Examination of the Efficacy and Cross-Reactivity of a Novel Polyclonal Antibody Targeting the Disintegrin Domain in SVMPs to Neutralize Snake Venom
Abstract
:1. Introduction
2. Results
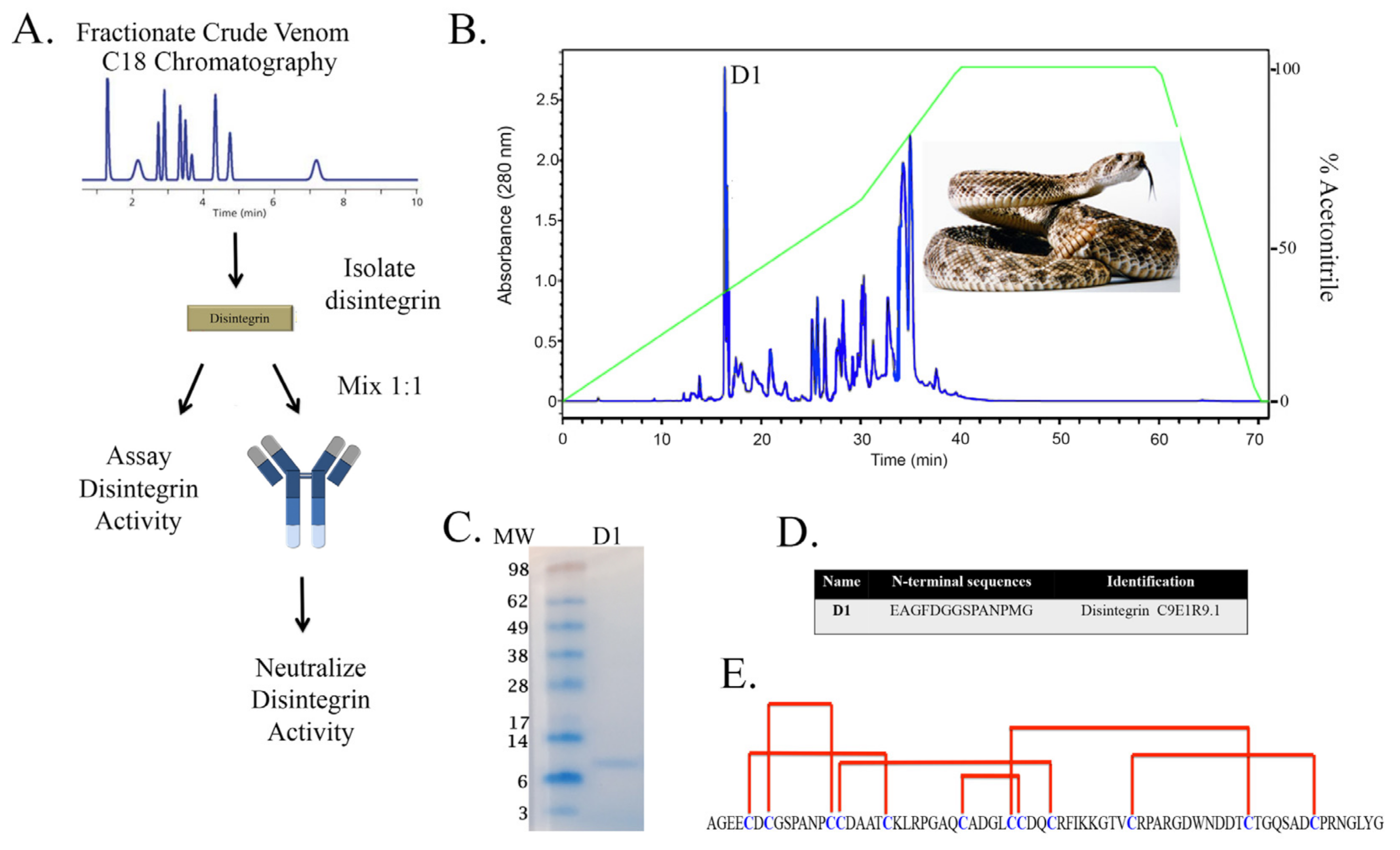
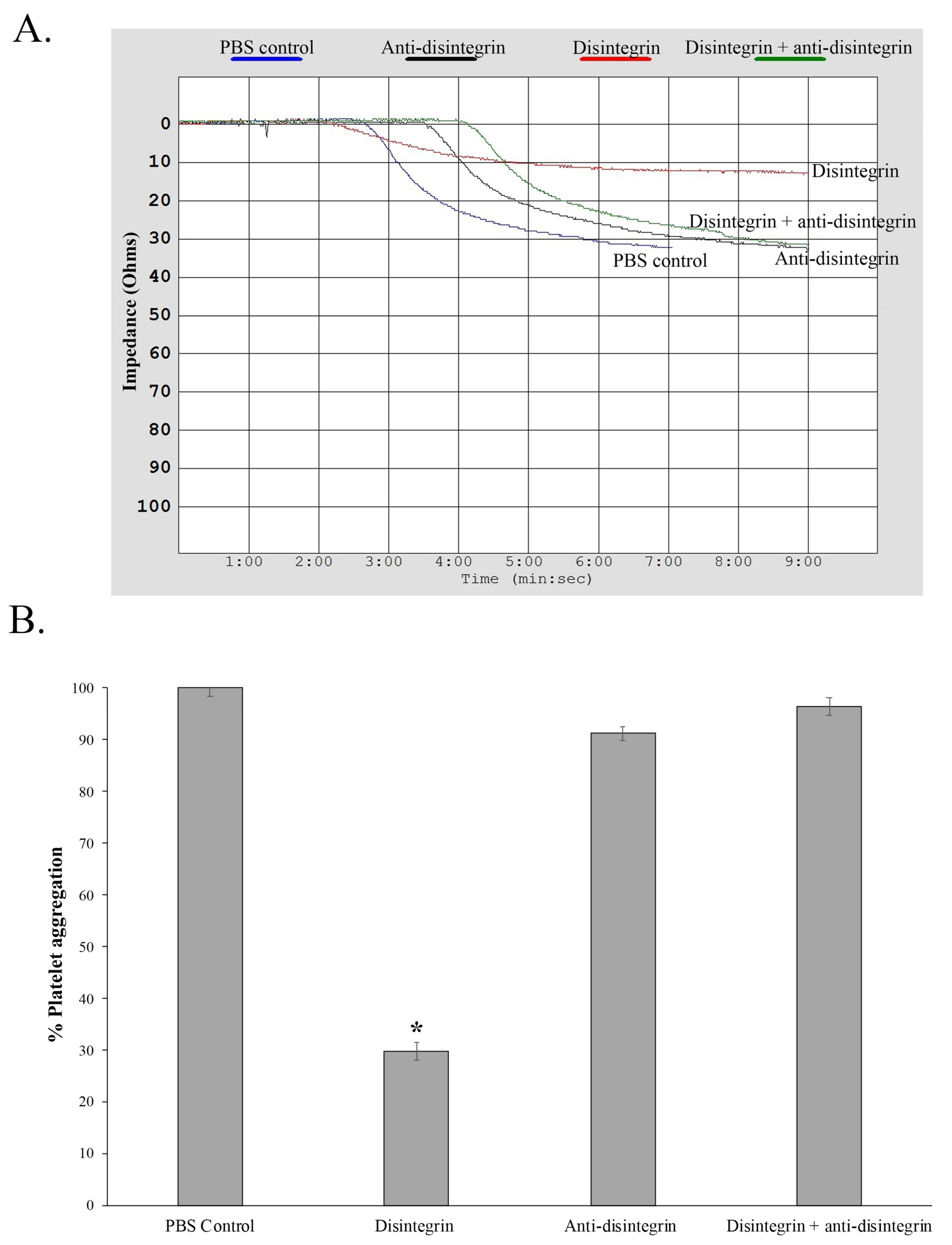
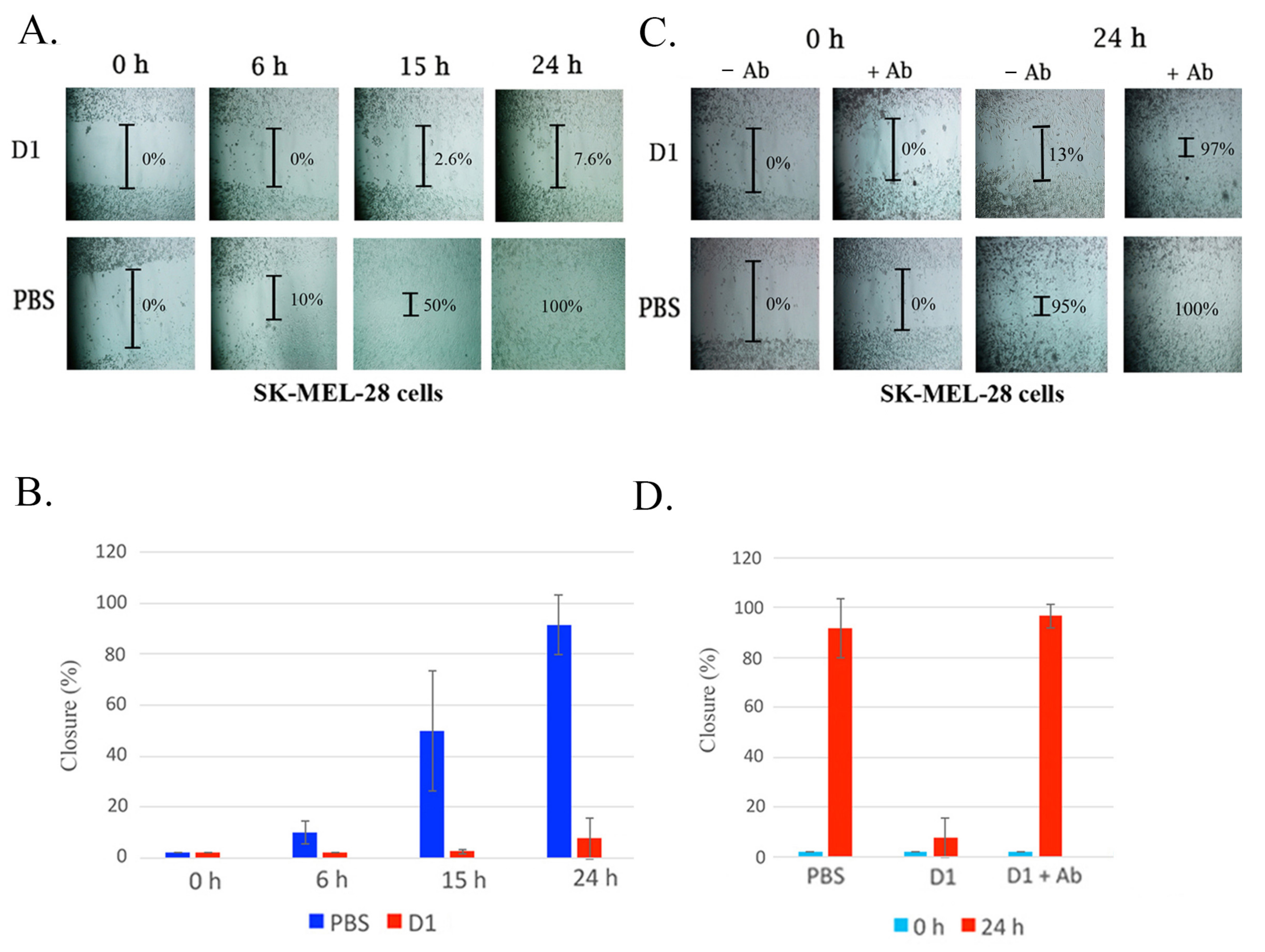
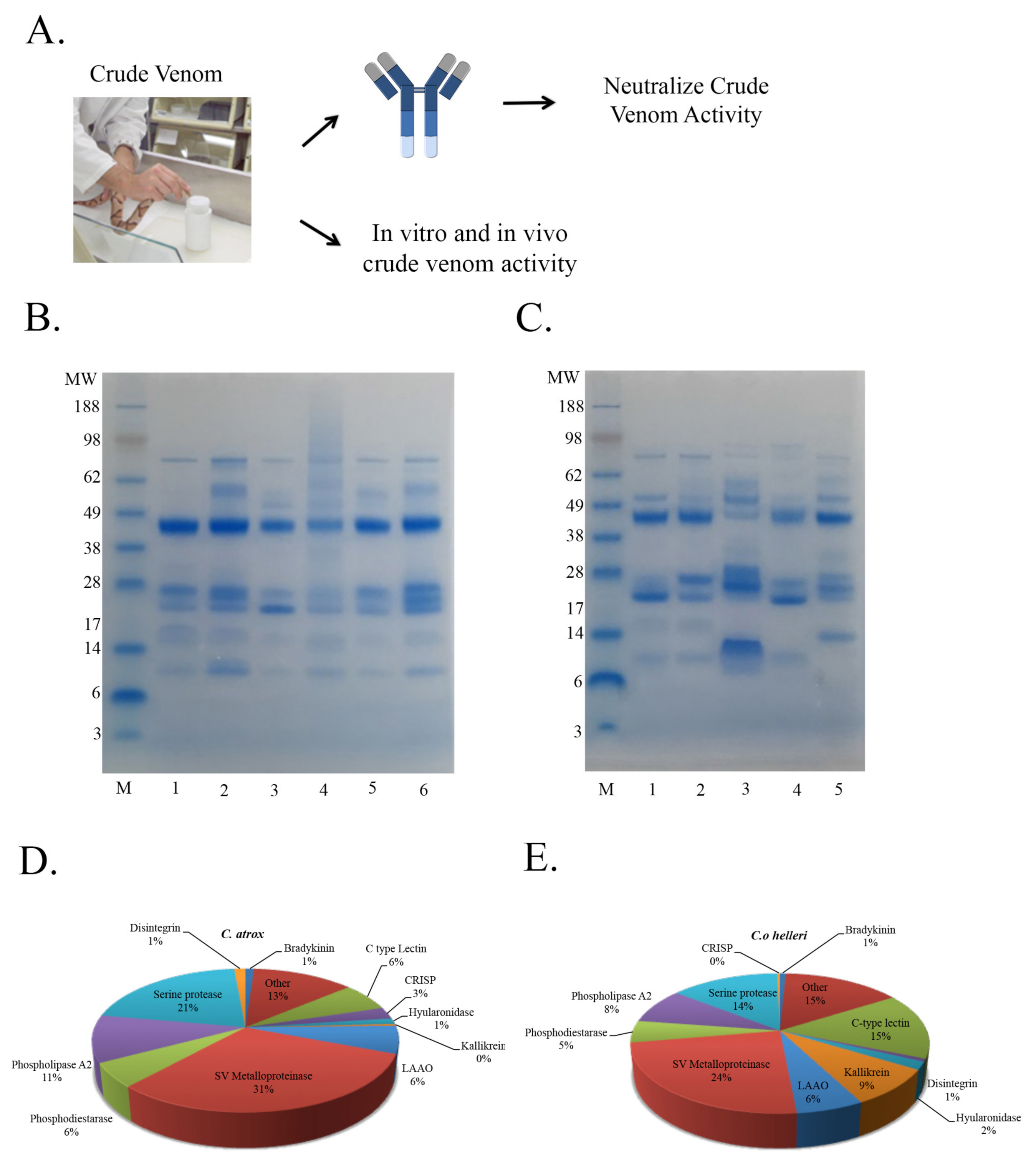
2.1. Rationale and Scheme for Disintegrin Neutralization
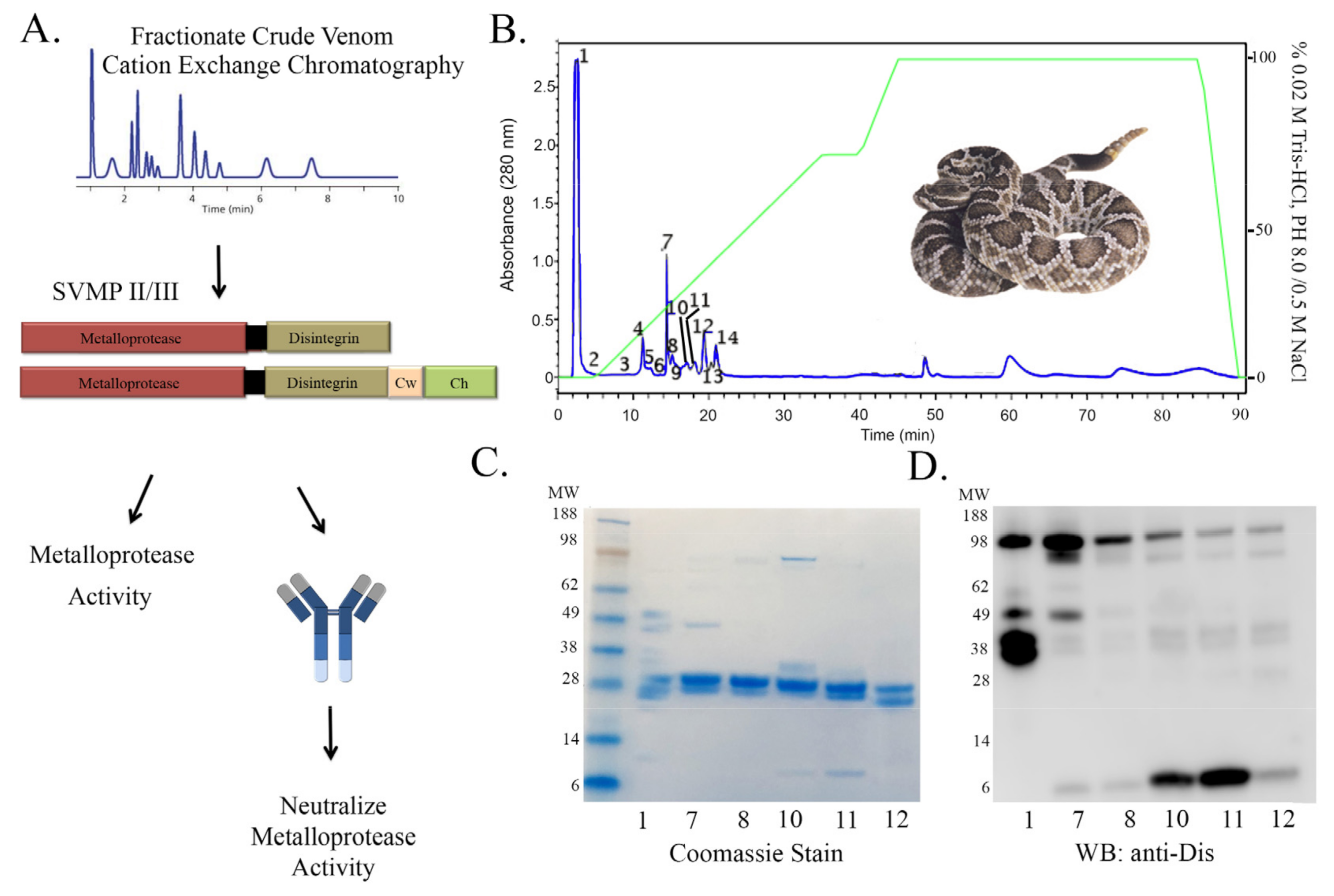
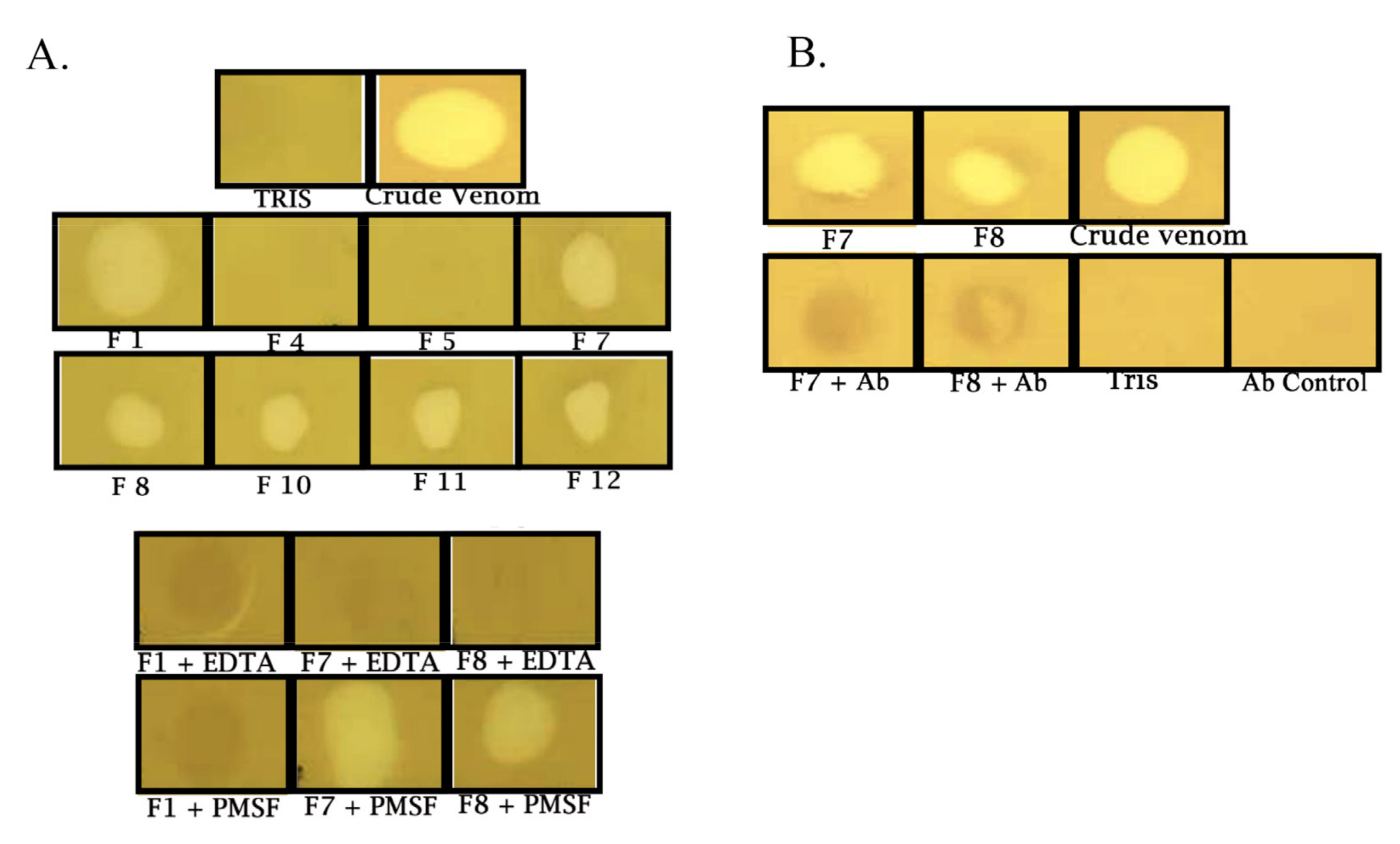
2.2. Rationale and Scheme for Partially Purified Metalloprotease Neutralization
2.3. Rationale and Scheme for Crude Venom Neutralization
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Collection of Crude Venoms
5.2. Venoms/Anti-Disintegrin
5.3. Purification of Native Disintegrins by C18-Reverse Phase Chromatography
5.4. SDS Polyacrylamide Gel Electrophoresis and Immunoblot Analysis
5.5. N-Terminal Sequencing
5.6. Platelet Aggregation
5.7. Cell Migration Assay
5.8. Cell Proliferation Assay
5.9. SVMP Purification by Cation Exchange Chromatography
5.10. Gelatinase Assay
5.11. Proteomic Analysis
5.12. Hemorrhagic Analysis
5.13. Ethical Procedures
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calvete, J.J. Venomics: Digging into the evolution of venomous systems and learning to twist nature to fight pathology. J. Proteom. 2009, 72, 121–126. [Google Scholar] [CrossRef]
- Calvete, J.J.; Fasoli, E.; Sanz, L.; Boschetti, E.; Righetti, P.G. Exploring the venom proteome of the Western Diamondback Rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res. 2009, 8, 3055–3067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, S.A.; Boyer, L.V.; Benson, B.E.; Rogers, J.J. AAPCC database characterization of native U.S. venomous snake exposures, 2001–2005. Clin. Toxicol. 2009, 47, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.H.; Ernst, E.M. Venomous Reptiles of the United States, Canada, and Northern Mexico; John Hopkins University Press: Baltimore, MD, USA, 2012. [Google Scholar]
- Campbell, J.; Lamar, W.W. Venomous Reptiles of the Western Hemisphere; Cornell University Press: Ithaca, NY, USA, 2004. [Google Scholar]
- Lavonas, E.J.; Khatri, V.; Daugherty, C.; Bucher-Bartelson, B.; King, T.; Dart, R.C. Medically significant late bleeding after treated crotaline envenomation: A systematic review. Ann. Emerg. Med. 2014, 63, 71–78.e1. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.V.; Seifert, S.A.; Clark, R.F.; McNally, J.T.; Williams, S.R.; Nordt, S.P.; Walter, F.G.; Dart, R.C. Recurrent and persistent coagulopathy following pit viper envenomation. Arch. Intern. Med. 1999, 159, 706–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dart, R.C.; McNally, J. Efficacy, safety, and use of snake antivenoms in the United States. Ann. Emerg. Med. 2001, 37, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Laustsen, A.H. Snakebites: Costing recombinant antivenoms. Nature 2016, 538, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laustsen, A.; Engmark, M.; Milbo, C.; Johannesen, J.; Lomonte, B.; Gutiérrez, J.; Lohse, B. From fangs to pharmacology: The future of snakebite envenoming therapy. Curr. Pharm. Des. 2016, 22, 5270–5293. [Google Scholar] [CrossRef]
- Rodríguez, C.; Estrada, R.; Herrera, M.; Gómez, A.; Segura, Á.; Vargas, M.; Villalta, M.; León, G. Bothrops asper envenoming in cattle: Clinical features and management using equine-derived whole IgG antivenom. Vet. J. 2016, 207, 160–163. [Google Scholar] [CrossRef]
- Segura, Á.; Herrera, M.; Villalta, M.; Vargas, M.; Gutiérrez, J.M.; León, G. Assessment of snake antivenom purity by comparing physicochemical and immunochemical methods. Biologicals 2013, 41, 93–97. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Gutiérrez, J.M.; Rasmussen, A.R.; Engmark, M.; Gravlund, P.; Sanders, K.L.; Lohse, B.; Lomonte, B. Danger in the reef: Proteome, toxicity, and neutralization of the venom of the olive sea snake, Aipysurus laevis. Toxicon 2015, 107, 187–196. [Google Scholar] [CrossRef] [Green Version]
- León, G.; Herrera, M.; Segura, Á.; Villalta, M.; Vargas, M.; Gutiérrez, J.M. Pathogenic mechanisms underlying adverse reactions induced by intravenous administration of snake antivenoms. Toxicon 2013, 76, 63–76. [Google Scholar] [CrossRef] [PubMed]
- da Silva, D.C.; de Medeiros, W.A.; Batista Ide, F.; Pimenta, D.C.; Lebrun, I.; Abdalla, F.M.; Sandoval, M.R. Characterization of a new muscarinic toxin from the venom of the Brazilian coral snake Micrurus lemniscatus in rat hippocampus. Life Sci. 2011, 89, 931–938. [Google Scholar] [CrossRef] [Green Version]
- Isbister, G.K.; Brown, S.G.; Macdonald, E.; White, J.; Currie, B.J. Current use of Australian snake antivenoms and frequency of immediate-type hypersensitivity reactions and anaphylaxis. Med. J. Aust. 2008, 188, 473–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantú, E.; Mallela, S.; Nyguen, M.; Báez, R.; Parra, V.; Johnson, R.; Wilson, K.; Suntravat, M.; Lucena, S.; Rodríguez-Acosta, A.; et al. The binding effectiveness of anti-r-disintegrin polyclonal antibodies against disintegrins and PII and PIII metalloproteases: An immunological survey of type A, B and A+B venoms from Mohave rattlesnakes. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2017, 191, 168–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juarez, P.; Comas, I.; González-Candelas, F.; Calvete, J.J. Evolution of snake venom disintegrins by positive darwinian selection. Mol. Biol. Evol. 2008, 25, 2391–2407. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Primers 2017, 3, 17063. [Google Scholar] [CrossRef]
- Adukauskienė, D.; Varanauskienė, E.; Adukauskaitė, A. Venomous snakebites. Medicina 2011, 47, 461. [Google Scholar] [CrossRef]
- Gutierrez, J.M.; Rucavado, A. Snake venom metalloproteinases: Their role in the pathogenesis of local tissue damage. Biochimie 2000, 82, 841–850. [Google Scholar] [CrossRef]
- Juárez, P.; Sanz, L.; Calvete, J.J. Snake venomics: Characterization of protein families in Sistrurus barbouri venom by cysteine mapping, N-terminal sequencing, and tandem mass spectrometry analysis. Proteomics 2004, 4, 327–338. [Google Scholar] [CrossRef]
- Asita de Silva, H.; Ryan, N.M.; Janaka de Silva, H. Adverse reactions to snake antivenom and their prevention treatment. Br. J. Clin. Pharmacol. 2016, 81, 446–452. [Google Scholar] [CrossRef] [Green Version]
- Lauridsen, L.P.; Laustsen, A.H.; Lomonte, B.; Gutiérrez, J.M. Exploring the venom of the forest cobra snake: Toxicovenomics and antivenom profiling of Naja melanoleuca. J. Proteom. 2017, 150, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Bazaa, A.; Marrakchi, N.; El Ayeb, M.; Sanz, L.; Calvete, J.J. Snake venomics: Comparative analysis of the venom proteomes of the Tunisian snakesCerastes cerastes, Cerastes vipera and Macrovipera lebetina. Proteomics 2005, 5, 4223–4235. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M.T. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef] [PubMed]
- Moura-Da-Silva, A.; Ramos, O.; Baldó, C.; Niland, S.; Hansen, U.; Ventura, J.; Furlan, S.; Butera, D.; Della-Casa, M.; Tanjoni, I.; et al. Collagen binding is a key factor for the hemorrhagic activity of snake venom metalloproteinases. Biochimie 2008, 90, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, E.E.; Lucena, S.E.; Reyes, S.; Soto, J.G.; Cantu, E.; Lopez-Johnston, J.C.; Guerrero, B.; Salazar, A.M.; Rodríguez-Acosta, A.; Galan, J.A.; et al. Cloning, expression, and hemostatic activities of a disintegrin, r-mojastin 1, from the mohave rattlesnake (Crotalus scutulatus scutulatus). Thromb. Res. 2010, 126, e211–e219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvete, J.J. Structure-function correlations of snake venom disintegrins. Curr. Pharm. Des. 2005, 11, 829–835. [Google Scholar] [CrossRef]
- Calvete, J.J.; Marcinkiewicz, C.; Monleón, D.; Esteve, V.; Celda, B.; Juárez, P.; Sanz, L. Snake venom disintegrins: Evolution of structure and function. Toxicon 2005, 45, 1063–1074. [Google Scholar] [CrossRef]
- Huang, T.-F.; Wu, Y.-J.; Ouyang, C. Characterization of a potent platelet aggregation inhibitor from Agkistrodon rhodostoma snake venom. Biochim. Biophys. Acta 1987, 925, 248–257. [Google Scholar] [CrossRef]
- Suntravat, M.; Jia, Y.; Lucena, S.E.; Sánchez, E.E.; Pérez, J.C. cDNA cloning of a snake venom metalloproteinase from the eastern diamondback rattlesnake (Crotalus adamanteus), and the expression of its disintegrin domain with anti-platelet effects. Toxicon 2013, 64, 43–54. [Google Scholar] [CrossRef] [Green Version]
- Lucena, S.E.; Jia, Y.; Soto, J.G.; Parral, J.; Cantu, E.; Brannon, J.; Lardner, K.; Ramos, C.J.; Seoane, A.I.; Sánchez, E.E. Anti-invasive and anti-adhesive activities of a recombinant disintegrin, r-viridistatin 2, derived from the Prairie rattlesnake (Crotalus viridis viridis). Toxicon 2012, 60, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Carey, C.M.; Bueno, R.; Gutierrez, D.A.; Petro, C.; Lucena, S.E.; Sánchez, E.E.; Soto, J.G. Recombinant rubistatin (r-Rub), an MVD disintegrin, inhibits cell migration and proliferation, and is a strong apoptotic inducer of the human melanoma cell line SK-Mel-28. Toxicon 2012, 59, 241–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niit, M.; Hoskin, V.; Carefoot, E.; Geletu, M.; Arulanandam, R.; Elliott, B.; Raptis, L. Cell-cell and cell-matrix adhesion in survival and metastasis: Stat3 versus Akt. Biomol. Concepts 2015, 6, 383–399. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Calvete, J.J. The continuing saga of snake venom disintegrins. Toxicon 2013, 62, 40–49. [Google Scholar] [CrossRef]
- McLane, M.A.; Sanchez, E.E.; Wong, A.; Paquette-Straub, C.; Perez, J.C. Disintegrins. Curr. Drug Targets Cardiovasc. Haematol. Disord. 2004, 4, 327–355. [Google Scholar] [CrossRef]
- Glenn, J.; Straight, R. Mojave rattlesnake Crotalus scutulatus scutulatus venom: Variation in toxicity with geographical origin. Toxicon 1978, 16, 81–84. [Google Scholar] [CrossRef]
- Salazar, A.M.; Aguilar, I.; Guerrero, B.; Girón, M.E.; Lucena, S.; Sánchez, E.E.; Rodríguez-Acosta, A. Intraspecies differences in hemostatic venom activities of the South American rattlesnakes, Crotalus durissus cumanensis, as revealed by a range of protease inhibitors. Blood Coagul. Fibrinolysis 2008, 19, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Strickland, J.L.; Mason, A.J.; Rokyta, D.R.; Parkinson, C.L. Phenotypic variation in Mojave Rattlesnake (Crotalus scutulatus) venom is driven by four toxin families. Toxins 2018, 10, 135. [Google Scholar] [CrossRef] [Green Version]
- Fry, B.G.; Winkel, K.D.; Wickramaratna, J.C.; Hodgson, W.C.; Wüster, W. Effectiveness of snake antivenom: Species and regional venom variation and its clinical impact. J. Toxicol. 2003, 22, 23–34. [Google Scholar] [CrossRef]
- Harrison, R.A.; Moura-Da-Silva, A.M.; Laing, G.D.; Wu, Y.; Richards, A.; Broadhead, A.; Bianco, A.E.; Theakston, R.D.G. Antibody from mice immunized with DNA encoding the carboxyl-disintegrin and cysteine-rich domain (JD9) of the haemorrhagic metalloprotease, Jararhagin, inhibits the main lethal component of viper venom. Clin. Exp. Immunol. 2000, 121, 358–363. [Google Scholar] [CrossRef]
- Tanjoni, I.; Butera, D.; Spencer, P.J.; Takehara, H.A.; Fernandes, I.; Moura-Da-Silva, A.M. Phylogenetic conservation of a snake venom metalloproteinase epitope recognized by a monoclonal antibody that neutralizes hemorrhagic activity. Toxicon 2003, 42, 809–816. [Google Scholar] [CrossRef]
- Tanjoni, I.; Butera, D.; Bento, L.; Della Casa, M.S.; Marques-Porto, R.; Takehara, H.A.; Gutiérrez, J.M.; Fernandes, I.; Da Silva, A.M.M. Snake venom metalloproteinases: Structure/function relationships studies using monoclonal antibodies. Toxicon 2003, 42, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Morine, N.; Matsuda, S.; Terada, K.; Eto, A.; Ishida, I.; Oku, H. Neutralization of hemorrhagic snake venom metalloproteinase HR1a from Protobothrops flavoviridis by human monoclonal antibody. Toxicon 2008, 51, 345–352. [Google Scholar] [CrossRef]
- Saxena, A.; Hur, R.; Doctor, B.P. Allosteric control of acetylcholinesterase activity by monoclonal antibodies. Biochemistry 1998, 37, 145–154. [Google Scholar] [CrossRef]
- Roguin, L.P.; Retegui, L.A. Monoclonal antibodies inducing conformational changes on the antigen molecule. Scand. J. Immunol. 2003, 58, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, E.E.; Rodríguez-Acosta, A.; Palomar, R.; Lucena, S.E.; Bashir, S.; Soto, J.G.; Pérez, J.C. Colombistatin: A disintegrin isolated from the venom of the South American snake (Bothrops colombiensis) that effectively inhibits platelet aggregation and SK-Mel-28 cell adhesion. Arch. Toxicol. 2008, 83, 271–279. [Google Scholar] [CrossRef]
- Sánchez, E.E.; Galan, J.A.; Powell, R.L.; Reyes, S.R.; Soto, J.G.; Russell, W.K.; Russell, D.H.; Pérez, J.C. Disintegrin, hemorrhagic, and proteolytic activities of Mohave rattlesnake, Crotalus scutulatus scutulatus venoms lacking Mojave toxin. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2005, 141, 124–132. [Google Scholar] [CrossRef]
- Umthong, S.; Puthong, S.; Chanchao, C. Trigona laeviceps propolis from Thailand: Antimicrobial, antiproliferative and cytotoxic activities. Am. J. Chin. Med. 2009, 37, 855–865. [Google Scholar] [CrossRef]
- Huang, S.-Y.; Pérez, J.C. Comparative study on hemorrhagic and proteolytic activities of snake venoms. Toxicon 1980, 18, 421–426. [Google Scholar] [CrossRef]
- Sánchez, E.E.; Galán, J.A.; Perez, J.C.; Rodrıguez-Acosta, A.; Chase, P.B. The efficacy of two antivenoms against the venom of North American snakes. Toxicon 2003, 41, 357–365. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szteiter, S.S.; Diego, I.N.; Ortegon, J.; Salinas, E.M.; Cirilo, A.; Reyes, A.; Sanchez, O.; Suntravat, M.; Salazar, E.; Sánchez, E.E.; et al. Examination of the Efficacy and Cross-Reactivity of a Novel Polyclonal Antibody Targeting the Disintegrin Domain in SVMPs to Neutralize Snake Venom. Toxins 2021, 13, 254. https://doi.org/10.3390/toxins13040254
Szteiter SS, Diego IN, Ortegon J, Salinas EM, Cirilo A, Reyes A, Sanchez O, Suntravat M, Salazar E, Sánchez EE, et al. Examination of the Efficacy and Cross-Reactivity of a Novel Polyclonal Antibody Targeting the Disintegrin Domain in SVMPs to Neutralize Snake Venom. Toxins. 2021; 13(4):254. https://doi.org/10.3390/toxins13040254
Chicago/Turabian StyleSzteiter, Shelby S., Ilse N. Diego, Jonathan Ortegon, Eliana M. Salinas, Abcde Cirilo, Armando Reyes, Oscar Sanchez, Montamas Suntravat, Emelyn Salazar, Elda E. Sánchez, and et al. 2021. "Examination of the Efficacy and Cross-Reactivity of a Novel Polyclonal Antibody Targeting the Disintegrin Domain in SVMPs to Neutralize Snake Venom" Toxins 13, no. 4: 254. https://doi.org/10.3390/toxins13040254





