [Google Scholar] [CrossRef] [Green Version]
- Lord, M.S.; Melrose, J.; Day, A.J.; Whitelock, J.M. The inter-α-trypsin inhibitor family: Versatile molecules in biology and pathology. J. Histochem. Cytochem. 2020, 68, 907–927. [Google Scholar] [CrossRef]
- Fries, E.; Blom, A. Bikunin—Not just a plasma proteinase inhibitor. Int. J. Biochem. Cell Biol. 2000, 32, 125–137. [Google Scholar] [CrossRef]
- Bost, F.; Diarra-Mehrpour, M.; Martin, J.-P. Inter-alpha-trypsin inhibitor proteoglycan family. A group of proteins binding and stabilizing the extracellular matrix. Eur. J. Biochem. 1998, 252, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, L.; Kimata, K. Structure and function of inter-α-trypsin inhibitor heavy chains. Connect. Tissue Res. 2008, 49, 311–320. [Google Scholar] [CrossRef]
- He, H.; Zhang, S.; Tighe, S.; Son, J.; Tseng, S.C.G. Immobilized heavy chain-hyaluronic acid polarizes lipopolysaccharide-activated macrophages toward M2 phenotype. J. Biol. Chem. 2013, 288, 25792–25803. [Google Scholar] [CrossRef] [Green Version]
- Catanese, J.J.; Kress, L.F. Enzymatic digestion of human plasma inter-α-trypsin inhibitor by snake venom metalloproteinases. Comp. Biochem. Physiol. Part B Comp. Biochem. 1985, 80, 507–512. [Google Scholar] [CrossRef]
- Jones, A.L.; Hulett, M.; Parish, C.R. Histidine-rich glycoprotein: A novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol. Cell Biol. 2005, 83, 106–118. [Google Scholar] [CrossRef]
- Poon, I.; Patel, K.K.; Davis, D.A.S.; Parish, C.; Hulett, M. Histidine-rich glycoprotein: The Swiss Army knife of mammalian plasma. Blood 2011, 117, 2093–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, L.; Saigo, K.; Grant, D. Heparin binds to human monocytes and modulates their procoagulant activities and secretory phenotypes. Effects of histidine-rich glycoprotein. Blood 1989, 73, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Borza, D.-B.; Morgan, W.T. Acceleration of plasminogen activation by tissue plasminogen activator on surface-bound histidine-proline-rich glycoprotein. J. Biol. Chem. 1997, 272, 5718–5726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lijnen, H.; Hoylaerts, M.; Collen, D. Isolation and characterization of a human plasma protein with affinity for the lysine binding sites in plasminogen. Role in the regulation of fibrinolysis and identification as histidine-rich glycoprotein. J. Biol. Chem. 1980, 255, 10214–10222. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Mortz, E.; Krogh, T.N.; Vorum, H.; Görg, A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 2001, 1, 1359–1363. [Google Scholar] [CrossRef]
- Travis, J.; Pannell, R. Selective removal of albumin from plasma by affinity chromatography. Clin. Chim. Acta 1973, 49, 49–52. [Google Scholar] [CrossRef]
- Subramanian, S.; Ross, P.D. Dye-ligand affinity chromatography: The interaction of cibacron blue F3GA® with Proteins and Enzyme. Crit. Rev. Biochem. 1984, 16, 169–205. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Ríos, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef]



| Number of Spectra * (exp. 1/exp. 2/exp. 3) | |||
|---|---|---|---|
| Identification | Protein | Mascot/TPP | PEAKS |
| ACTB_HUMAN | Actin cytoplasmic 1 | 0/0/0 | 7/3/0 |
| A1AT_HUMAN | Alpha-1-antitrypsin | 1/0/1 | 6/10/5 |
| A2AP_HUMAN | Alpha-2-antiplasmin | 12/13/13 | 20/24/18 |
| FETUA_HUMAN | Alpha-2-HS-glycoprotein | 75/94/92 | 270/437/391 |
| A2MG_HUMAN | Alpha-2-macroglobulin | 2/0/2 | 9/4/8 |
| APOA1_HUMAN | Apolipoprotein A-I | 9/25/14 | 43/58/50 |
| APOA2_HUMAN | Apolipoprotein A-II | 13/30/19 | 63/70/60 |
| APOA4_HUMAN | Apolipoprotein A-IV | 5/6/7 | 23/21/16 |
| APOC1_HUMAN | Apolipoprotein C-I | 0/0/0 | 4/5/0 |
| APOC2_HUMAN | Apolipoprotein C-II | 7/17/11 | 30/41/30 |
| APOC3_HUMAN | Apolipoprotein C-III | 9/19/13 | 34/43/30 |
| APOE_HUMAN | Apolipoprotein E | 3/4/3 | 10/9/10 |
| APOF_HUMAN | Apolipoprotein F | 2/5/3 | 7/11/12 |
| APOL1_HUMAN | Apolipoprotein L1 | 4/9/5 | 36/38/35 |
| CLUS_HUMAN | Clusterin | 17/42/29 | 89/108/90 |
| FA5_HUMAN | Coagulation factor V | 0/0/0 | 3/0/3 |
| C1R_HUMAN | Complement C1r subcomponent | 1/2/2 | 18/15/15 |
| C1S_HUMAN | Complement C1s subcomponent | 3/3/2 | 26/20/16 |
| CO3_HUMAN | Complement C3 | 9/18/16 | 20/26/21 |
| CO4A_HUMAN | Complement C4-A | 19/30/24 | 61/76/67 |
| CO9_HUMAN | Complement component C9 | 0/2/1 | 3/4/0 |
| CFAH_HUMAN | Complement factor H | 1/2/0 | 3/2/0 |
| FIBA_HUMAN | Fibrinogen alpha chain | 126/221/163 | 369/602/462 |
| FIBB_HUMAN | Fibrinogen beta chain | 16/52/32 | 39/53/45 |
| FINC_HUMAN | Fibronectin | 7/12/6 | 29/29/22 |
| GELS_HUMAN | Gelsolin | 5/8/7 | 28/21/25 |
| HRG_HUMAN | Histidine-rich glycoprotein | 10/10/8 | 24/55/39 |
| H2BFS_HUMAN | Histone H2B type F-S | 0/0/0 | 4/1/0 |
| IGHG1_HUMAN | Ig gamma-1 chain C region | 0/5/1 | 6/8/7 |
| IGHM_HUMAN | Ig mu chain C region | 2/5/2 | 6/6/6 |
| ITIH1_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H1 | 6/5/6 | 17/17/19 |
| ITIH2_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H2 | 51/92/79 | 204/230/224 |
| ITIH3_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H3 | 1/2/1 | 6/4/5 |
| ITIH4_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H4 | 24/37/28 | 93/103/94 |
| KNG1_HUMAN | Kininogen-1 | 8/8/10 | 33/21/30 |
| PEDF_HUMAN | Pigment epithelium-derived factor | 3/4/3 | 5/5/4 |
| PZP_HUMAN | Pregnancy zone protein | 0/1/2 | 5/5/5 |
| THRB_HUMAN | Prothrombin | 17/25/20 | 63/62/65 |
| ALBU_HUMAN | Serum albumin | 0/0/0 | 0/3/2 |
| TTHY_HUMAN | Transthyretin | 6/17/17 | 43/43/43 |
| VTNC_HUMAN | Vitronectin | 18/31/24 | 74/78/71 |
| Number of Spectra * (exp. 1/exp. 2/exp. 3) | |||
|---|---|---|---|
| Identification | Protein | Mascot/TPP | PEAKS |
| ACTG_HUMAN | Actin cytoplasmic 1 | 0/0/0 | 1/0/1 |
| A1AT_HUMAN | Alpha-1-antitrypsin | 1/10/11 | 16/24/25 |
| A2AP_HUMAN | Alpha-2-antiplasmin | 10/12/11 | 17/18/20 |
| FETUA_HUMAN | Alpha-2-HS-glycoprotein | 84/109/117 | 219/226/230 |
| APOA1_HUMAN | Apolipoprotein A-I | 13/0/1 | 33/8/6 |
| APOA2_HUMAN | Apolipoprotein A-II | 5/3/1 | 17/13/11 |
| APOA4_HUMAN | Apolipoprotein A-IV | 8/5/7 | 13/13/15 |
| APOC2_HUMAN | Apolipoprotein C-II | 4/3/2 | 11/8/8 |
| APOC3_HUMAN | Apolipoprotein C-III | 6/12/10 | 21/25/27 |
| APOE_HUMAN | Apolipoprotein E | 3/2/2 | 2/4/4 |
| APOL1_HUMAN | Apolipoprotein L1 | 0/4/1 | 4/8/10 |
| CERU_HUMAN | Ceruloplasmin | 0/2/3 | 2/6/9 |
| CLUS_HUMAN | Clusterin | 1/2/2 | 5/16/12 |
| CO3_HUMAN | Complement C3 | 0/2/4 | 0/2/8 |
| FIBA_HUMAN | Fibrinogen alpha chain | 0/3/14 | 2/13/41 |
| FIBB_HUMAN | Fibrinogen beta chain | 7/14/20 | 13/20/28 |
| FINC_HUMAN | Fibronectin | 0/1/2 | 0/0/0 |
| GPX3_HUMAN | Glutathione peroxidase 3 | 0/0/0 | 5/7/0 |
| IGHG1_HUMAN | Ig gamma-1 chain C region | 2/1/0 | 0/0/0 |
| ITIH2_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H2 | 2/7/18 | 7/26/42 |
| ITIH4_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H4 | 0/4/4 | 0/8/7 |
| MGAP_HUMAN | Isoform 3 of MAX gene-associated protein | 4/2/0 | 0/0/0 |
| KNG1_HUMAN | Kininogen-1 | 1/2/1 | 2/8/6 |
| THRB_HUMAN | Prothrombin | 1/2/5 | 2/6/12 |
| PON1_HUMAN | Serum paraoxonase/arylesterase 1 | 7/15/21 | 27/36/52 |
| TTHY_HUMAN | Transthyretin | 27/33/34 | 44/57/68 |
| Number of Spectra * (exp. 1/exp. 2/exp. 3) | |||
|---|---|---|---|
| Identification | Protein | Mascot/TPP | PEAKS |
| ACTB_HUMAN | Actin cytoplasmic 1 | 6/0/2 | 18/0/2 |
| A2AP_HUMAN | Alpha-2-antiplasmin | 70/28/20 | 81/39/30 |
| FETUA_HUMAN | Alpha-2-HS-glycoprotein | 270/150/134 | 454/232/245 |
| A2MG_HUMAN | Alpha-2-macroglobulin | 5/4/3 | 7/6/4 |
| APOA1_HUMAN | Apolipoprotein A-I | 69/39/36 | 91/58/62 |
| APOA2_HUMAN | Apolipoprotein A-II | 39/19/21 | 56/30/37 |
| APOA4_HUMAN | Apolipoprotein A-IV | 28/48/15 | 56/71/31 |
| APOC1_HUMAN | Apolipoprotein C-I | 0/0/0 | 2/2/0 |
| APOC2_HUMAN | Apolipoprotein C-II | 39/4/10 | 46/6/18 |
| APOC3_HUMAN | Apolipoprotein C-III | 55/9/10 | 68/14/17 |
| APOE_HUMAN | Apolipoprotein E | 4/9/2 | 8/11/7 |
| APOF_HUMAN | Apolipoprotein F | 17/7/2 | 26/4/0 |
| APOL1_HUMAN | Apolipoprotein L1 | 8/4/7 | 8/3/19 |
| CBPB2_HUMAN | Carboxypeptidase B2 | 0/0/0 | 1/0/4 |
| CLUS_HUMAN | Clusterin | 5/2/8 | 10/4/30 |
| F13A_HUMAN | Coagulation factor XIII A chain | 5/0/1 | 5/0/1 |
| C1R_HUMAN | Complement C1r subcomponent | 5/0/2 | 0/0/0 |
| CO3_HUMAN | Complement C3 | 5/2/3 | 5/2/6 |
| CO4A_HUMAN | Complement C4-A | 6/0/10 | 9/0/15 |
| ECM1_HUMAN | Extracellular matrix protein 1 | 0/0/0 | 4/3/0 |
| FIBA_HUMAN | Fibrinogen alpha chain | 159/50/83 | 162/0/122 |
| FIBB_HUMAN | Fibrinogen beta chain | 31/38/14 | 34/44/17 |
| FINC_HUMAN | Fibronectin | 4/1/8 | 6/5/22 |
| GELS_HUMAN | Gelsolin | 49/18/14 | 68/23/42 |
| HRG_HUMAN | Histidine-rich glycoprotein | 2/6/14 | 5/7/29 |
| IGHG1_HUMAN | Ig gamma-1 chain C region | 4/0/9 | 4/0/12 |
| ITIH2_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H2 | 20/18/34 | 31/23/69 |
| ITIH3_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H3 | 0/0/0 | 2/0/9 |
| ITIH4_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H4 | 131/44/38 | 174/56/80 |
| FIBG_HUMAN | Isoform Gamma-A of Fibrinogen gamma chain | 2/19/0 | 3/19/0 |
| KNG1_HUMAN | Kininogen-1 | 7/1/3 | 17/3/9 |
| THRB_HUMAN | Prothrombin | 1/2/23 | 1/2/61 |
| SAA4_HUMAN | Serum amyloid A-4 protein | 0/0/0 | 7/1/0 |
| TETN_HUMAN | Tetranectin | 5/2/5 | 5/2/6 |
| TTHY_HUMAN | Transthyretin | 9/2/10 | 10/2/15 |
| VTNC_HUMAN | Vitronectin | 33/9/11 | 43/12/15 |
| Number of Spectra * (exp 1/exp 2/exp 3) | |||
|---|---|---|---|
| Identification | Protein | Mascot/TPP | PEAKS |
| A1AT_HUMAN | Alpha-1-antitrypsin | 3/15/1 | 5/18/3 |
| A2AP_HUMAN | Alpha-2-antiplasmin | 1/2/0 | 2/4/0 |
| FETUA_HUMAN | Alpha-2-HS-glycoprotein | 32/31/30 | 51/60/49 |
| APOA1_HUMAN | Apolipoprotein A-I | 80/127/17 | 171/258/37 |
| APOA2_HUMAN | Apolipoprotein A-II | 37/69/20 | 56/132/26 |
| APOA4_HUMAN | Apolipoprotein A-IV | 52/86/11 | 124/201/20 |
| APOC1_HUMAN | Apolipoprotein C-I | 11/17/0 | 34/45/2 |
| APOC2_HUMAN | Apolipoprotein C-II | 22/21/5 | 27/32/9 |
| APOC3_HUMAN | Apolipoprotein C-III | 19/51/5 | 43/88/11 |
| APOE_HUMAN | Apolipoprotein E | 29/9/4 | 57/30/13 |
| APOF_HUMAN | Apolipoprotein F | 1/2/0 | 1/2/0 |
| APOL1_HUMAN | Apolipoprotein L1 | 0/0/0 | 0/3/2 |
| C4BPA_HUMAN | C4b-binding protein alpha chain | 0/0/0 | 3/5/0 |
| CBPB2_HUMAN | Carboxypeptidase B2 | 1/0/1 | 1/0/1 |
| CLUS_HUMAN | Clusterin | 1/18/0 | 7/49/0 |
| CO1A2_HUMAN | Collagen alpha-2(I) chain | 0/0/0 | 1/0/1 |
| COL11_HUMAN | Collectin-11 | 0/2/1 | 3/5/2 |
| C1QB_HUMAN | Complement C1q subcomponent subunit B | 0/0/0 | 1/0/1 |
| C1S_HUMAN | Complement C1s subcomponent | 0/0/0 | 1/3/0 |
| CO3_HUMAN | Complement C3 | 8/9/7 | 13/22/21 |
| CO4A_HUMAN | Complement C4-A | 4/14/4 | 17/30/11 |
| CFAH_HUMAN | Complement factor H | 0/3/2 | 0/7/2 |
| FHR1_HUMAN | Complement factor H-related protein 1 | 0/0/0 | 3/8/0 |
| FIBA_HUMAN | Fibrinogen alpha chain | 82/141/40 | 149/241/63 |
| FIBB_HUMAN | Fibrinogen beta chain | 160/29/9 | 84/50/7 |
| FINC_HUMAN | Fibronectin | 4/7/6 | 4/10/15 |
| FCN2_HUMAN | Ficolin-2 | 1/2/0 | 2/4/0 |
| LG3BP_HUMAN | Galectin-3-binding protein | 0/0/0 | 2/2/0 |
| GELS_HUMAN | Gelsolin | 3/2/1 | 5/6/2 |
| GPX3_HUMAN | Glutathione peroxidase 3 | 1/0/1 | 1/2/1 |
| HRG_HUMAN | Histidine-rich glycoprotein | 0/0/0 | 1/5/0 |
| HABP2_HUMAN | Hyaluronan-binding protein 2 | 2/3/0 | 2/6/0 |
| IGHG1_HUMAN | Ig gamma-1 chain C region | 0/2/4 | 0/6/10 |
| IGKC_HUMAN | Ig kappa chain C region | 1/9/0 | 3/14/1 |
| KV309_HUMAN | Ig kappa chain V-III region VG (Fragment) | 0/0/0 | 1/6/0 |
| KV402_HUMAN | Ig kappa chain V-IV region Len | 0/0/0 | 1/2/0 |
| LV101_HUMAN | Ig lambda chain V-I region V | 0/0/0 | 1/3/2 |
| LV403_HUMAN | Ig lambda chain V-IV region Hil | 0/0/0 | 0/3/1 |
| LAC2_HUMAN | Ig lambda-2 chain C regions | 3/4/0 | 8/12/9 |
| IGLL5_HUMAN | Immunoglobulin lambda-like polypeptide 5 | 0/0/0 | 0/8/12 |
| ITIH1_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H1 | 2/4/0 | 3/8/0 |
| ITIH2_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H2 | 23/24/2 | 35/42/3 |
| ITIH4_HUMAN | Inter-alpha-trypsin inhibitor heavy chain H4 | 10/7/2 | 21/20/8 |
| FIBG_HUMAN | Isoform Gamma-A of fibrinogen gamma chain | 8/9/0 | 19/25/0 |
| KNG1_HUMAN | Kininogen-1 | 2/3/0 | 6/6/3 |
| PLMN_HUMAN | Plasminogen | 0/0/0 | 1/6/0 |
| PRAP1_HUMAN | Proline-rich acidic protein 1 | 0/0/0 | 4/1/2 |
| THRB_HUMAN | Prothrombin | 7/20/5 | 21/43/12 |
| ALBU_HUMAN | Serum albumin | 1/10/1 | 2/25/6 |
| SAA4_HUMAN | Serum amyloid A-4 protein | 4/3/0 | 14/11/1 |
| TTHY_HUMAN | Transthyretin | 2/5/0 | 7/13/3 |
| PROC_HUMAN | Vitamin K-dependent protein C | 0/0/0 | 0/1/1 |
| PROS_HUMAN | Vitamin K-dependent protein S | 2/1/0 | 4/5/1 |
| VTNC_HUMAN | Vitronectin | 19/43/13 | 41/85/20 |
| Actin Cytoplasmic-1 | Fibrinogen |
|---|---|
| Alpha-1-antitrypsin | Fibronectin |
| Alpha-2-antiplasmin | Ficolin-2 |
| Alpha-2-HS-glycoprotein | Galectin-3-binding protein |
| Apha-2-macroglobulin | Gelsolin |
| Apolipoprotein A-I | Glutathione peroxidase 3 |
| Apolipoprotein A-II | Histidine-rich glycoprotein |
| Apolipoprotein A-IV | Histone H2B type F-S |
| Apolipoprotein C-I | Hyaluronan-binding protein 2 |
| Apolipoprotein C-II | Ig gamma |
| Apolipoprotein C-IIII | Ig kappa |
| Apolipoprotein E | Ig lambda |
| Apolipoprotein F | Ig mu chain C region |
| Apolipoprotein L1 | Ig lambda-like polypeptide 5 |
| C4b-binding protein alpha chain | Inter-alpha-trypsin inhibitor H1−H4 chains |
| Carboxypeptidade B2 | Isoform 3 of MAX gene-associated protein |
| Ceruloplasmin | Kininogen |
| Clusterin | Pigment ephitelium-derived factor |
| Coagulation factor V | Plasminogen |
| Coagulation factor XIII A chain | Pregnancy zone protein |
| Collagen alpha-2(I) chain | Proline-rich acidic protein 1 |
| Collectin 11 | Prothrombin |
| Complement C1q subcomponent subunit B | Serum albumin |
| Complement C1r subcomponent | Serum amyloid A-4 protein |
| Complement C1s subcomponent | Serum paraoxonase/arylesterase 1 |
| Complement component C3 | Tetranectin |
| Complement component C4-A | Transthyretin |
| Complement component C9 | Vitamin K-dependent protein C |
| Complement factor H | Vitamin K-dependent protein S |
| Complement factor H-related protein | Vitronectin |
| Extracellular matrix protein 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertholim, L.; Chaves, A.F.A.; Oliveira, A.K.; Menezes, M.C.; Asega, A.F.; Tashima, A.K.; Zelanis, A.; Serrano, S.M.T. Systemic Effects of Hemorrhagic Snake Venom Metalloproteinases: Untargeted Peptidomics to Explore the Pathodegradome of Plasma Proteins. Toxins 2021, 13, 764. https://doi.org/10.3390/toxins13110764
Bertholim L, Chaves AFA, Oliveira AK, Menezes MC, Asega AF, Tashima AK, Zelanis A, Serrano SMT. Systemic Effects of Hemorrhagic Snake Venom Metalloproteinases: Untargeted Peptidomics to Explore the Pathodegradome of Plasma Proteins. Toxins. 2021; 13(11):764. https://doi.org/10.3390/toxins13110764
Chicago/Turabian StyleBertholim, Luciana, Alison F. A. Chaves, Ana K. Oliveira, Milene C. Menezes, Amanda F. Asega, Alexandre K. Tashima, Andre Zelanis, and Solange M. T. Serrano. 2021. "Systemic Effects of Hemorrhagic Snake Venom Metalloproteinases: Untargeted Peptidomics to Explore the Pathodegradome of Plasma Proteins" Toxins 13, no. 11: 764. https://doi.org/10.3390/toxins13110764
APA StyleBertholim, L., Chaves, A. F. A., Oliveira, A. K., Menezes, M. C., Asega, A. F., Tashima, A. K., Zelanis, A., & Serrano, S. M. T. (2021). Systemic Effects of Hemorrhagic Snake Venom Metalloproteinases: Untargeted Peptidomics to Explore the Pathodegradome of Plasma Proteins. Toxins, 13(11), 764. https://doi.org/10.3390/toxins13110764





