Detoxification and Excretion of Trichothecenes in Transgenic Arabidopsis thaliana Expressing Fusarium graminearum Trichothecene 3-O-acetyltransferase
Abstract
:1. Introduction
2. Results
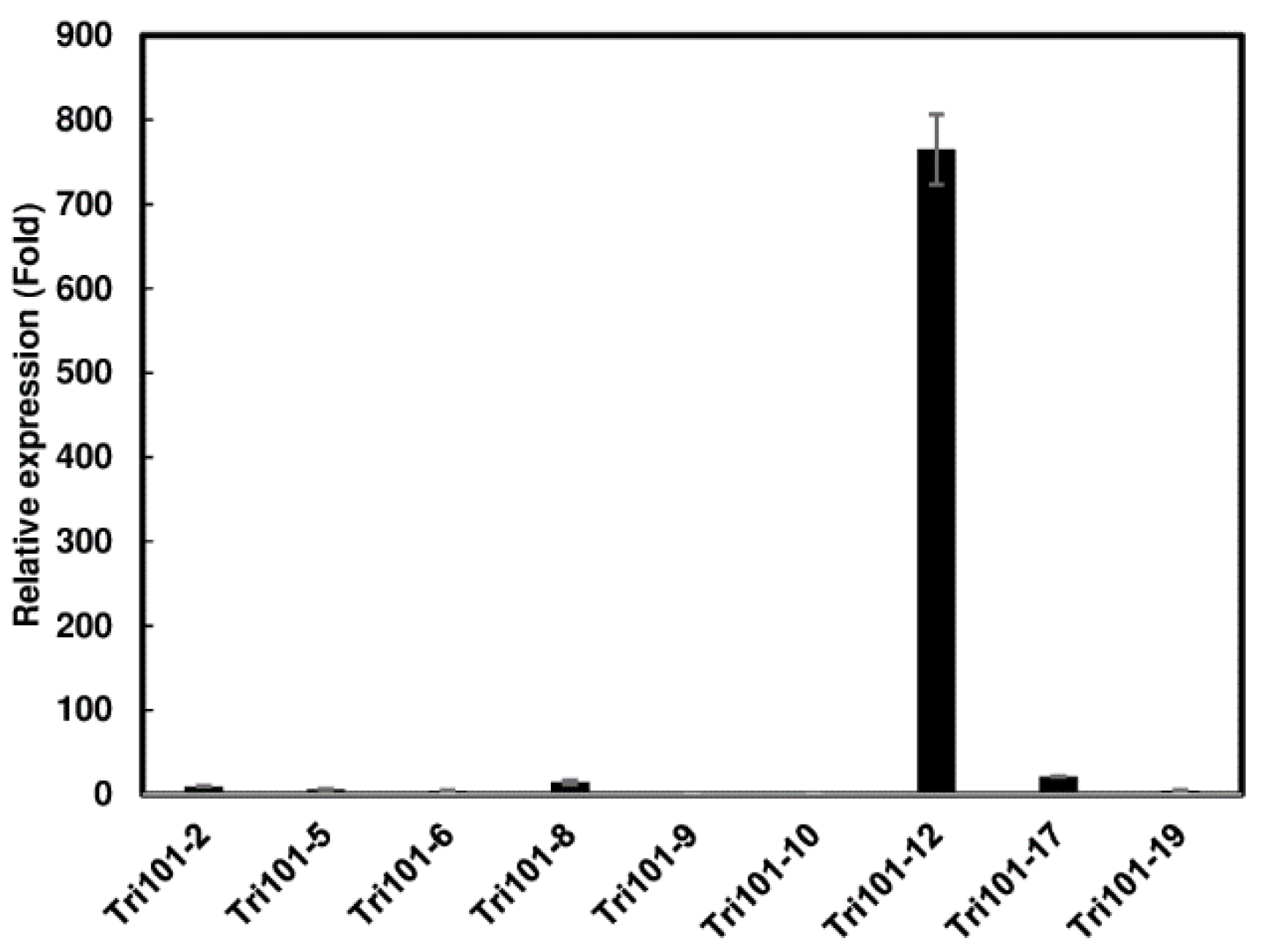
2.1. Generation of Transgenic Arabidopsis Expressing FgTri101
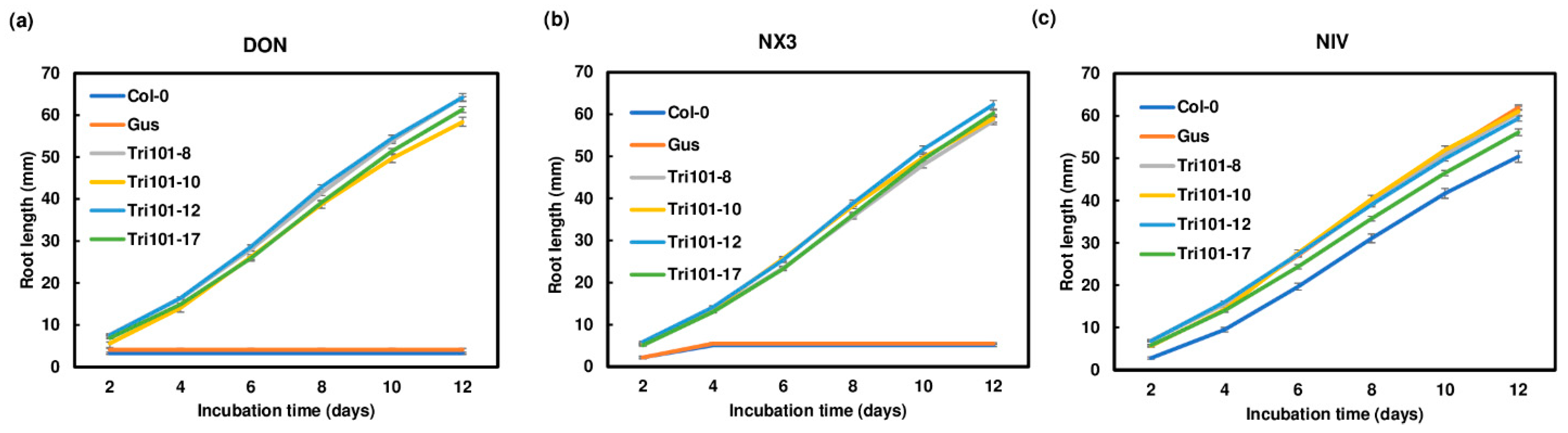
2.2. Enhanced Root Growth of Transgenic Arabidopsis Plants Expressing FgTri101 on Media Containing Trichothecenes
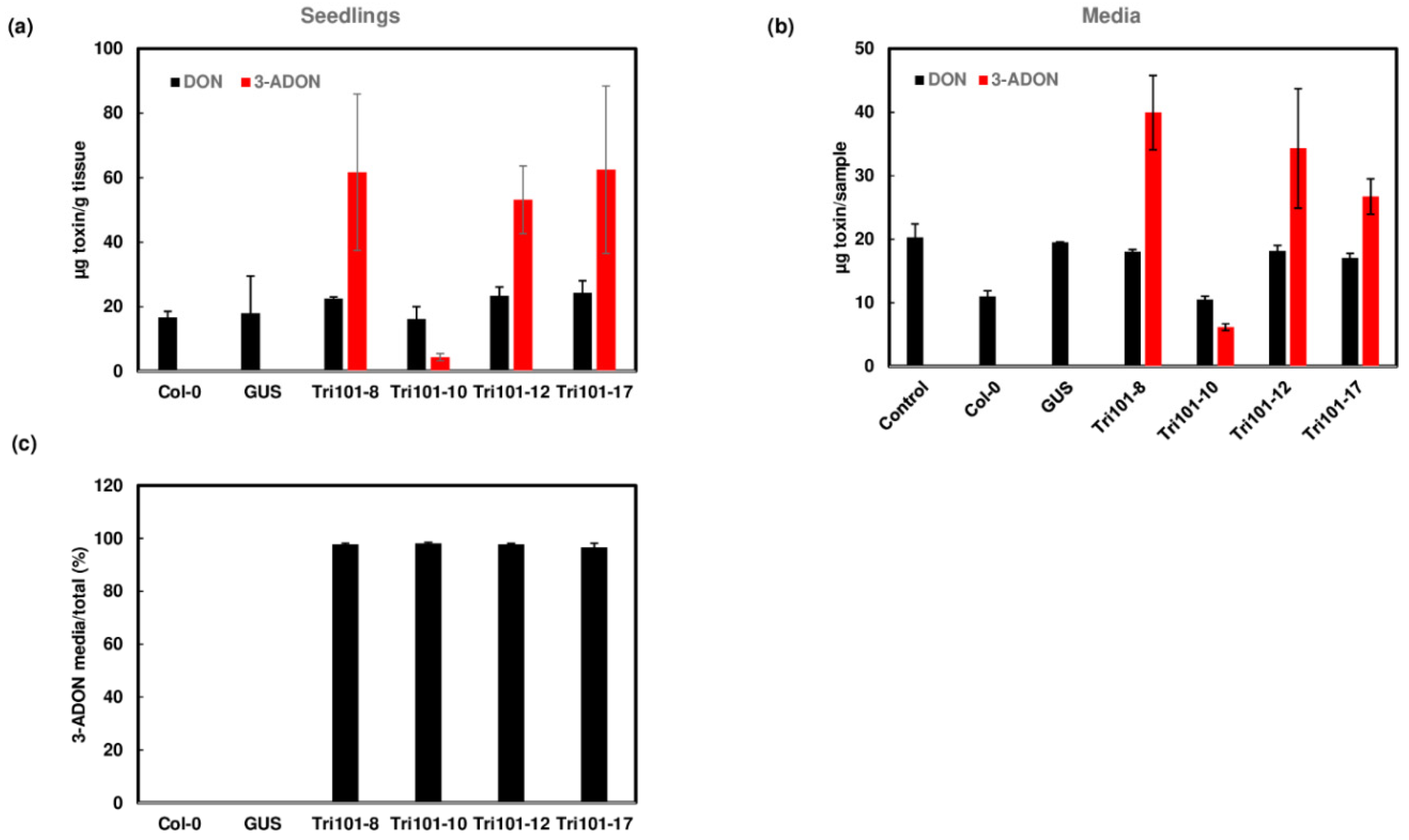
2.3. Acetylation and Excretion of Trichothecenes by Transgenic Arabidopsis Seedlings Expressing FgTri101
2.4. Transgenic Arabidopsis Expressing FgTri101 Accumulated Higher Weight with Addition of Trichothecenes
2.5. Morphology of Transgenic Arabidopsis Plants Expressing FgTri101
3. Discussion
4. Materials and Methods
4.1. Construction of FgTri101 Vector and Arabidopsis Transformation
4.2. Molecular Analysis of Transgenic Arabidopsis
4.3. Mycotoxins
4.4. Root Growth Assays
4.5. Conversion of Trichothecenes by Transgenic Arabidopsis Seedlings
4.6. Mycotoxin Extraction and Quantification
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rocha, O.; Ansari, K.; Doohan, F.M. Effects of trichothecene mycotoxins on eukaryotic cells: A review. Food Addit. Contam. 2005, 22, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Bin-Umer, M.A.; McLaughlin, J.E.; Basu, D.; McCormick, S.; Tumer, N.E. Trichothecene Mycotoxins Inhibit Mitochondrial Translation—Implication for the Mechanism of Toxicity. Toxins 2011, 3, 1484–1501. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H. Reduced Virulence ofGibberella zeaeCaused by Disruption of a TrichotheceneToxin Biosynthetic Gene. Mol. Plant Microbe Interact. 1995, 8, 593–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, C.; Von Wettstein, D.; Schäfer, W.; Kogel, K.-H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.; Torres-Acosta, J.A.; Heinen, S.J.; McCormick, S.; Lemmens, M.; Paris, M.P.K.; Berthiller, F.; Adam, G.; Muehlbauer, G.J. Transgenic Arabidopsis thaliana expressing a barley UDP-glucosyltransferase exhibit resistance to the mycotoxin deoxynivalenol. J. Exp. Bot. 2012, 63, 4731–4740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Shin, S.; Heinen, S.; Dill-Macky, R.; Berthiller, F.; Nersesian, N.; Clemente, T.; McCormick, S.; Muehlbauer, G.J. Transgenic Wheat Expressing a Barley UDP-Glucosyltransferase Detoxifies Deoxynivalenol and Provides High Levels of Resistance to Fusarium graminearum. Mol. Plant Microbe Interact. 2015, 28, 1237–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef]
- Proctor, R.H.; McCormick, S.P.; Kim, H.-S.; Cardoza, R.E.; Stanley, A.M.; Lindo, L.; Kelly, A.; Brown, D.W.; Lee, T.; Vaughan, M.M.; et al. Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PLoS Pathog. 2018, 14, e1006946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCormick, S.P.; Alexander, N.J.; Trapp, S.E.; Hohn, T.M. Disruption of TRI101, the Gene Encoding Trichothecene 3-O-Acetyltransferase, fromFusarium sporotrichioides. Appl. Environ. Microbiol. 1999, 65, 5252–5256. [Google Scholar] [CrossRef] [Green Version]
- Muhitch, M.J.; McCormick, S.P.; Alexander, N.J.; Hohn, T.M. Transgenic expression of the TRI101 or PDR5 gene increases resistance of tobacco to the phytotoxic effects of the trichothecene 4,15-diacetoxyscirpenol. Plant Sci. 2000, 157, 201–207. [Google Scholar] [CrossRef]
- Okubara, P.A.; Blechl, A.E.; McCormick, S.P.; Alexander, N.J.; Dill-Macky, R.; Hohn, T.M. Engineering deoxynivalenol metabolism in wheat through the expression of a fungal trichothecene acetyltransferase gene. Theor. Appl. Genet. 2002, 106, 74–83. [Google Scholar] [CrossRef]
- Manoharan, M.; Dahleen, L.S.; Hohn, T.M.; Neate, S.M.; Yu, X.-H.; Alexander, N.J.; McCormick, S.P.; Bregitzer, P.; Schwarz, P.B.; Horsley, R.D. Expression of 3-OH trichothecene acetyltransferase in barley (Hordeum vulgare L.) and effects on deoxynivalenol. Plant Sci. 2006, 171, 699–706. [Google Scholar] [CrossRef]
- Ohsato, S.; Ochiai-Fukuda, T.; Nishiuchi, T.; Takahashi-Ando, N.; Koizumi, S.; Hamamoto, H.; Kudo, T.; Yamaguchi, I.; Kimura, M. Transgenic rice plants expressing trichothecene 3-O-acetyltransferase show resistance to the Fusarium phytotoxin deoxynivalenol. Plant Cell Rep. 2006, 26, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Garvey, G.S.; McCormick, S.P.; Rayment, I. Structural and Functional Characterization of the TRI101 Trichothecene 3-O-Acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum. J. Biol. Chem. 2008, 283, 1660–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, A.C.; Ward, T.J. Population genomics of Fusarium graminearum reveals signatures of divergent evolution within a major cereal pathogen. PLoS ONE 2018, 13, e0194616. [Google Scholar] [CrossRef] [Green Version]
- Woelflingseder, L.; Gruber, N.; Adam, G.; Marko, D. Pro-Inflammatory Effects of NX-3 Toxin Are Comparable to Deoxynivalenol and not Modulated by the Co-Occurring Pro-Oxidant Aurofusarin. Microorganisms 2020, 8, 603. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Lakshman, D.K.; Galvez, L.C.; Mitra, S.; Baenziger, P.S.; Mitra, A. Transgenic expression of lactoferrin imparts enhanced resistance to head blight of wheat caused by Fusarium graminearum. BMC Plant Biol. 2012, 12, 33. [Google Scholar] [CrossRef] [Green Version]
- Masuda, D.; Ishida, M.; Yamaguchi, K.; Yamaguchi, I.; Kimura, M.; Nishiuchi, T. Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 1617–1626. [Google Scholar] [CrossRef] [Green Version]
- Gatti, M.; Choulet, F.; Macadré, C.; Guérard, F.; Seng, J.-M.; Langin, T.; Dufresne, M. Identification, Molecular Cloning, and Functional Characterization of a Wheat UDP-Glucosyltransferase Involved in Resistance to Fusarium Head Blight and to Mycotoxin Accumulation. Front. Plant Sci. 2018, 9, 1853. [Google Scholar] [CrossRef]
- Wipfler, R.; McCormick, S.P.; Proctor, R.H.; Teresi, J.M.; Hao, G.; Ward, T.J.; Alexander, N.J.; Vaughan, M.M. Ward Synergistic Phytotoxic Effects of Culmorin and Trichothecene Mycotoxins. Toxins 2019, 11, 555. [Google Scholar] [CrossRef] [Green Version]
- Eudes, F.; Comeau, A.; Rioux, S.; Collin, J. Phytotoxicité de huit mycotoxines associées à la fusariose de l’épi chez le blé. Can. J. Plant Pathol. 2000, 22, 286–292. [Google Scholar] [CrossRef]
- Cummins, I.; Dixon, D.P.; Freitag-Pohl, S.; Skipsey, M.; Edwards, R. Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab. Rev. 2011, 43, 266–280. [Google Scholar] [CrossRef]
- Walter, S.; Kahla, A.; Arunachalam, C.; Perochon, A.; Khan, M.R.; Scofield, S.R.; Doohan, F.M. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 2015, 66, 2583–2593. [Google Scholar] [CrossRef] [Green Version]
- Tommasini, R.; Vogt, E.; Fromenteau, M.; Hörtensteiner, S.; Matile, P.; Amrhein, N.; Martinoia, E. An ABC-transporter of Arabidopsis thalianahas both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J. 1998, 13, 773–780. [Google Scholar] [CrossRef]
- Szponarski, W.; Sommerer, N.; Boyer, J.-C.; Rossignol, M.; Gibrat, R. Large-scale characterization of integral proteins from Arabidopsis vacuolar membrane by two-dimensional liquid chromatography. Proteomics 2004, 4, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Balzi, E.; Wang, M.; Leterme, S.; Van Dyck, L.; Goffeau, A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 1994, 269, 2206–2214. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Hohn, T.M. TRI12, a trichothecene efflux pump from Fusarium sporotrichioides: Gene isolation and expression in yeast. Mol. Genet. Genom. 1999, 261, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arab. Book 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jasinski, M.; Stukkens, Y.; Degand, H.; Purnelle, B.; Marchand-Brynaert, J.; Boutry, M. A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 2001, 13, 1095–1107. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.; Lee, S.-H.; Shin, J.Y.; Kim, H.-K.; Yun, S.-H.; Kim, H.-Y.; Lee, S.; Ryu, J.-G. Comparison of Trichothecene Biosynthetic Gene Expression between Fusarium graminearum and Fusarium asiaticum. Plant Pathol. J. 2014, 30, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Alexander, N.J.; McCormick, S.P.; Ziegenhorn, S.L. Phytotoxicity of selected trichothecenes using Chlamydomonas rein-hardtii as a model systemt. Nat. Toxins 1999, 7, 265–269. [Google Scholar] [CrossRef]
- Desjardins, A.E.; McCormick, S.P.; Appell, M. Structure−Activity Relationships of Trichothecene Toxins in anArabidopsis thalianaLeaf Assay. J. Agric. Food Chem. 2007, 55, 6487–6492. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; Stover, E.; Gupta, G. Overexpression of a Modified Plant Thionin Enhances Disease Resistance to Citrus Canker and Huanglongbing (HLB). Front. Plant Sci. 2016, 7, 1078. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Hao, G.; Bakker, M.G.; Kim, H.S. Enhanced Resistance to Fusarium graminearum in Transgenic Arabidopsis Plants Expressing a Modified Plant Thionin. Phytopathology 2020. [Google Scholar] [CrossRef] [PubMed]
- Garcia, I.; Rodgers, M.; Pepin, R.; Hsieh, T.-F.; Matringe, M. Characterization and Subcellular Compartmentation of Recombinant 4-Hydroxyphenylpyruvate Dioxygenase from Arabidopsis in Transgenic Tobacco. Plant Physiol. 1999, 119, 1507–1516. [Google Scholar] [CrossRef] [Green Version]
- Hao, G.; McCormick, S.; Vaughan, M.M.; Naumann, T.A.; Kim, H.-S.; Proctor, R.; Kelly, A.; Ward, T.J. Fusarium graminearum arabinanase (Arb93B) Enhances Wheat Head Blight Susceptibility by Suppressing Plant Immunity. Mol. Plant Microbe Interact. 2019, 32, 888–898. [Google Scholar] [CrossRef] [PubMed]


| Genotypes | Flower Time (days) | Rosette Leaf Number a | Plant Height (cm) b | Shoot Number b |
|---|---|---|---|---|
| GUS | 27 | 5.4 ± 0.52 | 32.6 ± 2.29 | 5.6 ± 1.30 |
| Tri101-8 | 27 | 5.1 ± 0.35 | 29.5 ± 2.09 | 6.5 ± 1.41 |
| Tri101-12 | 27 | 4.6 * ± 0.50 | 31.5 ± 3.85 | 6.5 ± 1.69 |
| Tri101-17 | 27 | 5.0 ± 0.53 | 31.5 ± 2.19 | 5.6 ± 1.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, G.; McCormick, S.; Tiley, H.; Usgaard, T. Detoxification and Excretion of Trichothecenes in Transgenic Arabidopsis thaliana Expressing Fusarium graminearum Trichothecene 3-O-acetyltransferase. Toxins 2021, 13, 320. https://doi.org/10.3390/toxins13050320
Hao G, McCormick S, Tiley H, Usgaard T. Detoxification and Excretion of Trichothecenes in Transgenic Arabidopsis thaliana Expressing Fusarium graminearum Trichothecene 3-O-acetyltransferase. Toxins. 2021; 13(5):320. https://doi.org/10.3390/toxins13050320
Chicago/Turabian StyleHao, Guixia, Susan McCormick, Helene Tiley, and Thomas Usgaard. 2021. "Detoxification and Excretion of Trichothecenes in Transgenic Arabidopsis thaliana Expressing Fusarium graminearum Trichothecene 3-O-acetyltransferase" Toxins 13, no. 5: 320. https://doi.org/10.3390/toxins13050320
APA StyleHao, G., McCormick, S., Tiley, H., & Usgaard, T. (2021). Detoxification and Excretion of Trichothecenes in Transgenic Arabidopsis thaliana Expressing Fusarium graminearum Trichothecene 3-O-acetyltransferase. Toxins, 13(5), 320. https://doi.org/10.3390/toxins13050320





