Platelet Endothelial Cell Adhesion Molecule 1 (CD31) Is Essential for Clostridium perfringens Beta-Toxin Mediated Cytotoxicity in Human Endothelial and Monocytic Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Human Endothelial and Monocytic Cell Lines Express CD31 and Are Susceptible to CPB
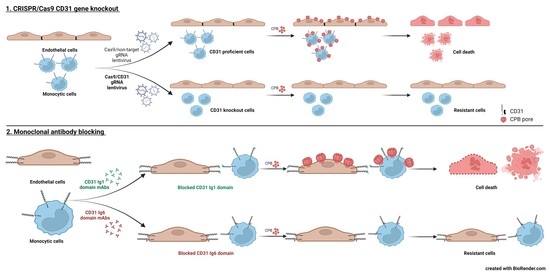
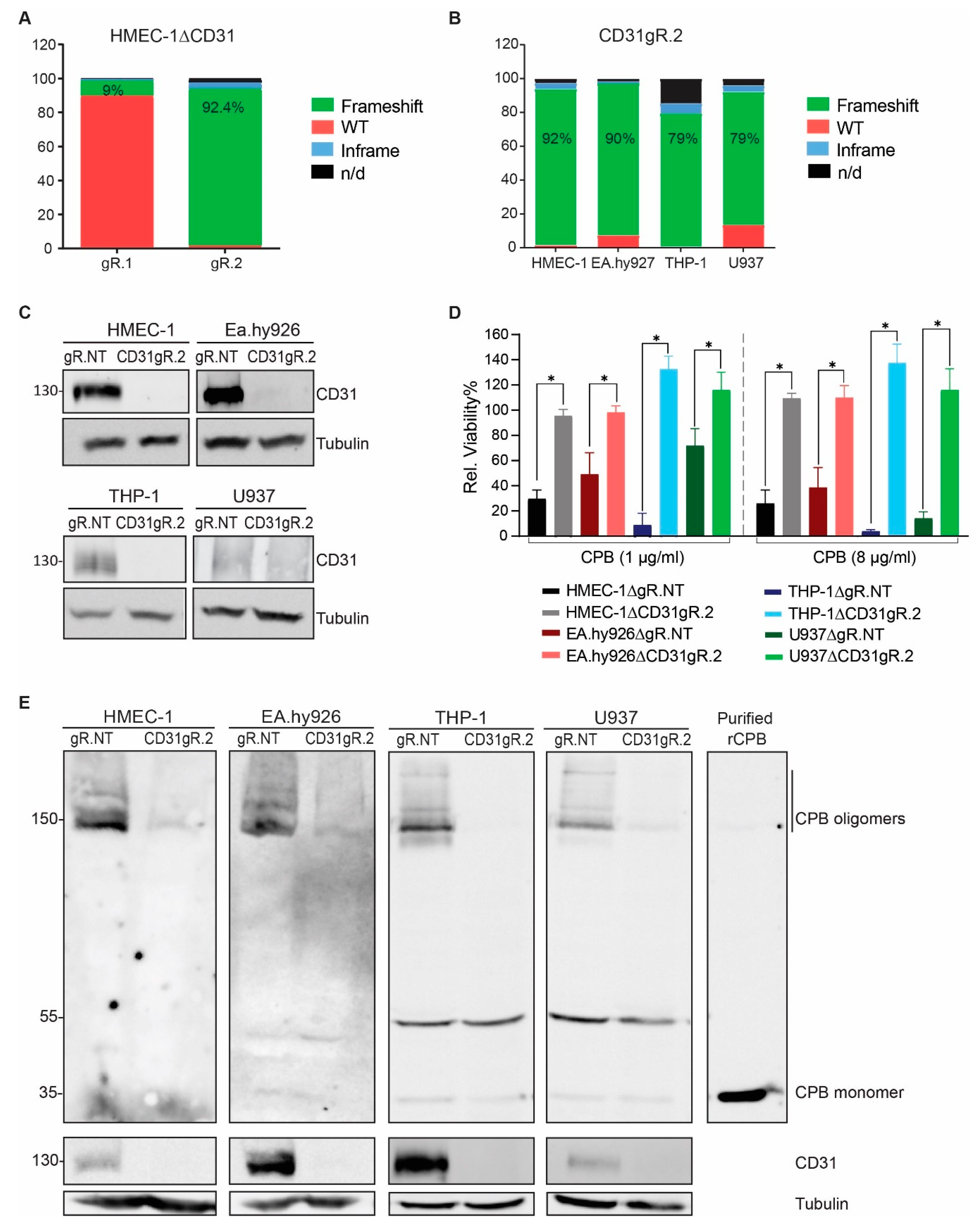
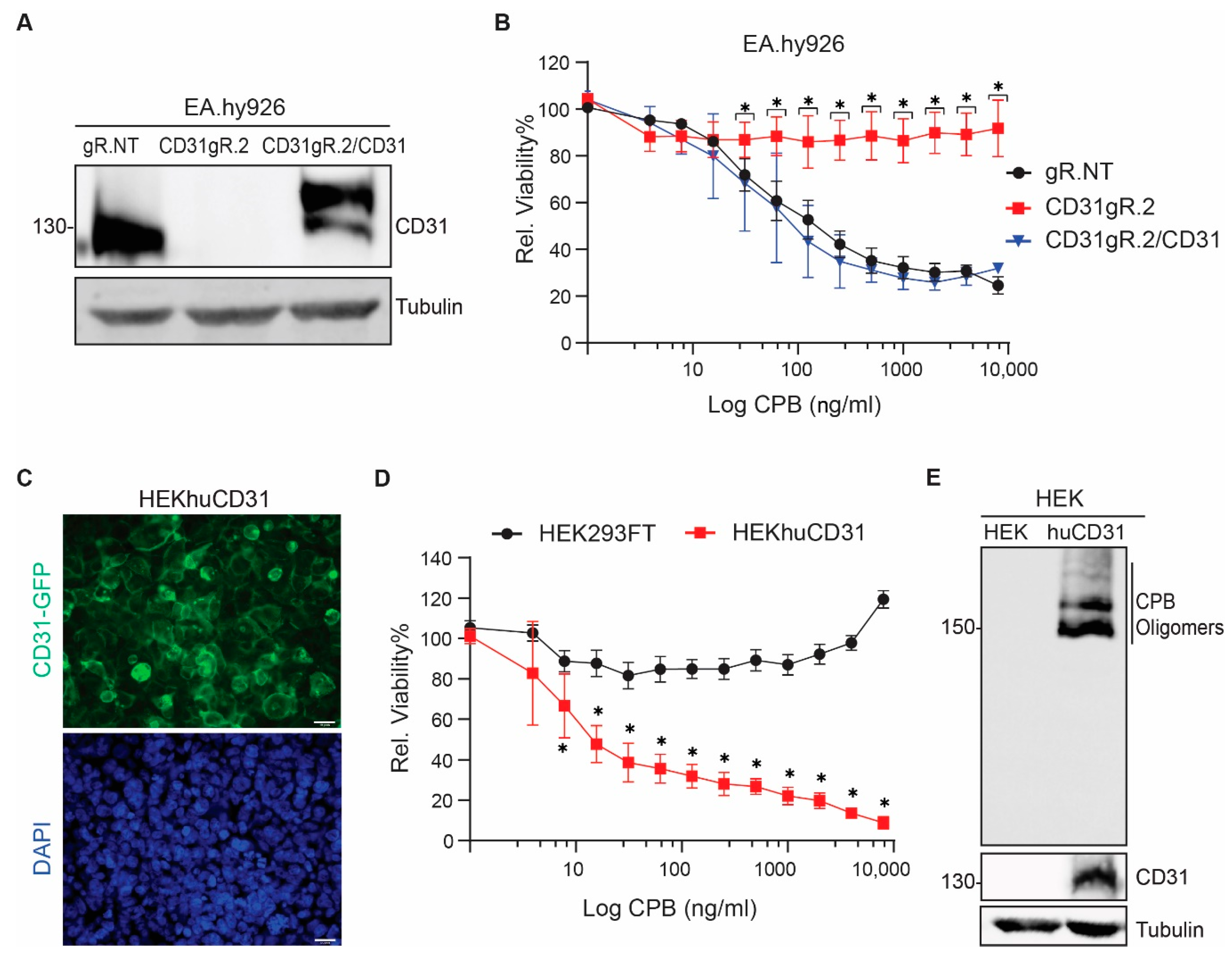
2.2. CD31 Is Essential for CPB-Mediated Cytotoxicity in Human Endothelial and Monocytic Cells
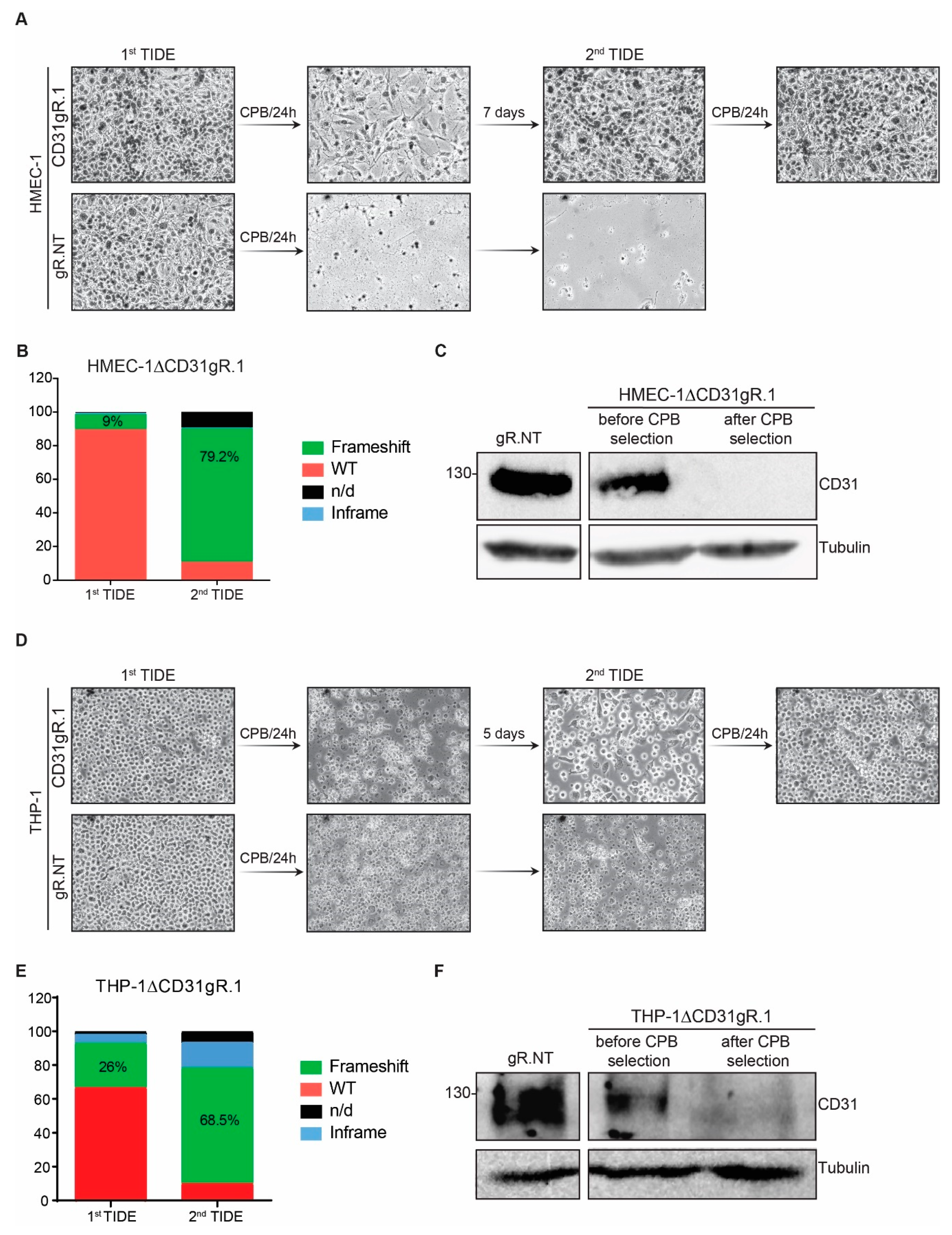
2.3. Selective Enrichment of CD31 Knockout Cells with CPB
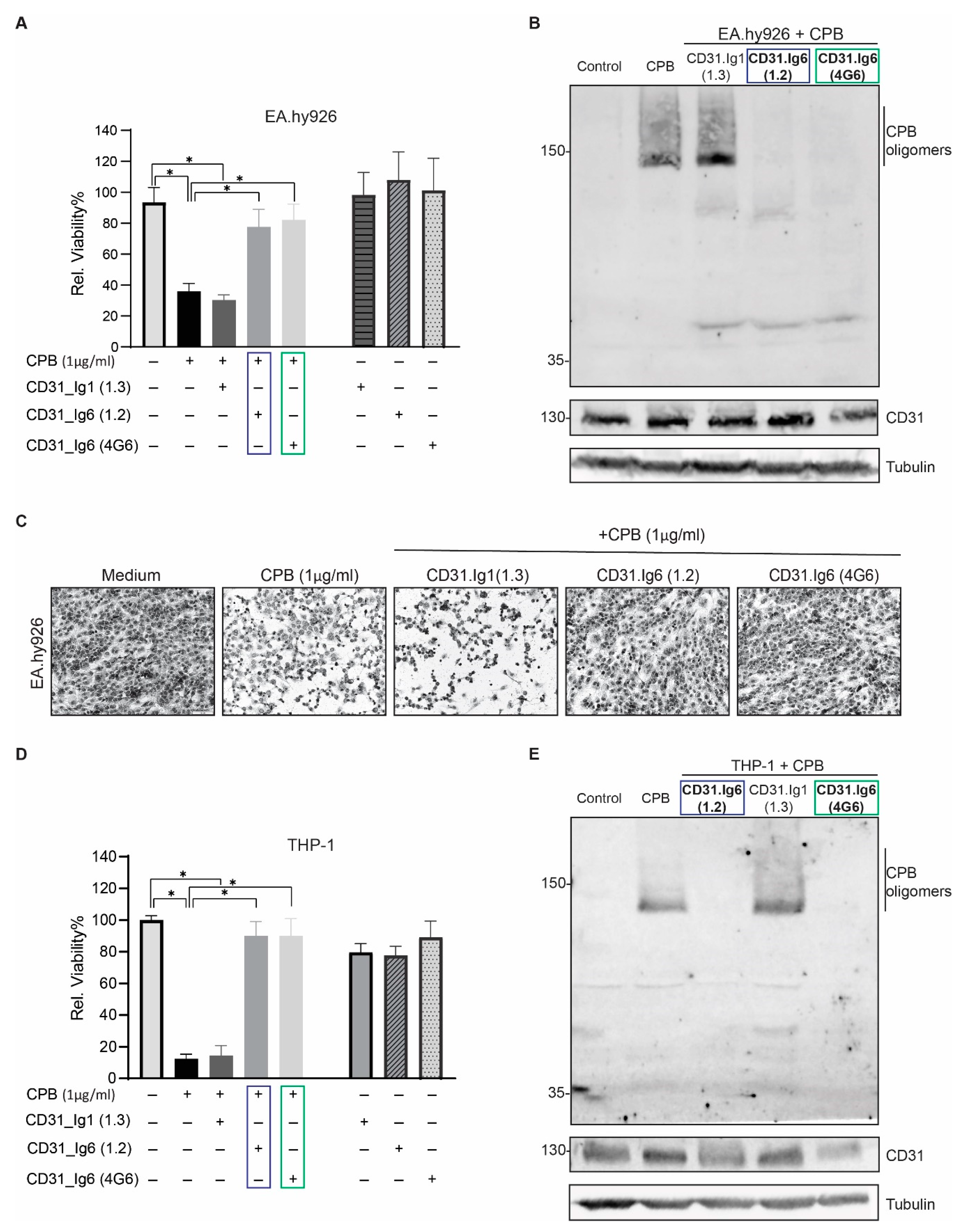
2.4. Blocking the CD31 Ig6 Domain Protects Sensitive Cells against CPB Induced Cytotoxicity
3. Conclusions
4. Materials and Methods
4.1. Cells and Reagents
4.2. Recombinant CPB Production
4.3. Western Blotting
4.4. SDS-Resistant Oligomer Formation
4.5. Cell Viability Assays
4.6. Generation of CD31 Knockout Cell Lines
4.7. Complementation and Overexpression of CD31
4.8. CD31 Extracellular Ig Domains Blocking Assay
4.9. Immunofluorescence
4.10. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Posthaus, H.; Kittl, S.; Tarek, B.; Bruggisser, J. Clostridium perfringens type C necrotic enteritis in pigs: Diagnosis, pathogenesis, and prevention. J. Vet. Diagn. Investig. 2020, 32, 203–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jäggi, M.; Wollschlager, N.; Abril, C.; Albini, S.; Brachelente, C.; Wyder, M.; Posthaus, H. Retrospective study on necrotizing enteritis in piglets in Switzerland. Schweiz. Arch. Tierheilkd. 2009, 151, 369–375. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, G.; Walker, P. Pathogenesis of enteritis necroticans in Papua New Guinea. Lancet 1976, 307, 125–126. [Google Scholar] [CrossRef]
- Gui, L.; Subramony, C.; Fratkin, J.; Hughson, M.D. Fatal enteritis necroticans (pigbel) in a diabetic adult. Mod. Pathol. 2002, 15, 66–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuda, T.; Okada, Y.; Inagi, E.; Tanabe, Y.; Shimizu, Y.; Nagashima, K.; Sakurai, J.; Nagahama, M.; Tanaka, S. Enteritis necroticans ‘pigbel’ in a Japanese diabetic adult. Pathol. Int. 2007, 57, 622–626. [Google Scholar] [CrossRef]
- Petrillo, T.M.; Beck-Sague, C.M.; Songer, J.G.; Abramowsky, C.; Fortenberry, J.D.; Meacham, L.; Dean, A.G.; Lee, H.; Bueschel, D.M.; Nesheim, S.R. Enteritis necroticans (pigbel) in a diabetic child. N. Engl. J. Med. 2000, 342, 1250–1253. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Daroca, P.J., Jr.; Crawford, B.E. Clostridial enteritis necroticans versus secondary clostridial infection superimposed upon ischemic bowel disease. J. La. State Med. Soc. 2002, 154, 251–255. [Google Scholar]
- Hunter, S.E.; Brown, J.E.; Oyston, P.C.; Sakurai, J.; Titball, R.W. Molecular genetic analysis of beta-toxin of Clostridium perfringens reveals sequence homology with alpha-toxin, gamma-toxin, and leukocidin of Staphylococcus aureus. Infect. Immun. 1993, 61, 3958–3965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popoff, M.R. Clostridial pore-forming toxins: Powerful virulence factors. Anaerobe 2014, 30, 220–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacovache, I.; van der Goot, F.G.; Pernot, L. Pore formation: An ancient yet complex form of attack. Biochim. Biophys. Acta 2008, 1778, 1611–1623. [Google Scholar] [CrossRef] [Green Version]
- Nagahama, M.; Hayashi, S.; Morimitsu, S.; Sakurai, J. Biological activities and pore formation of Clostridium perfringens beta toxin in HL 60 cells. J. Biol. Chem. 2003, 278, 36934–36941. [Google Scholar] [CrossRef] [Green Version]
- Nagahama, M.; Ochi, S.; Oda, M.; Miyamoto, K.; Takehara, M.; Kobayashi, K. Recent insights into Clostridium perfringens beta-toxin. Toxins 2015, 7, 396–406. [Google Scholar] [CrossRef] [Green Version]
- Miclard, J.; Jaggi, M.; Sutter, E.; Wyder, M.; Grabscheid, B.; Posthaus, H. Clostridium perfringens beta-toxin targets endothelial cells in necrotizing enteritis in piglets. Vet. Microbiol. 2009, 137, 320–325. [Google Scholar] [CrossRef]
- Miclard, J.; van Baarlen, J.; Wyder, M.; Grabscheid, B.; Posthaus, H. Clostridium perfringens beta-toxin binding to vascular endothelial cells in a human case of enteritis necroticans. J. Med. Microbiol. 2009, 58, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, V.L.; Martel, A.; Pasmans, F.; Van Immerseel, F.; Posthaus, H. Endothelial binding of beta toxin to small intestinal mucosal endothelial cells in early stages of experimentally induced Clostridium perfringens type C enteritis in pigs. Vet. Pathol. 2013, 50, 626–629. [Google Scholar] [CrossRef]
- Autheman, D.; Wyder, M.; Popoff, M.; D’Herde, K.; Christen, S.; Posthaus, H. Clostridium perfringens beta-toxin induces necrostatin-inhibitable, calpain-dependent necrosis in primary porcine endothelial cells. PLoS ONE 2013, 8, e64644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagahama, M.; Shibutani, M.; Seike, S.; Yonezaki, M.; Takagishi, T.; Oda, M.; Kobayashi, K.; Sakurai, J. The p38 MAPK and JNK pathways protect host cells against Clostridium perfringens beta-toxin. Infect. Immun. 2013, 81, 3703–3708. [Google Scholar] [CrossRef] [Green Version]
- Thiel, A.; Mogel, H.; Bruggisser, J.; Baumann, A.; Wyder, M.; Stoffel, M.H.; Summerfield, A.; Posthaus, H. Effect of Clostridium perfringens beta-Toxin on Platelets. Toxins 2017, 9, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruggisser, J.; Tarek, B.; Wyder, M.; Muller, P.; von Ballmoos, C.; Witz, G.; Enzmann, G.; Deutsch, U.; Engelhardt, B.; Posthaus, H. CD31 (PECAM-1) Serves as the Endothelial Cell-Specific Receptor of Clostridium perfringens beta-Toxin. Cell Host Microbe 2020, 28, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Popescu, F.; Wyder, M.; Gurtner, C.; Frey, J.; Cooke, R.A.; Greenhill, A.R.; Posthaus, H. Susceptibility of primary human endothelial cells to C. perfringens beta-toxin suggesting similar pathogenesis in human and porcine necrotizing enteritis. Vet. Microbiol. 2011, 153, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Steinthorsdottir, V.; Halldorsson, H.; Andresson, O.S. Clostridium perfringens beta-toxin forms multimeric transmembrane pores in human endothelial cells. Microb. Pathog. 2000, 28, 45–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLisser, H.M.; Newman, P.J.; Albelda, S.M. Molecular and functional aspects of PECAM-1/CD31. Immunol. Today 1994, 15, 490–495. [Google Scholar] [CrossRef]
- Albeda, F.W.; van der Meer, J.; Vellenga, E. Vascular proliferation as an unusual cause of hemorrhagic diathesis in myelofibrosis. Am. J. Clin. Pathol. 1991, 95, 564–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lertkiatmongkol, P.; Liao, D.; Mei, H.; Hu, Y.; Newman, P.J. Endothelial functions of platelet/endothelial cell adhesion molecule-1 (CD31). Curr. Opin. Hematol. 2016, 23, 253–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, D.E. The unfolding tale of PECAM-1. FEBS Lett. 2003, 540, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Shatursky, O.; Bayles, R.; Rogers, M.; Jost, B.H.; Songer, J.G.; Tweten, R.K. Clostridium perfringens beta-toxin forms potential-dependent, cation-selective channels in lipid bilayers. Infect. Immun. 2000, 68, 5546–5551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal, J.E.; McClane, B.A.; Saputo, J.; Parker, J.; Uzal, F.A. Effects of Clostridium perfringens beta-toxin on the rabbit small intestine and colon. Infect. Immun. 2008, 76, 4396–4404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roos, S.; Wyder, M.; Candi, A.; Regenscheit, N.; Nathues, C.; van Immerseel, F.; Posthaus, H. Binding studies on isolated porcine small intestinal mucosa and in vitro toxicity studies reveal lack of effect of C. perfringens beta-toxin on the porcine intestinal epithelium. Toxins 2015, 7, 1235–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagahama, M.; Seike, S.; Shirai, H.; Takagishi, T.; Kobayashi, K.; Takehara, M.; Sakurai, J. Role of P2X7 receptor in Clostridium perfringens beta-toxin-mediated cellular injury. Biochim. Biophys. Acta 2015, 1850, 2159–2167. [Google Scholar] [CrossRef] [PubMed]
- Newman, P.J.; Berndt, M.C.; Gorski, J.; White, G.C., 2nd; Lyman, S.; Paddock, C.; Muller, W.A. PECAM-1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily. Science 1990, 247, 1219–1222. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhang, H.; Liu, Q.; Hao, Q. Structural Basis for Human PECAM-1-Mediated Trans-homophilic Cell Adhesion. Sci. Rep. 2016, 6, 38655. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Williams, J.; Yan, H.C.; Amin, K.M.; Albelda, S.M.; DeLisser, H.M. Platelet endothelial cell adhesion molecule-1 (PECAM-1) homophilic adhesion is mediated by immunoglobulin-like domains 1 and 2 and depends on the cytoplasmic domain and the level of surface expression. J. Biol. Chem. 1996, 271, 18561–18570. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.H.; DeLisser, H.M.; Zukowski, M.M.; Paddock, C.; Albelda, S.M.; Newman, P.J. Individually distinct Ig homology domains in PECAM-1 regulate homophilic binding and modulate receptor affinity. J. Biol. Chem. 1996, 271, 11090–11098. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Lin, L.; Li, R.; Yuan, C.; Xu, M.; Huang, J.H.; Huang, M. Dimer conformation of soluble PECAM-1, an endothelial marker. Int. J. Biochem. Cell Biol. 2016, 77, 102–108. [Google Scholar] [CrossRef]
- Newton, J.P.; Buckley, C.D.; Jones, E.Y.; Simmons, D.L. Residues on both faces of the first immunoglobulin fold contribute to homophilic binding sites of PECAM-1/CD31. J. Biol. Chem. 1997, 272, 20555–20563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graesser, D.; Solowiej, A.; Bruckner, M.; Osterweil, E.; Juedes, A.; Davis, S.; Ruddle, N.H.; Engelhardt, B.; Madri, J.A. Altered vascular permeability and early onset of experimental autoimmune encephalomyelitis in PECAM-1-deficient mice. J. Clin. Investig. 2002, 109, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Deaglio, S.; Morra, M.; Mallone, R.; Ausiello, C.M.; Prager, E.; Garbarino, G.; Dianzani, U.; Stockinger, H.; Malavasi, F. Human CD38 (ADP-ribosyl cyclase) is a counter-receptor of CD31, an Ig superfamily member. J. Immunol. 1998, 160, 395–402. [Google Scholar] [PubMed]
- Sachs, U.J.; Andrei-Selmer, C.L.; Maniar, A.; Weiss, T.; Paddock, C.; Orlova, V.V.; Choi, E.Y.; Newman, P.J.; Preissner, K.T.; Chavakis, T.; et al. The neutrophil-specific antigen CD177 is a counter-receptor for platelet endothelial cell adhesion molecule-1 (CD31). J. Biol. Chem. 2007, 282, 23603–23612. [Google Scholar] [CrossRef] [Green Version]
- Piali, L.; Hammel, P.; Uherek, C.; Bachmann, F.; Gisler, R.H.; Dunon, D.; Imhof, B.A. CD31/PECAM-1 is a ligand for alpha v beta 3 integrin involved in adhesion of leukocytes to endothelium. J. Cell Biol. 1995, 130, 451–460. [Google Scholar] [CrossRef]
- Privratsky, J.R.; Newman, D.K.; Newman, P.J. PECAM-1: Conflicts of interest in inflammation. Life Sci. 2010, 87, 69–82. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.C.; Pilewski, J.M.; Zhang, Q.; DeLisser, H.M.; Romer, L.; Albelda, S.M. Localization of multiple functional domains on human PECAM-1 (CD31) by monoclonal antibody epitope mapping. Cell Adhes. Commun. 1995, 3, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Alzari, P.M. Encyclopedia of Immunology, 2nd ed.; Academic Press: San Diego, CA, USA, 1998. [Google Scholar]
- Gurtner, C.; Popescu, F.; Wyder, M.; Sutter, E.; Zeeh, F.; Frey, J.; von Schubert, C.; Posthaus, H. Rapid cytopathic effects of Clostridium perfringens beta-toxin on porcine endothelial cells. Infect. Immun. 2010, 78, 2966–2973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkman, E.K.; Chen, T.; Amendola, M.; van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014, 42, e168. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarek, B.; Bruggisser, J.; Cattalani, F.; Posthaus, H. Platelet Endothelial Cell Adhesion Molecule 1 (CD31) Is Essential for Clostridium perfringens Beta-Toxin Mediated Cytotoxicity in Human Endothelial and Monocytic Cells. Toxins 2021, 13, 893. https://doi.org/10.3390/toxins13120893
Tarek B, Bruggisser J, Cattalani F, Posthaus H. Platelet Endothelial Cell Adhesion Molecule 1 (CD31) Is Essential for Clostridium perfringens Beta-Toxin Mediated Cytotoxicity in Human Endothelial and Monocytic Cells. Toxins. 2021; 13(12):893. https://doi.org/10.3390/toxins13120893
Chicago/Turabian StyleTarek, Basma, Julia Bruggisser, Filippo Cattalani, and Horst Posthaus. 2021. "Platelet Endothelial Cell Adhesion Molecule 1 (CD31) Is Essential for Clostridium perfringens Beta-Toxin Mediated Cytotoxicity in Human Endothelial and Monocytic Cells" Toxins 13, no. 12: 893. https://doi.org/10.3390/toxins13120893
APA StyleTarek, B., Bruggisser, J., Cattalani, F., & Posthaus, H. (2021). Platelet Endothelial Cell Adhesion Molecule 1 (CD31) Is Essential for Clostridium perfringens Beta-Toxin Mediated Cytotoxicity in Human Endothelial and Monocytic Cells. Toxins, 13(12), 893. https://doi.org/10.3390/toxins13120893