Charged Residues Flanking the Transmembrane Domain of Two Related Toxin–Antitoxin System Toxins Affect Host Response
Abstract
1. Introduction
2. Results
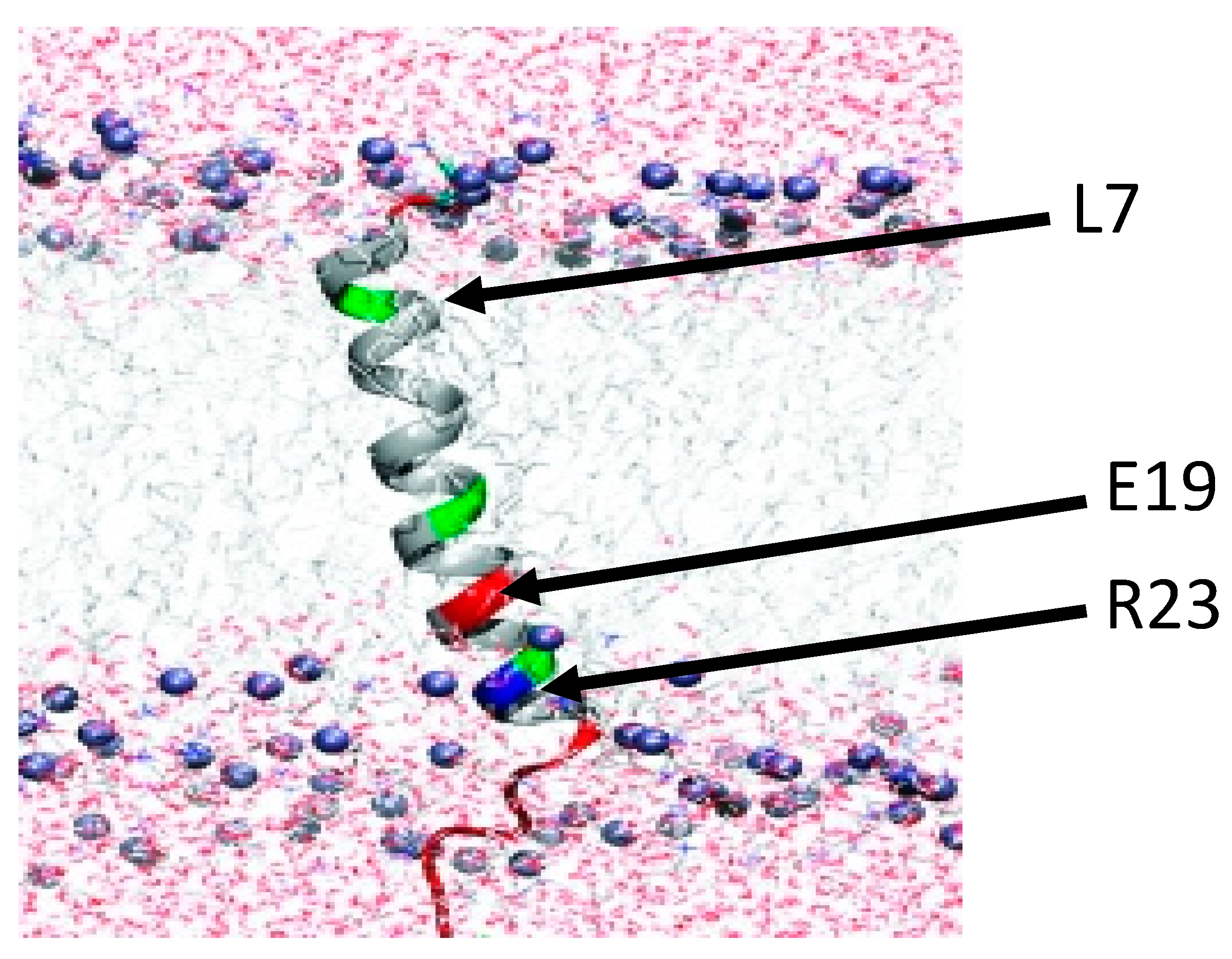
2.1. Importance of the Charged C-Terminal Tail to Fst Toxicity
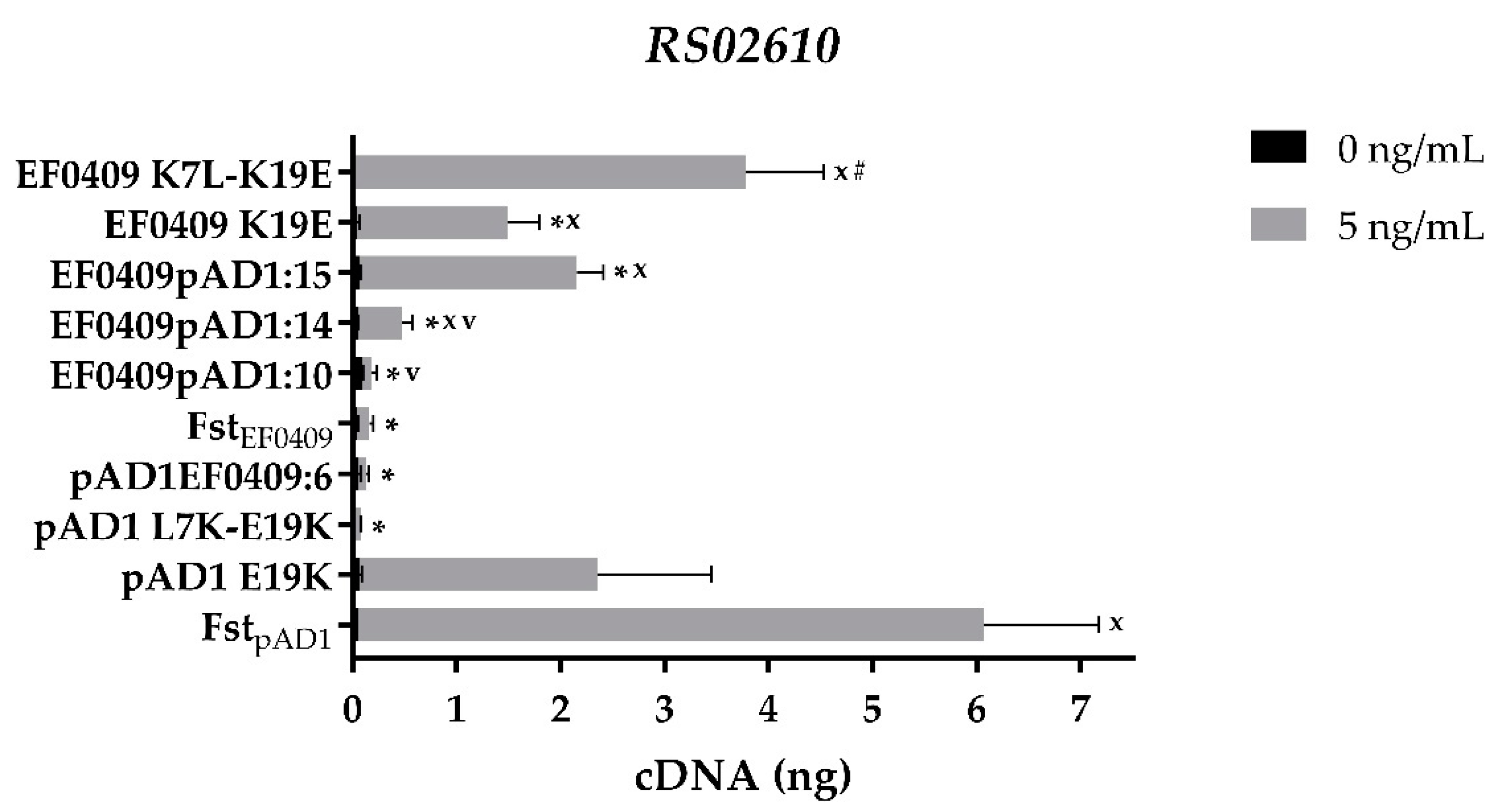
2.2. Identification of the Key Residues for the Differential Response of RS02610
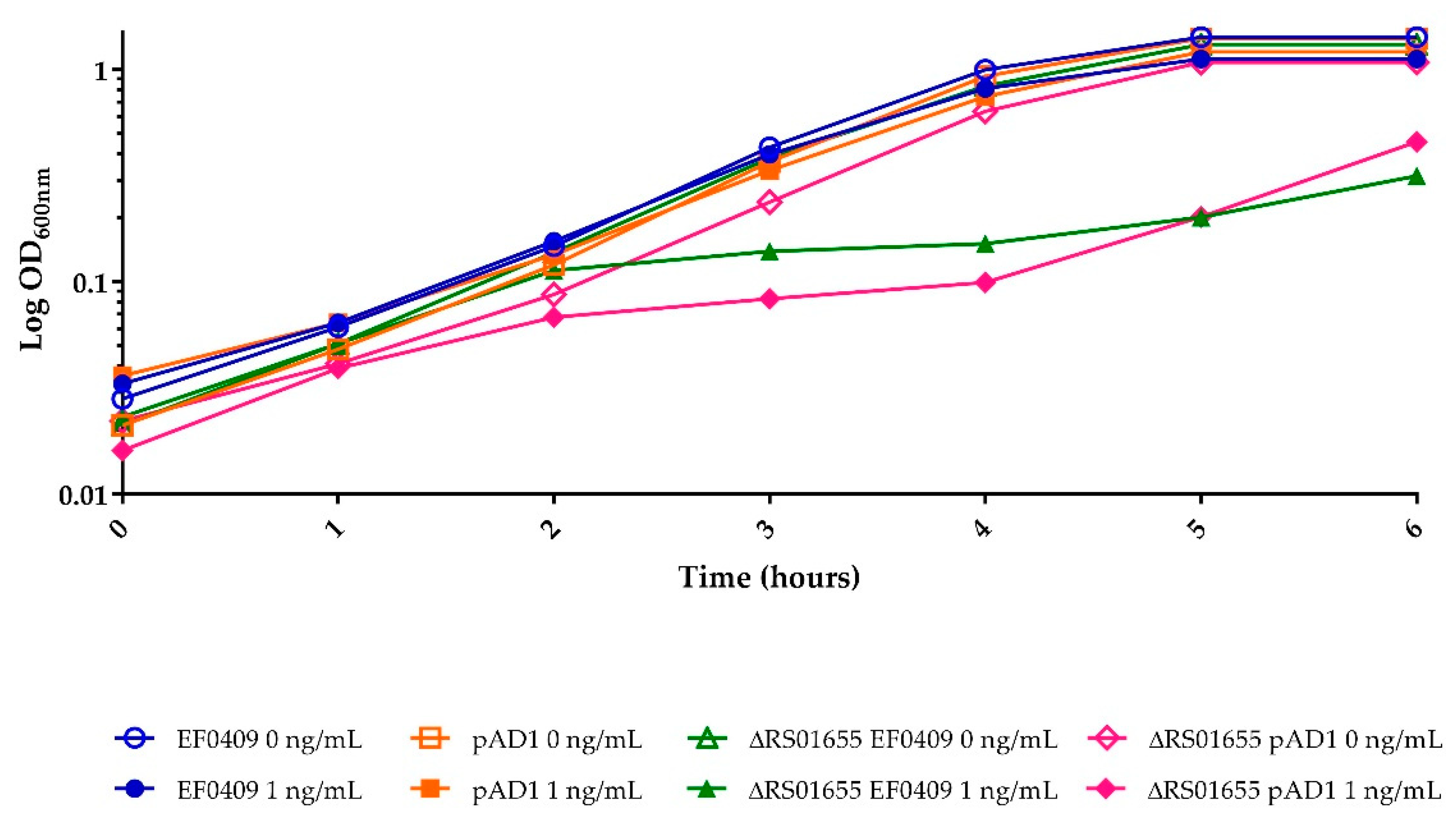
2.3. The Differential Response of RS01655 May Relate to Its Function as an Efflux Pump
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media, and Growth Conditions
4.2. Genetic Manipulations
4.3. RNA Purification, Manipulation, and Statistical Methodology
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef]
- Unterholzner, S.J.; Poppenberger, B.; Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elem. 2013, 3, e26219. [Google Scholar] [CrossRef]
- Brantl, S.; Müller, P. Toxin–Antitoxin Systems in Bacillus subtilis. Toxins 2019, 11, 262. [Google Scholar] [CrossRef]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Van Melderen, L. Bacterial toxin-antitoxin systems: Translation inhibitors everywhere. Mob. Genet. Elem. 2011, 1, 283–306. [Google Scholar] [CrossRef]
- Pandey, D.P.; Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wood, T.K. Toxin/Antitoxin System Paradigms: Toxins Bound to Antitoxins Are Not Likely Activated by Preferential Antitoxin Degradation. Adv. Biosyst. 2020, 4, e1900290. [Google Scholar] [CrossRef]
- Van Melderen, L.; De Bast, M.S. Bacterial Toxin–Antitoxin Systems: More Than Selfish Entities? PLoS Genet. 2009, 5, e1000437. [Google Scholar] [CrossRef]
- Masachis, S.; Darfeuille, F. Type I Toxin-Antitoxin Systems: Regulating Toxin Expression via Shine-Dalgarno Sequence Sequestration and Small RNA Binding. In Regulating with RNA in Bacteria and Archaea; American Society of Microbiology: Washington, DC, USA, 2019. [Google Scholar]
- Brantl, S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012, 9, 1488–1490. [Google Scholar] [CrossRef]
- Fozo, E.M.; Hemm, M.R.; Storz, G. Small Toxic Proteins and the Antisense RNAs That Repress Them. Microbiol. Mol. Biol. Rev. 2008, 72, 579–589. [Google Scholar] [CrossRef]
- Weaver, K. The Fst/Ldr Family of Type I TA System Toxins: Potential Roles in Stress Response, Metabolism and Pathogenesis. Toxins 2020, 12, 474. [Google Scholar] [CrossRef]
- Durand, S.; Jahn, N.; Condon, C.; Brantl, S. Type I toxin-antitoxin systems inBacillus subtilis. RNA Biol. 2012, 9, 1491–1497. [Google Scholar] [CrossRef]
- Brielle, R.; Pinel-Marie, M.-L.; Felden, B. Linking bacterial type I toxins with their actions. Curr. Opin. Microbiol. 2016, 30, 114–121. [Google Scholar] [CrossRef]
- Gurnev, P.A.; Ortenberg, R.; Dörr, T.; Lewis, K.; Bezrukov, S.M. Persister-promoting bacterial toxin TisB produces anion-selective pores in planar lipid bilayers. FEBS Lett. 2012, 586, 2529–2534. [Google Scholar] [CrossRef]
- Patel, S.; Weaver, K.E. Addiction Toxin Fst Has Unique Effects on Chromosome Segregation and Cell Division in Enterococcus faecalis and Bacillussubtilis. J. Bacteriol. 2006, 188, 5374–5384. [Google Scholar] [CrossRef] [PubMed]
- Jahn, N.; Brantl, S.; Strahl, H. Against the mainstream: The membrane-associated type I toxin BsrG fromBacillus subtilisinterferes with cell envelope biosynthesis without increasing membrane permeability. Mol. Microbiol. 2015, 98, 651–666. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wood, T.K. Post-segregational Killing and Phage Inhibition are not mediated by Cell Death through Toxin/Antitoxin Systems. Front. Microbiol. 2018, 9, 814. [Google Scholar] [CrossRef]
- Storz, G.; Wolf, Y.I.; Ramamurthi, K.S. Small Proteins Can No Longer Be Ignored. Annu. Rev. Biochem. 2014, 83, 753–777. [Google Scholar] [CrossRef] [PubMed]
- Kosfeld, A.; Jahreis, K. Characterization of the Interaction between the Small Regulatory Peptide SgrT and the EIICBGlc of the Glucose-Phosphotransferase System of E. coli K-12. Metabolites 2012, 2, 756–774. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Orr, M.W.; Wang, H.; Hobbs, E.C.; Shabalina, S.A.; Storz, G. The small protein MgtS and small RNA MgrR modulate the PitA phosphate symporter to boost intracellular magnesium levels. Mol. Microbiol. 2019, 111, 131–144. [Google Scholar] [CrossRef]
- Yeom, J.; Shao, Y.; Groisman, E.A. Small proteins regulateSalmonellasurvival inside macrophages by controlling degradation of a magnesium transporter. Proc. Natl. Acad. Sci. USA 2020, 117, 20235–20243. [Google Scholar] [CrossRef] [PubMed]
- Michaux, C.; Hartke, A.; Martini, C.; Reiss, S.; Albrecht, D.; Budin-Verneuil, A.; Sanguinetti, M.; Engelmann, S.; Hain, T.; Verneuil, N.; et al. Involvement of Enterococcus Faecalis Small RNAs in Stress Response and Virulence. Infect. Immun. 2014, 82, 3599–3611. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, T.J.; Ehli, E.; Kirshenmann, T.; Franch, T.; Gerdes, K.; Weaver, K.E. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol. Microbiol. 2002, 37, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Weaver, K.E.; Jensen, K.D.; Colwell, A.; Sriram, S. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 1996, 20, 53–63. [Google Scholar] [CrossRef]
- Weaver, K.E.; Weaver, D.M.; Wells, C.L.; Waters, C.M.; Gardner, M.E.; Ehli, E.A. Enterococcus faecalis Plasmid pAD1-Encoded Fst Toxin Affects Membrane Permeability and Alters Cellular Responses to Lantibiotics. J. Bacteriol. 2003, 185, 2169–2177. [Google Scholar] [CrossRef]
- Weaver, K.E.; Reddy, S.G.; Brinkman, C.L.; Patel, S.; Bayles, K.W.; Endres, J.L. Identification and characterization of a family of toxin–antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 2009, 155, 2930–2940. [Google Scholar] [CrossRef]
- Kwong, S.M.; Jensen, S.O.; Firth, N. Prevalence of Fst-like toxin–antitoxin systems. Microbiology 2010, 156, 975–977. [Google Scholar] [CrossRef]
- Fozo, E.M.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Storz, G. Abundance of type I toxin–antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef]
- Göbl, C.; Kosol, S.; Stockner, T.; Rückert, H.M.; Zangger, K. Solution Structure and Membrane Binding of the Toxin Fst of theparAddiction Module. Biochemistry 2010, 49, 6567–6575. [Google Scholar] [CrossRef] [PubMed]
- Sayed, N.; Nonin-Lecomte, S.; Réty, S.; Felden, B. Functional and Structural Insights of a Staphylococcus aureus Apoptotic-like Membrane Peptide from a Toxin-Antitoxin Module. J. Biol. Chem. 2012, 287, 43454–43463. [Google Scholar] [CrossRef]
- Weaver, K.E.; Chen, Y.; Miiller, E.M.; Johnson, J.N.; Dangler, A.A.; Manias, D.A.; Clem, A.M.; Schjodt, D.J.; Dunny, G.M. Examination of Enterococcus faecalis Toxin-Antitoxin System Toxin Fst Function Utilizing a Pheromone-Inducible Expression Vector with Tight Repression and Broad Dynamic Range. J. Bacteriol. 2017, 199, e00065-17. [Google Scholar] [CrossRef]
- Weaver, K.E. Thepartoxin-antitoxin system fromEnterococcus faecalisplasmid pAD1 and its chromosomal homologs. RNA Biol. 2012, 9, 1498–1503. [Google Scholar] [CrossRef]
- Maggi, S.; Yabre, K.; Ferrari, A.; Lazzi, C.; Kawano, M.; Rivetti, C.; Folli, C. Functional characterization of the type I toxin Lpt from Lactobacillus rhamnosus by fluorescence and atomic force microscopy. Sci. Rep. 2019, 9, 15208. [Google Scholar] [CrossRef] [PubMed]
- Ban, X.; Lahiri, P.; Dhoble, A.S.; Li, D.; Gu, Z.; Li, C.; Cheng, L.; Hong, Y.; Li, Z.; Kaustubh, B. Evolutionary Stability of Salt Bridges Hints Its Contribution to Stability of Proteins. Comput. Struct. Biotechnol. J. 2019, 17, 895–903. [Google Scholar] [CrossRef]
- Walker, K.D.; Causgrove, T.P. Contribution of arginine-glutamate salt bridges to helix stability. J. Mol. Model. 2009, 15, 1213–1219. [Google Scholar] [CrossRef]
- Donald, J.E.; Kulp, D.W.; DeGrado, W.F. Salt bridges: Geometrically specific, designable interactions. Proteins: Struct. Funct. Bioinform. 2010, 79, 898–915. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Mukharjee, D. Salt-bridge networks within globular and disordered proteins: Characterizing trends for designable interactions. J. Mol. Model. 2017, 23, 206. [Google Scholar] [CrossRef] [PubMed]
- Dunny, G.M.; Brown, B.L.; Clewell, D.B. Induced cell aggregation and mating in Streptococcus faecalis: Evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 1978, 75, 3479–3483. [Google Scholar] [CrossRef] [PubMed]
- Dunny, G.M.; Zimmerman, D.L.; Tortorello, M.L. Induction of surface exclusion (entry exclusion) by Streptococcus faecalis sex pheromones: Use of monoclonal antibodies to identify an inducible surface antigen involved in the exclusion process. Proc. Natl. Acad. Sci. USA 1985, 82, 8582–8586. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Cruz-Rodz, A.L.; Gilmore, M.S. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 1990, 224, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, C.L.; Bumgarner, R.; Kittichotirat, W.; Dunman, P.M.; Kuechenmeister, L.J.; Weaver, K.E. Characterization of the Effects of an rpoC Mutation That Confers Resistance to the Fst Peptide Toxin-Antitoxin System Toxin. J. Bacteriol. 2013, 195, 156–166. [Google Scholar] [CrossRef]
- Johnson, C.M.; Manias, D.A.; Haemig, H.A.H.; Shokeen, S.; Weaver, K.E.; Henkin, T.M.; Dunny, G.M. Direct Evidence for Control of the Pheromone-Inducible prgQ Operon of Enterococcus faecalis Plasmid pCF10 by a Countertranscript-Driven Attenuation Mechanism. J. Bacteriol. 2010, 192, 1634–1642. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, S.L.; Little, J.L.; Hoff, J.S.; Kristich, C.J. Requirement of the CroRS Two-Component System for Resistance to Cell Wall-Targeting Antimicrobials in Enterococcus faecium. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Thurlow, L.R.; Thomas, V.C.; Hancock, L.E. Capsular Polysaccharide Production in Enterococcus faecalis and Contribution of CpsF to Capsule Serospecificity. J. Bacteriol. 2009, 191, 6203–6210. [Google Scholar] [CrossRef] [PubMed]



| Mutant Fst toxin | Sequence a |
|---|---|
| FstpAD1 | VKDLMSLVIAPIFVGLVLEMISRVLDEEDDSRK |
| pAD1ter2 | VKDLMSLVIAPIFVGLVLEMISRVLDEEDDS |
| pAD1ter5 | VKDLMSLVIAPIFVGLVLEMISRVLDEE |
| pAD1ter7 | VKDLMSLVIAPIFVGLVLEMISRVLD |
| FstEF0409 | MYEIVTKILVPIFVGIVLKLVTIWLEKQNEE |
| EF0409ter2 | MYEIVTKILVPIFVGIVLKLVTIWLEKQN |
| EF0409ter5 | MYEIVTKILVPIFVGIVLKLVTIWLE |
| pAD1EF0409:6 | VKDLMSLVIAPIFVGLVLEMISRVLEKQNEE |
| EF0409pAD1:8 | MYEIVTKILVPIFVGIVLKLVTIWLDEEDDSRK |
| EF0409pAD1:10 | MYEIVTKILVPIFVGIVLKLVTIVLDEEDDSRK |
| EF0409pAD1:12 | MYEIVTKILVPIFVGIVLKLVSRVLDEEDDSRK |
| EF0409pAD1:14 | MYEIVTKILVPIFVGIVLKMISRVLDEEDDSRK |
| EF0409pAD1:15 | MYEIVTKILVPIFVGIVLEMISRVLDEEDDSRK |
| EF0409 K19E | MYEIVTKILVPIFVGIVLELVTIWLEKQNEE |
| pAD1 E19K | VKDLMSLVIAPIFVGLVLKMISRVLDEEDDSRK |
| EF0409pAD1N6-K19E | MKDLMSLILVPIFVGIVLELVTIWLEKQNEE |
| EF0409 K7L-K19E | MYEIVTLILVPIFVGIVLELVTIWLEKQNEE |
| pAD1 L7K-E19K | VKDLMSKVIAPIFVGLVLKMISRVLDEEDDSRK |
| EF0409pAD1FL | MYEIVTKILVPIFVGIVLKLVFLVLDEEDDSRK |
| Primer | Sequence |
|---|---|
| pCIE-EF0409 FWD | GTATACAGTTCATGTATATGTTCCC |
| pCIE-EF0409 REV | TGTGATGCACCTCCTTTC |
| RS02610 FWD | CAGATGACGGCTCAATTCAAAC |
| RS02610 REV | CAGCGGTACTTCCTTCAATCA |
| RS01655 FWD | GCACGATGTCTGGTGATGAT |
| RS01655 REV | CTTCGCTCCTAAATCCGCTAAG |
| mgtA FWD | AAAGGTGCGGTTGAAGAAATG |
| mgtA REV | TGACGCAGTGTCTCTGTTAAG |
| celA3 FWD | AGAAGATCGTGGCATGGAAG |
| celA3 REV | TGAAACGAACTTGTGGACCTAA |
| GyrB FWD | ACCAACACCGTGCAAGCC |
| GyrB REV | CAAGCCAAAACAGGTCGCC |
| Delta01655 FWD | GGGTCCTTCTGTGTGTGTAAATA |
| Delta01655 REV | GTCCACTCGGCTAAACGTATAAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holmes, A.; Sadlon, J.; Weaver, K. Charged Residues Flanking the Transmembrane Domain of Two Related Toxin–Antitoxin System Toxins Affect Host Response. Toxins 2021, 13, 329. https://doi.org/10.3390/toxins13050329
Holmes A, Sadlon J, Weaver K. Charged Residues Flanking the Transmembrane Domain of Two Related Toxin–Antitoxin System Toxins Affect Host Response. Toxins. 2021; 13(5):329. https://doi.org/10.3390/toxins13050329
Chicago/Turabian StyleHolmes, Andrew, Jessie Sadlon, and Keith Weaver. 2021. "Charged Residues Flanking the Transmembrane Domain of Two Related Toxin–Antitoxin System Toxins Affect Host Response" Toxins 13, no. 5: 329. https://doi.org/10.3390/toxins13050329
APA StyleHolmes, A., Sadlon, J., & Weaver, K. (2021). Charged Residues Flanking the Transmembrane Domain of Two Related Toxin–Antitoxin System Toxins Affect Host Response. Toxins, 13(5), 329. https://doi.org/10.3390/toxins13050329




