New Knowledge on Distribution and Abundance of Toxic Microalgal Species and Related Toxins in the Northwestern Black Sea
Abstract
1. Introduction
2. Results
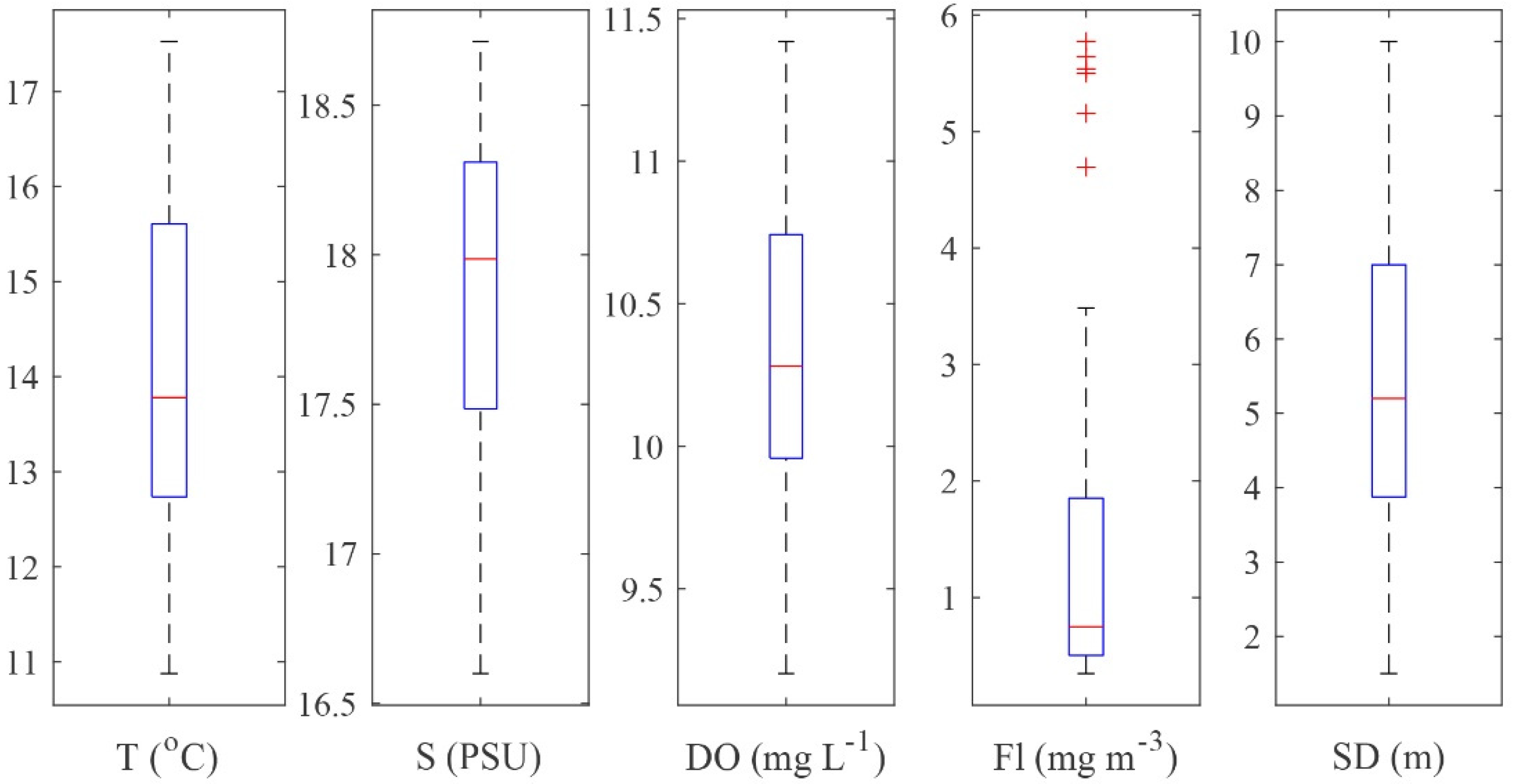
2.1. Environmental Characteristics of the Sampling Sites
2.2. Microscopy Identification and Enumeration of Potentially Toxic Microalgae
2.3. Potentially Toxic Species Detected with Metabarcoding
2.4. qPCR Analyses of Field Samples
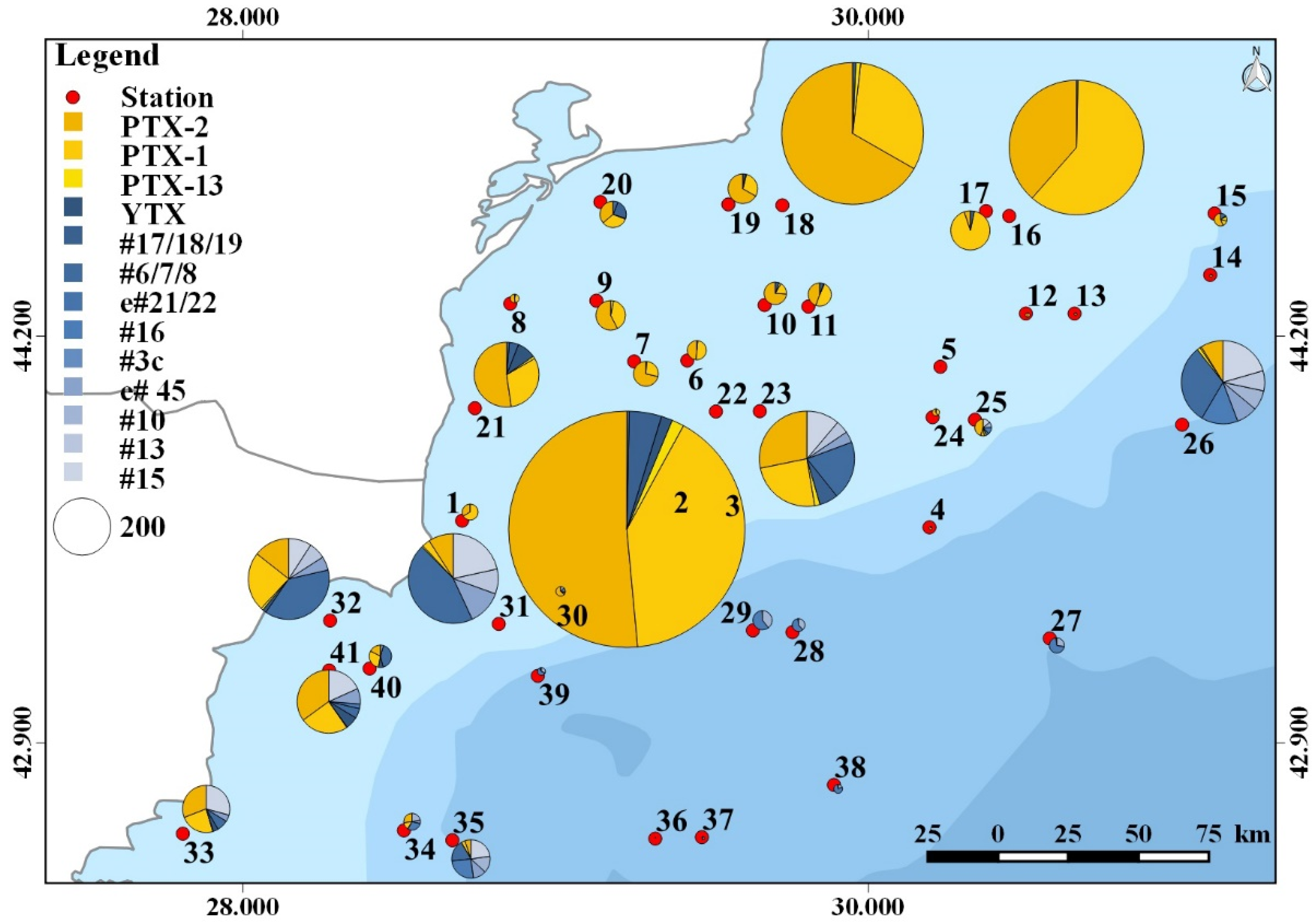
2.5. Toxin Distribution
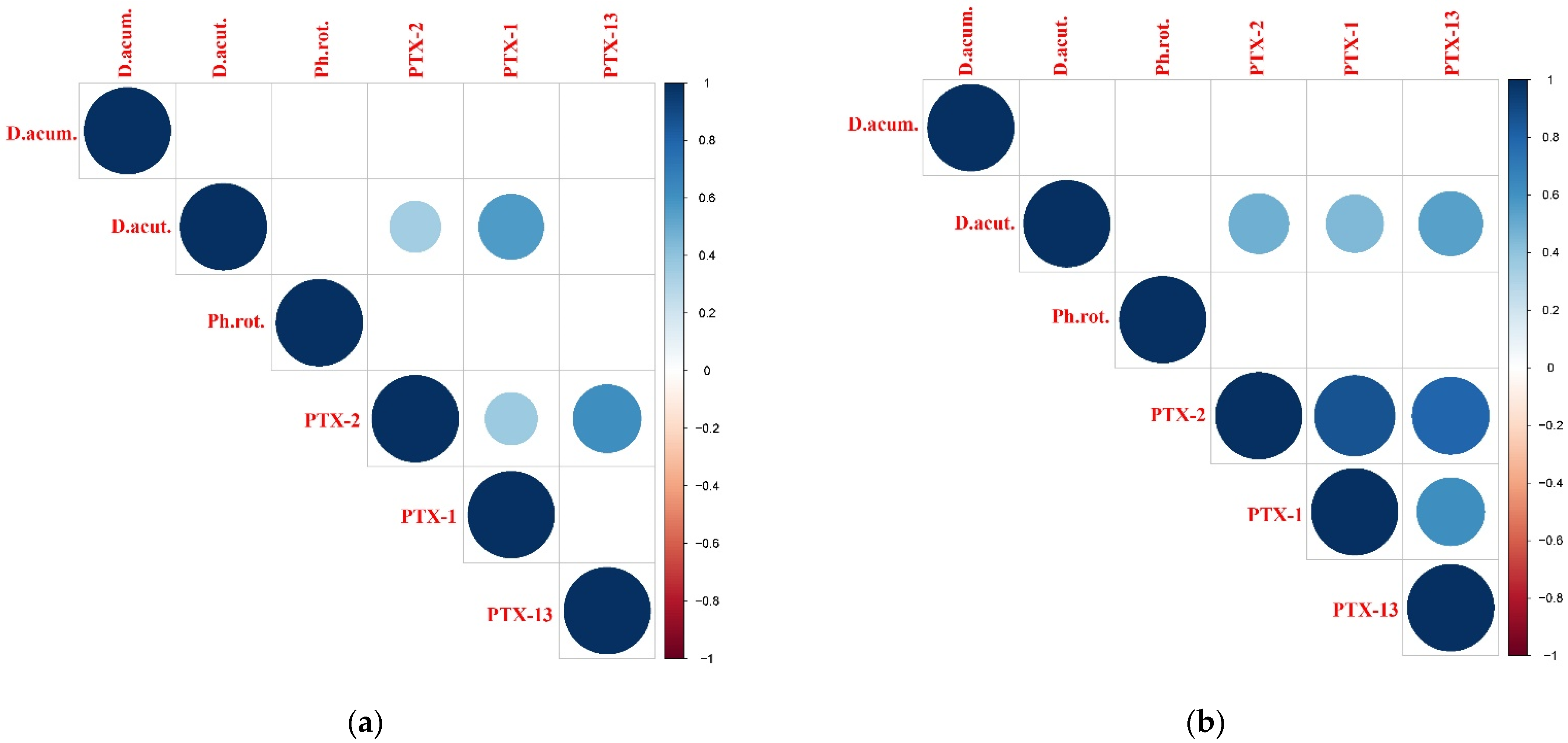
2.6. Identified Potentially Toxic Plankton Species and Correspondence with Detected Phycotoxins
3. Discussion
3.1. Pseudo-nitzschia and DA
3.2. Alexandrium, Gymnodinium catenatum, and PSP
3.3. Dinophysis, Phalacroma rotundatum, and OA/DTX/PTX Distribution
3.4. Protoceratium reticulatum, Lingulodinium polyedra, Gonyaulax spinifera, and YTX Profiles
3.5. Amphidomataceae and AZA
3.6. Karlodinium veneficum and KmTx
3.7. Other Potentially Toxic Species Identified in the Study
4. Conclusions
5. Materials and Methods
5.1. Study Area and Sampling
5.2. Plankton Sampling
5.3. Microscopy
5.4. DNA (metabarcoding) Analysis
5.5. DNA Extraction and qPCR
5.6. Toxin Extraction
5.7. LC-FLD and LC-MS/MS Analysis
5.8. Statistical Analysis
5.9. Data Used for Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, D.M.; Cembella, A.D.; Hallegraeff, G.M. Progress in understanding harmful algal blooms: Paradigm shifts and new technologies for research, monitoring, and management. Annu. Rev. Mar. Sci. 2012, 4, 143–176. [Google Scholar] [CrossRef]
- Gobler, C.J. Climate change and harmful algal blooms: Insights and perspective. Harmful Algae 2019, 91, 101731. [Google Scholar] [CrossRef]
- Hallegraeff, G.; Enevoldsen, H.; Zingone, A. Global harmful algal bloom status reporting. Harmful Algae 2021, 102, 101992. [Google Scholar] [CrossRef]
- Landsberg, J.H. The effects of harmful algal blooms on aquatic organisms. Rev. Fish. Sci. Aquac. 2002, 10, 113–390. [Google Scholar] [CrossRef]
- Vilariño, N.; Louzao, M.C.; Abal, P.; Cagide, E.; Carrera, C.; Vieytes, M.R.; Botana, L.M. Human poisoning from marine toxins: Unknowns for optimal consumer protection. Toxins 2018, 10, 324. [Google Scholar] [CrossRef]
- Lassus, P.; Chomérat, N.; Hess, P.; Nézan, E. Toxic and Harmful Microalgae of the World Ocean/Micro-Algues Toxiques et Nuisibles de L’océan Mondial; IOC Manuals and Guides, 68. (Bilingual English/French); International Society for the Study of Harmful Algae/Intergovernmental Oceanographic Commission of UNESCO: Copenhagen, Denmark, 2016; p. 523. [Google Scholar]
- Ferrante, M. Harmful algal blooms in the Mediterranean Sea: Effects on human health. Sci. Rep. 2013, 2, 25–34. [Google Scholar] [CrossRef]
- Shumway, S.E.; Allen, S.M.; Dee Boersma, P. Marine birds and harmful algal blooms: Sporadic victims or under-reported events? Harmful Algae 2003, 2, 1–17. [Google Scholar] [CrossRef]
- Richlen, M.L.; Morton, S.L.; Jamali, E.A.; Rajan, A.; Anderson, D.M. The catastrophic 2008–2009 red tide in the Arabian Gulf region, with observations on the identification and phylogeny of the fish-killing dinoflagellate Cochlodinium polykrikoides. Harmful Algae 2010, 9, 163–172. [Google Scholar] [CrossRef]
- Amaya, O.; Quintanilla, R.; Stacy, B.A.; Dechraoui Bottein, M.-Y.; Flewelling, L.; Hardy, R.; Dueñas, C.; Ruiz, G. Large-Scale Sea Turtle mortality events in El Salvador attributed to paralytic shellfish toxin-producing algae blooms. Front. Mar. Sci. 2018, 5, 411. [Google Scholar] [CrossRef]
- Broadwater, M.H.; Van Dolah, F.M.; Fire, S.E. Vulnerabilities of marine mammals to harmful algal blooms. In Harmful Algae: A Compendium Desk Reference; Shumway, S.E., Burkholder, J.M., Morton, S.L., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 191–222. [Google Scholar]
- Petroff, R.; Richards, T.; Crouthamel, B.; McKain, N.; Stanley, C.; Grant, K.S.; Shum, S.; Jing, J.; Isoherranen, N.; Burbacher, T.M. Chronic, low-level oral exposure to marine toxin, domoic acid, alters whole brain morphometry in nonhuman primates. NeuroToxicology 2019, 72, 114–124. [Google Scholar] [CrossRef]
- Farabegoli, F.; Blanco, L.; Rodríguez, L.; Vieites, J.; Cabado, A. Phycotoxins in marine shellfish: Origin, occurrence and effects on humans. Mar. Drugs 2018, 16, 188. [Google Scholar] [CrossRef]
- Zaitsev, Y.; Mamaev, V. Biological Diversity in the Black Sea: A Study of Change and Decline; Black Sea Environmental Series; United Nations Publishing: New York, NY, USA, 1997; Volume 3, p. 208. [Google Scholar]
- Bakan, G.; Büyükgüngör, H. The Black Sea. Mar. Pollut. Bull. 2000, 41, 24–43. [Google Scholar] [CrossRef]
- Moncheva, S.; Gotsis-Skretas, O.; Pagou, K.; Krastev, A. Phytoplankton blooms in Black Sea and Mediterranean coastal ecosystems subjected to anthropogenic eutrophication: Similarities and differences. Estuar. Coast. Shelf Sci. 2001, 53, 281–295. [Google Scholar] [CrossRef]
- Nesterova, D.; Moncheva, S.; Mikaelyan, A.; Vershinin, A.; Akatov, V.; Boicenco, L.; Aktan, Y.; Sahin, F.; Gvarishvili, T. Chapter 5. The State of Phytoplankton. In State of the Environment of the Black Sea (2001–2006/7); Oguz, T., Ed.; Black Sea Commission Publications 2008-3: Istanbul, Turkey, 2008; pp. 133–167. [Google Scholar]
- Kideys, A.E. ECOLOGY: Enhanced: Fall and rise of the Black Sea ecosystem. Science 2002, 297, 1482–1484. [Google Scholar] [CrossRef]
- Moncheva, S.; Boicenco, L.; Mikaelyan, A.; Zotov, A.; Dereziuk, N.; Gvarishvili, C.; Slabakova, N.; Mavrodieva, R.; Vlas, O.; Pautova, L.; et al. 1.3.2 Phytoplankton. In State of the Environment of the Black Sea (2009-2014/5); Krutov, A., Ed.; Publications of the Commission on the Protection of the Black Sea Against Pollution (BSC): Istanbul, Turkey, 2019; pp. 225–285. [Google Scholar]
- Ryabushko, L.I. Atlas of Toxic Microalgae of the Black Sea and the Sea of Azov; Ministry of Defence of Ukraine, National Academy of Sciences of Ukraine, Scientific Center of Armed Forces of Ukraine: Sevastopol, Ukraine, 2003; p. 140. [Google Scholar]
- Dzhembekova, N.; Urusizaki, S.; Moncheva, S.; Ivanova, P.; Nagai, S. Applicability of massively parallel sequencing on monitoring harmful algae at Varna Bay in the Black Sea. Harmful Algae 2017, 68, 40–51. [Google Scholar] [CrossRef]
- Dzhembekova, N.; Moncheva, S.; Ivanova, P.; Slabakova, N.; Nagai, S. Biodiversity of phytoplankton cyst assemblages in surface sediments of the Black Sea based on metabarcoding. Biotechnol. Biotechnol. Equip. 2018, 32, 1507–1513. [Google Scholar] [CrossRef]
- Peneva, V.; Gogov, Y.; Kalinova, G.; Slavova, A. Application of HPLC method for determination of ASP toxins in bivalve mulluscs. In Proceedings of the Jubilee Scientific Session—110 years National Diagnostic Science-and-Research Veterinary Medical Institute, Sofia, Bulgaria, 8–9 November 2011. [Google Scholar]
- Peteva, Z.; Krock, B.; Georgieva, S.; Stancheva, M. Occurrence and variability of marine biotoxins in mussel (Mytillus galloprovincialis) and in plankton samples from Bulgarian coast in spring 2017. IJAES 2018, 5, 1–11. [Google Scholar] [CrossRef]
- Peteva, Z.V.; Krock, B.; Stancheva, M.D.; Georgieva, S.K. Comparison of seasonal and spatial phycotoxin profiles of mussels from South Bulgarian coast. Bulg. Chem. Commun. 2020, 52, 16–21. [Google Scholar]
- Besiktepe, S.; Ryabushko, L.; Ediger, D.; Yilmaz, D.; Zenginer, A.; Ryabushko, V.; Lee, R. Domoic acid production by Pseudo-nitzschia calliantha Lundholm, Moestrup et Hasle (Bacillariophyta) isolated from the Black Sea. Harmful Algae 2008, 7, 438–442. [Google Scholar] [CrossRef]
- Vershinin, A.; Kamnev, A. Harmful algae in Russian European coastal waters. In Proceedings of the Ninth International Conference on Harmful Algal Blooms, Hobart, Australia, 7–11 February 2000. [Google Scholar]
- Morton, S.L.; Vershinin, A.; Smith, L.L.; Leighfield, T.A.; Pankov, S.; Quilliam, M.A. Seasonality of Dinophysis spp. and Prorocentrum lima in Black Sea phytoplankton and associated shellfish toxicity. Harmful Algae 2009, 8, 629–636. [Google Scholar] [CrossRef]
- Morton, S.L.; Vershinin, A.; Leighfield, T.; Smith, L.; Quilliam, M. Identification of yessotoxin in mussels from the Caucasian Black Sea Coast of the Russian Federation. Toxicon 2007, 50, 581–584. [Google Scholar] [CrossRef]
- Vershinin, A.; Morton, S.; Leighfield, T.; Pankov, S.; Smith, L.; Quilliam, M.; Ramsdell, J. Alexandrium in the Black Sea—Identity, ecology and PSP toxicity. Afr. J. Mar. Sci. 2006, 28, 209–213. [Google Scholar] [CrossRef]
- Kalinova, G. A Study of paralytic toxins in cultured mussels from Bulgarian Black Sea. TJS 2015, 13, 303–308. [Google Scholar] [CrossRef]
- Krumova-Valcheva, G.; Kalinova, G. Escherichia coli and paralytic shellfish poisoning toxins contamination of mussels farmed in Bulgarian Black Sea coast. Acta Microbiol. Bulg. 2017, 33, 30–35. [Google Scholar]
- Peteva, Z.V.; Kalinova, G.N.; Krock, B.; Stancheva, M.D.; Georgieva, S.K. Evaluation of paralytic shellfish poisoning toxin profile of mussels from Bulgarian North Black Sea coast by HPLC-FlD with post and pre-column derivatization. Bulg. Chem. Commun. 2019, 51, 233–240. [Google Scholar]
- Moncheva, S.; Petrova-Karadjova, V.; Palasov, A. Harmful algal blooms along the Bulgarian Black Sea coast and possible patterns of fish and zoobenthic mortalities. In Proceedings of the Sixth International Conference on Toxic Marine Phytoplankton, Nantes, France, 18–22 October 1993. [Google Scholar]
- Zaitsev, Y.P.; Garkavaya, G.P.; Nesterova, D.A.; Polischuk, L.N. The Danube—Main source of eutrophication of the Black Sea. Gidrobiologycheskyi. Zhurnal 1989, 25, 21–24. [Google Scholar]
- Bresnan, E.; Arévalo, F.; Belin, C.; Branco, M.A.C.; Cembella, A.D.; Clarke, D.; Correa, J.; Davidson, K.; Dhanji-Rapkova, M.; Lozano, R.F.; et al. Diversity and regional distribution of harmful algal events along the Atlantic margin of Europe. Harmful Algae 2021, 102, 101976. [Google Scholar] [CrossRef]
- Miles, C.O.; Samdal, I.A.; Aasen, J.A.G.; Jensen, D.J.; Quilliam, M.A.; Petersen, D.; Briggs, L.M.; Wilkins, A.L.; Rise, F.; Cooney, J.M.; et al. Evidence for numerous analogs of yessotoxin in Protoceratium reticulatum. Harmful Algae 2005, 4, 1075–1091. [Google Scholar] [CrossRef]
- Miles, C.O.; Wilkins, A.L.; Hawkes, A.D.; Selwood, A.I.; Jensen, D.J.; Cooney, J.M.; Beuzenberg, V.; MacKenzie, A. Lincoln. Identification of 45-Hydroxy-46,47-Dinoryessotoxin, 44-Oxo-45,46,47-Trinoryessotoxin, and 9-Methyl-42,43,44,45,46,47,55-Heptanor-38-En-41-Oxoyessotoxin, and partial characterization of some minor yessotoxins, from Protoceratium reticulatum. Toxicon 2006, 47, 229–240. [Google Scholar] [CrossRef]
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R; Springer: New York, NY, USA, 2011; p. 688. [Google Scholar]
- Dzhembekova, N.; Rubino, F.; Nagai, S.; Zlateva, I.; Slabakova, N.; Ivanova, P.; Slabakova, V.; Moncheva, S. Comparative analysis of morphological and molecular approaches integrated into the study of the dinoflagellate biodiversity within the recently deposited Black Sea sediments—Benefits and drawbacks. Biodivers. Data J. 2020, 8, e55172. [Google Scholar] [CrossRef]
- Dzhembekova, N.; Slabakova, N.; Slabakova, V.; Zlateva, I.; Moncheva, S. Long-term Trends in Pseudo-nitzschia complex blooms in the Black Sea—Is there a potential risk for ecological and human hazards. Ecol. Balk. 2021, 13, 55–75. [Google Scholar]
- Terenko, L.; Terenko, G. Dominant Pseudo-nitzschia (Bacillariophyta) species in the Black Sea (Ukraine). Bot. Lith. 2012, 18, 27–34. [Google Scholar] [CrossRef]
- Dzhembekova, N.; Atanasov, I.; Ivanova, P.; Moncheva, S. New potentially toxic Pseudo-nitzschia species (Bacillariophyceae) identified by molecular approach in the Black Sea (Varna Bay). In Proceedings of the 17th International Multidisciplinary Scientific GeoConference SGEM2017, Water Resources. Forest, Marine and Ocean Ecosystems, Albena, Bulgaria, 29 June–5 July 2017. [Google Scholar]
- Trainer, V.L.; Bates, S.S.; Lundholm, N.; Thessen, A.E.; Cochlan, W.P.; Adams, N.G.; Trick, C.G. Pseudo-nitzschia physiological ecology, phylogeny, toxicity, monitoring and impacts on ecosystem health. Harmful Algae 2012, 14, 271–300. [Google Scholar] [CrossRef]
- Bates, S.S.; Hubbard, K.A.; Lundholm, N.; Montresor, M.; Leaw, C.P. Pseudo-nitzschia, Nitzschia, and domoic acid: New research since 2011. Harmful Algae 2018, 79, 3–43. [Google Scholar] [CrossRef]
- Bates, S.S.; Lundholm, N.; Hubbard, K.A.; Montresor, M.; Leaw, C.P. Toxic and harmful marine diatoms. In Diatoms: Fundamentals and Applications; Seckbach, J., Gordon, R., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 389–434. [Google Scholar]
- Pednekar, S.M.; Bates, S.S.; Kerkar, V.; Matondkar, S.G.P. Environmental factors affecting the distribution of Pseudo-nitzschia in two monsoonal estuaries of western India and effects of salinity on growth and domoic acid production by P. pungens. Estuaries Coasts 2018, 41, 1448–1462. [Google Scholar] [CrossRef]
- Doucette, G.J.; King, K.L.; Thessen, A.E.; Dortch, Q. The effect of salinity on domoic acid production by the diatom Pseudo-nitzschia multiseries. Nova Hedwig. 2008, 133, 31–46. [Google Scholar]
- Hällfors, G. Checklist of Baltic Sea Phytoplankton Species (Including Some Heterotrophic Protistan Groups); Baltic Sea Environment Proceedings 95; Helsinki Commission, Baltic Marine Environment Protection Commission: Helsinki, Finland, 2004; p. 208. [Google Scholar]
- Wasmund, N.; Göbel, J.; Bodungen, B.V. 100-years-changes in the phytoplankton community of Kiel Bight (Baltic Sea). J. Mar. Syst. 2008, 73, 300–322. [Google Scholar] [CrossRef]
- Wasmund, N.; Dutz, J.; Pollehne, F.; Siegel, H.; Zettler, M.L. Biological assessment of the Baltic Sea 2015. Meereswiss. Ber. 2016, 102, 96. [Google Scholar] [CrossRef]
- Lelong, A.; Hégaret, H.; Soudant, P.; Bates, S.S. Pseudo-nitzschia (Bacillariophyceae) species, domoic acid and amnesic shellfish poisoning: Revisiting previous paradigms. Phycologia 2012, 51, 168–216. [Google Scholar] [CrossRef]
- Bates, S.S.; de Freitas, A.S.W.; Milley, J.E.; Pocklington, R.; Quilliam, M.A.; Smith, J.C.; Worms, J. Controls on domoic acid production by the diatom Nitzschia pungens f. multiseries in culture: Nutrients and irradïance. Can. J. Fish. Aquat. Sci. 1991, 48, 1136–1144. [Google Scholar] [CrossRef]
- Pan, Y.; Parsons, M.; Busman, M.; Moeller, P.; Dortch, Q.; Powell, C.; Doucette, G. Pseudo-nitzschia sp. cf. pseudodelicatissima—A confirmed producer of domoic acid from the Northern Gulf of Mexico. Mar. Ecol. Prog. Ser. 2001, 220, 83–92. [Google Scholar] [CrossRef]
- Black Sea Phytoplankton Checklist. Available online: http://phyto.bss.plankton.kiev.ua (accessed on 5 May 2022).
- Zhang, Y.; Pavlovska, M.; Stoica, E.; Prekrasna, I.; Yang, J.; Slobodnik, J.; Zhang, X.; Dykyi, E. Holistic pelagic biodiversity monitoring of the Black Sea via eDNA metabarcoding approach: From bacteria to marine mammals. Environ. Int. 2020, 135, 105307. [Google Scholar] [CrossRef]
- Mavrodieva, R.; Moncheva, S.; Hiebaum, G. Abnormal outburst of the dinoflagellate Alexandrium ostenfeldii (Paulsen) Balech et Tangen along the Bulgarian Black sea coast (the Bay of Sozopol) in winter—Ecological surprise or ecological concern? BDUA—J. Biol. 2007, II, 84. [Google Scholar]
- Kremp, A.; Lindholm, T.; Dreßler, N.; Erler, K.; Gerdts, G.; Eirtovaara, S.; Leskinen, E. Bloom forming Alexandrium ostenfeldii (Dinophyceae) in shallow waters of the Åland Archipelago, Northern Baltic Sea. Harmful Algae 2009, 8, 318–328. [Google Scholar] [CrossRef]
- Setälä, O.; Lehtinen, S.; Kremp, A.; Hakanen, P.; Kankaanpää, H.; Erler, K.; Suikkanen, S. Bioaccumulation of PSTs produced by Alexandrium ostenfeldii in the Northern Baltic Sea. Hydrobiologia 2013, 726, 143–154. [Google Scholar] [CrossRef]
- Martens, H.; Van de Waal, D.B.; Brandenburg, K.M.; Krock, B.; Tillmann, U. Salinity effects on growth and toxin production in an Alexandrium ostenfeldii (Dinophyceae) isolate from the Netherlands. J. Plankton Res. 2016, 38, 1302–1316. [Google Scholar] [CrossRef]
- Coolen, M.J.L.; Orsi, W.D.; Balkema, C.; Quince, C.; Harris, K.; Sylva, S.P.; Filipova-Marinova, M.; Giosan, L. Evolution of the plankton paleome in the Black Sea from the deglacial to anthropocene. Proc. Natl. Acad. Sci. USA 2013, 110, 8609–8614. [Google Scholar] [CrossRef] [PubMed]
- CABI. Gymnodinium catenatum. In Invasive Species Compendium; CAB International: Wallingford, UK, 2021; Available online: www.cabi.org/isc (accessed on 6 April 2022).
- Likumahua, S.; de Boer, M.K.; Krock, B.; Hehakaya, S.; Imu, L.; Müller, A.; Max, T.; Buma, A.G.J. Variability of dinoflagellates and their associated toxins in relation with environmental drivers in Ambon Bay, Eastern Indonesia. Mar. Pollut. Bull. 2020, 150, 110778. [Google Scholar] [CrossRef]
- Moreno-Pino, M.; Krock, B.; De la Iglesia, R.; Echenique-Subiabre, I.; Pizarro, G.; Vásquez, M.; Trefault, N. Next generation sequencing and mass spectrometry reveal high taxonomic diversity and complex phytoplankton-phycotoxins patterns in southeastern Pacific Fjords. Toxicon 2018, 151, 5–14. [Google Scholar] [CrossRef]
- González-Gil, S.; Pizarro, G.; Paz, B.; Velo-Suárez, L.; Reguera, B. Considerations on the toxigenic nature and prey sources of Phalacroma rotundatum. Aquat. Microb. Ecol. 2011, 64, 197–203. [Google Scholar] [CrossRef]
- Reguera, B.; Riobó, P.; Rodríguez, F.; Díaz, P.; Pizarro, G.; Paz, B.; Franco, J.; Blanco, J. Dinophysis toxins: Causative organisms, distribution and fate in shellfish. Mar. Drugs 2014, 12, 394–461. [Google Scholar] [CrossRef]
- Miles, C.O.; Wilkins, A.L.; Munday, R.; Dines, M.H.; Hawkes, A.D.; Briggs, L.R.; Sandvik, M.; Jensen, D.J.; Cooney, J.M.; Holland, P.T.; et al. Isolation of Pectenotoxin-2 from Dinophysis acuta and its conversion to pectenotoxin-2 seco acid, and preliminary assessment of their acute toxicities. Toxicon 2004, 43, 1–9. [Google Scholar] [CrossRef]
- MacKenzie, L.; Beuzenberg, V.; Holland, P.; McNabb, P.; Suzuki, T.; Selwood, A. Pectenotoxin and okadaic acid-based toxin profiles in Dinophysis acuta and Dinophysis acuminata from New Zealand. Harmful Algae 2005, 4, 75–85. [Google Scholar] [CrossRef]
- Fernández, M.L.; Reguera, B.; González-Gil, S.; Míguez, A. Pectenotoxin-2 in single-cell isolates of Dinophysis caudata and Dinophysis acuta from the Galician Rías (NW Spain). Toxicon 2006, 48, 477–490. [Google Scholar] [CrossRef]
- Kuuppo, P.; Uronen, P.; Petermann, A.; Tamminen, T.; Granéli, E. Pectenotoxin-2 and dinophysistoxin-1 in suspended and sedimenting organic matter in the Baltic Sea. Limnol. Oceanogr. 2006, 51, 2300–2307. [Google Scholar] [CrossRef]
- Blanco, J.; Álvarez, G.; Uribe, E. Identification of pectenotoxins in plankton, filter feeders, and isolated cells of a Dinophysis acuminata with an atypical toxin profile, from Chile. Toxicon 2007, 49, 710–716. [Google Scholar] [CrossRef]
- Suzuki, T.; Miyazono, A.; Baba, K.; Sugawara, R.; Kamiyama, T. LC–MS/MS analysis of okadaic acid analogues and other lipophilic toxins in single-cell isolates of several Dinophysis species collected in Hokkaido, Japan. Harmful Algae 2009, 8, 233–238. [Google Scholar] [CrossRef]
- Nielsen, L.T.; Krock, B.; Hansen, P.J. Production and excretion of okadaic acid, pectenotoxin-2 and a novel dinophysistoxin from the DSP-causing marine dinoflagellate Dinophysis acuta—Effects of light, food availability and growth phase. Harmful Algae 2013, 23, 34–45. [Google Scholar] [CrossRef]
- Riobó, P.; Reguera, B.; Franco, J.M.; Rodríguez, F. First report of the toxin profile of Dinophysis sacculus Stein from LC–MS analysis of laboratory cultures. Toxicon 2013, 76, 221–224. [Google Scholar] [CrossRef]
- Krock, B.; Tillmann, U.; Selwood, A.I.; Cembella, A.D. Unambiguous identification of pectenotoxin-1 and distribution of pectenotoxins in plankton from the North Sea. Toxicon 2008, 52, 927–935. [Google Scholar] [CrossRef]
- Miles, C.O.; Wilkins, A.L.; Hawkes, A.D.; Jensen, D.J.; Selwood, A.I.; Beuzenberg, V.; Lincoln MacKenzie, A.; Cooney, J.M.; Holland, P.T. Isolation and identification of pectenotoxins-13 and -14 from Dinophysis acuta in New Zealand. Toxicon 2006, 48, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Petrova, D.; Velikova, V. A report of potentially toxic species Dinophysis in Bulgarian Black Sea. In Proceedings of the 4th IWA Specialized Conference on Assessment and Control of Hazardous Substances in Water (ECOHAZARD2003), Aachen, Germany, 14–17 September 2003. [Google Scholar]
- Teren’ko, L.M. The genus Dinophysis Ehrenb. (Dinophyta) in the Ukrainian Black Sea coastal waters: Species composition, distribution, dynamics. Int. J. Algae 2011, 13, 341–351. [Google Scholar] [CrossRef]
- Dzhembekova, N.; Moncheva, S. Recent trends of potentially toxic phytoplankton species along the Bulgarian Black Sea area. In Proceedings of the Twelfth International Conference on Marine Sciences and Technologies, Varna, Bulgaria, 25–27 September 2014. [Google Scholar]
- Yasumoto, T.; Murata, M.; Oshima, Y.; Sano, M.; Matsumoto, G.K.; Clardy, J. Diarrhetic shellfish toxins. Tetrahedron 1985, 41, 1019–1025. [Google Scholar] [CrossRef]
- Reguera, B.; Velo-Suárez, L.; Raine, R.; Park, M.G. Harmful Dinophysis species: A review. Harmful Algae 2012, 14, 87–106. [Google Scholar] [CrossRef]
- Nielsen, L.; Krock, B.; Hansen, P. Effects of light and food availability on toxin production, growth and photosynthesis in Dinophysis acuminata. Mar. Ecol. Prog. Ser. 2012, 471, 37–50. [Google Scholar] [CrossRef]
- Basti, L.; Uchida, H.; Matsushima, R.; Watanabe, R.; Suzuki, T.; Yamatogi, T.; Nagai, S. Influence of temperature on growth and production of pectenotoxin-2 by a monoclonal culture of Dinophysis caudata. Mar. Drugs 2015, 13, 7124–7137. [Google Scholar] [CrossRef]
- Ajani, P.; Larsson, M.E.; Rubio, A.; Bush, S.; Brett, S.; Farrell, H. Modelling bloom formation of the toxic dinoflagellates Dinophysis acuminata and Dinophysis caudata in a highly modified estuary, south eastern Australia. Estuar. Coast. Shelf Sci. 2016, 183, 95–106. [Google Scholar] [CrossRef]
- Kamiyama, T.; Nagai, S.; Suzuki, T.; Miyamura, K. Effect of Temperature on production of okadaic acid, dinophysistoxin-1, and pectenotoxin-2 by Dinophysis acuminata in culture experiments. Aquat. Microb. Ecol. 2010, 60, 193–202. [Google Scholar] [CrossRef]
- Boundy, M.J.; Harwood, D.T.; Kiermeier, A.; McLeod, C.; Nicolas, J.; Finch, S. Risk assessment of pectenotoxins in New Zealand bivalve molluscan shellfish, 2009–2019. Toxins 2020, 12, 776. [Google Scholar] [CrossRef]
- Peteva, Z.; Georgieva, S.; Krock, B.; Gerasimova, A.; Stancheva, M.; Merdzhanova, A. Lipophilic marine biotoxins in mussels from Bulgarian coast and dietary intake of different population groups. Proc. Nutr. Soc. 2020, 79, OCE2. [Google Scholar] [CrossRef]
- Fabro, E.; Krock, B.; Torres, A.; Paparazzo, F.; Schloss, I.; Ferreyra, G.; Almandoz, G. Toxigenic dinoflagellates and associated toxins in San Jorge Gulf, Argentina. Oceanography 2018, 31, 145–153. [Google Scholar] [CrossRef]
- Paz, B. Yessotoxins, a group of marine polyether toxins: An overview. Mar. Drugs 2008, 6, 73–102. [Google Scholar] [CrossRef] [PubMed]
- Paz, B.; Riobó, P.; Ramilo, I.; Franco, J.M. Yessotoxins profile in strains of Protoceratium reticulatum from Spain and USA. Toxicon 2007, 50, 1–17. [Google Scholar] [CrossRef]
- Krock, B.; Alpermann, T.; Tillmann, U.; Pitcher, G.C.; Cembella, A.D. Yessotoxin profiles of the marine dinoflagellates Protoceratium reticulatum and Gonyaulax spinifera. In Proceedings of the 12th International Conference on Harmful Algae. International Society for the Study of Harmful Algae (ISSHA) and Intergovernmental Oceanographic Commission of UNESCO, Copenhagen, Denmark, 4–8 September 2006. [Google Scholar]
- Röder, K.; Hantzsche, F.M.; Gebühr, C.; Miene, C.; Helbig, T.; Krock, B.; Hoppenrath, M.; Luckas, B.; Gerdts, G. Effects of salinity, temperature and nutrients on growth, cellular characteristics and yessotoxin production of Protoceratium reticulatum. Harmful Algae 2012, 15, 59–70. [Google Scholar] [CrossRef]
- Sala-Pérez, M.; Alpermann, T.J.; Krock, B.; Tillmann, U. Growth and Bioactive secondary metabolites of arctic Protoceratium reticulatum (Dinophyceae). Harmful Algae 2016, 55, 85–96. [Google Scholar] [CrossRef]
- Peter, C.; Krock, B.; Cembella, A. Effects of salinity variation on growth and yessotoxin composition in the marine dinoflagellate Lingulodinium polyedra from a Skagerrak Fjord System (Western Sweden). Harmful Algae 2018, 78, 9–17. [Google Scholar] [CrossRef]
- Wang, N.; Mertens, K.N.; Krock, B.; Luo, Z.; Derrien, A.; Pospelova, V.; Liang, Y.; Bilien, G.; Smith, K.F.; De Schepper, S.; et al. Cryptic speciation in Protoceratium reticulatum (Dinophyceae): Evidence from morphological, molecular and ecophysiological data. Harmful Algae 2019, 88, 101610. [Google Scholar] [CrossRef]
- Tubaro, A.; Sosa, S.; Carbonatto, M.; Altinier, G.; Vita, F.; Melato, M.; Satake, M.; Yasumoto, T. Oral and intraperitoneal acute toxicity studies of yessotoxin and homoyessotoxins in mice. Toxicon 2003, 41, 783–792. [Google Scholar] [CrossRef]
- Tubaro, A.; Sosa, S.; Altinier, G.; Soranzo, M.R.; Satake, M.; Della Loggia, R.; Yasumoto, T. Short-term oral toxicity of homoyessotoxins, yessotoxin and okadaic acid in mice. Toxicon 2004, 43, 439–445. [Google Scholar] [CrossRef]
- Franchini, A.; Marchesini, E.; Poletti, R.; Ottaviania, E. Swiss mice CD1 fed on mussels contaminated by okadaic acid and yessotoxins: Effects on thymus and spleen. Eur. J. Histochem. 2005, 49, 179–188. [Google Scholar]
- Tillmann, U.; Elbrächter, M.; Krock, B.; John, U.; Cembella, A. Azadinium spinosum gen. et sp. nov. (Dinophyceae) identified as a primary producer of azaspiracid toxins. Eur. J. Phycol. 2009, 44, 63–79. [Google Scholar] [CrossRef]
- Krock, B.; Tillmann, U.; Voß, D.; Koch, B.P.; Salas, R.; Witt, M.; Potvin, É.; Jeong, H.J. New azaspiracids in Amphidomataceae (Dinophyceae). Toxicon 2012, 60, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Rossi, R.; Dell’Aversano, C.; Krock, B.; Ciminiello, P.; Percopo, I.; Tillmann, U.; Soprano, V.; Zingone, A. Mediterranean Azadinium dexteroporum (Dinophyceae) produces six novel azaspiracids and azaspiracid-35: A structural study by a multi-platform mass spectrometry approach. Anal. Bioanal. Chem. 2016, 409, 1121–1134. [Google Scholar] [CrossRef] [PubMed]
- Tillmann, U.; Salas, R.; Jauffrais, T.; Hess, P.; Silke, J. AZA–the producing organisms–biology and trophic transfer. In Seafood and Freshwater Toxins: Pharmacology, Physiology, and Detection, 3rd ed.; Botana, L.M., Ed.; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar] [CrossRef]
- Bachvaroff, T.R.; Adolf, J.E.; Place, A.R. Strain variation in Karlodinium veneficum (Dinophyceae): Toxin profiles, pigments, and growth characteristics. J. Phycol. 2009, 45, 137–153. [Google Scholar] [CrossRef]
- Place, A.R.; Bowers, H.A.; Bachvaroff, T.R.; Adolf, J.E.; Deeds, J.R.; Sheng, J. Karlodinium veneficum—The little dinoflagellate with a big bite. Harmful Algae 2012, 14, 179–195. [Google Scholar] [CrossRef]
- Krock, B.; Busch, J.; Tillmann, U.; García-Camacho, F.; Sánchez-Mirón, A.; Gallardo-Rodríguez, J.; López-Rosales, L.; Andree, K.; Fernández-Tejedor, M.; Witt, M.; et al. LC-MS/MS detection of karlotoxins reveals new variants in strains of the marine dinoflagellate Karlodinium veneficum from the Ebro Delta (NW Mediterranean). Mar. Drugs 2017, 15, 391. [Google Scholar] [CrossRef]
- Gong, W.; Marchetti, A. Estimation of 18S Gene copy number in marine eukaryotic plankton using a next-generation sequencing approach. Front. Mar. Sci. 2019, 6, 219. [Google Scholar] [CrossRef]
- Yarimizu, K.; Sildever, S.; Hamamoto, Y.; Tazawa, S.; Oikawa, H.; Yamaguchi, H.; Basti, L.; Mardones, J.I.; Paredes-Mella, J.; Nagai, S. Development of an absolute quantification method for ribosomal RNA gene copy numbers per eukaryotic single cell by digital PCR. Harmful Algae 2021, 103, 102008. [Google Scholar] [CrossRef]
- Goncharenko, I.; Krakhmalnyi, M.; Velikova, V.; Ascencio, E.; Krakhmalnyi, A. Ecological niche modeling of toxic dinoflagellate Prorocentrum cordatum in the Black Sea. Ecohydrol. Hydrobiol. 2021, 21, 747–759. [Google Scholar] [CrossRef]
- Petrova-Karadjova, V. Red-tide of Prorocentrum mıcans (Ehr) and Exuvıaella cordata (Ost.) in Varna Bay and along the coast in November, 1984. Hydrobıology 1985, 26, 70–78. [Google Scholar]
- Moncheva, S. Ecology of Marine Phytoplankton from the Black Sea in Conditions of Anthropogenic Eutrophication. Doctoral Dissertation, Institute of Oceanology—Bulgarian Academy of Sciences, Varna, Bulgaria, 30 May 1991. [Google Scholar]
- Tang, Y.Z.; Harke, M.J.; Gobler, C.J. Morphology, phylogeny, dynamics, and ichthyotoxicity of Pheopolykrikos hartmannii (Dinophyceae) isolates and blooms from New York, USA. J. Phycol. 2013, 49, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Gobler, C.J.; Lonsdale, D.J.; Boyer, G.L. A Review of the causes, effects, and potential management of harmful brown tide blooms caused by Aureococcus anophagefferens (Hargraves et Sieburth). Estuaries 2005, 28, 726–749. [Google Scholar] [CrossRef]
- Moncheva, S.; Parr, B. Phytoplankton Manual—Manual for Phytoplankton Sampling and Analysis in the Black Sea; Black Sea Commission: Istanbul, Türkiye, 2010; p. 46. Available online: http://www.blacksea-commission.org/Downloads/Phytoplankton_%20Mannual-Final-1.pdf (accessed on 15 June 2020).
- Hasle, G.R.; Syvertsen, E.E. Marine diatoms. In Identifying Marine Phytoplankton; Tomas, C.R., Ed.; Academic Press: San Diego, CA, USA, 1997; pp. 5–385. [Google Scholar]
- Moncheva, S. Guidelines for Quality Control of Biological Data-Phytoplankton; Black Sea Commission: Istanbul, Türkiye, 2010; p. 19. Available online: http://www.blacksea-commission.org/Downloads/Guidlines-Phytoplankton-QC-QA.pdf (accessed on 23 October 2020).
- Tanabe, A.S.; Nagai, S.; Hida, K.; Yasuike, M.; Fujiwara, A.; Nakamura, Y.; Takano, Y.; Katakura, S. Comparative study of the validity of three regions of the 18S-rRNA gene for massively parallel sequencing-based monitoring of the planktonic eukaryote community. Mol Ecol Resour. 2015, 16, 402–414. [Google Scholar] [CrossRef]
- IOC-UNESCO Taxonomic Reference List of Harmful Micro Algae. Available online: https://www.marinespecies.org/hab (accessed on 15 June 2022). [CrossRef]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 April 2022).
- Smith, K.F.; Rhodes, L.; Harwood, D.T.; Adamson, J.; Moisan, C.; Munday, R.; Tillmann, U. Detection of Azadinium poporum in New Zealand: The use of molecular tools to assist with species isolations. J. Appl. Phycol. 2015, 28, 1125–1132. [Google Scholar] [CrossRef]
- Tillmann, U.; Wietkamp, S.; Krock, B.; Tillmann, A.; Voss, D.; Gu, H. Amphidomataceae (Dinophyceae) in the Western Greenland area, including description of Azadinium perforatum sp. nov. Phycologia 2019, 59, 63–88. [Google Scholar] [CrossRef]
- Toebe, K.; Joshi, A.R.; Messtorff, P.; Tillmann, U.; Cembella, A.; John, U. Molecular discrimination of taxa within the dinoflagellate genus Azadinium, the source of azaspiracid toxins. J. Plankton Res. 2012, 35, 225–230. [Google Scholar] [CrossRef]
- Wietkamp, S.; Tillmann, U.; Clarke, D.; Toebe, K. Molecular detection and quantification of the azaspiracid-producing dinoflagellate Amphidoma languida (Amphidomataceae, Dinophyceae). J. Plankton Res. 2019, 41, 101–113. [Google Scholar] [CrossRef]
- Wietkamp, S.; Krock, B.; Gu, H.; Voß, D.; Klemm, K.; Tillmann, U. Occurrence and distribution of Amphidomataceae (Dinophyceae) in Danish coastal waters of the North Sea, the Limfjord and the Kattegat/Belt Area. Harmful Algae 2019, 88, 101637. [Google Scholar] [CrossRef]
- Forootan, A.; Sjöback, R.; Björkman, J.; Sjögreen, B.; Linz, L.; Kubista, M. Methods to determine limit of detection and limit of quantification in quantitative Real-Time PCR (QPCR). Biomol. Detect. Quantif. 2017, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Krock, B.; Tillmann, U.; Wen, Y.; Hansen, P.J.; Larsen, T.O.; Andersen, A.J.C. Development of a LC-MS/MS method for the quantification of goniodomins a and B and its application to Alexandrium pseudogonyaulax strains and plankton field samples of Danish coastal waters. Toxicon 2018, 155, 51–60. [Google Scholar] [CrossRef]
- The Math Works, Inc. MATLAB, version 2020a; The Math Works, Inc.: Natick, MA, USA, 2020; Available online: https://www.mathworks.com/ (accessed on 7 June 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 12 January 2022).
- Harrell, F.E., Jr. Hmisc: Harrell Miscellaneous (Version 4.1-1). Cran. 2018. Available online: http://CRAN.R-project.org/package=Hmisc.2018 (accessed on 12 January 2022).
- Oksanen, J. Vegan: An Introduction to Ordination. 2015. Available online: http://cran.r-project.org/web/packages/vegan/vignettes/intro-vegan.pdf (accessed on 15 January 2022).
- Miles, C.O.; Wilkins, A.L.; Hawkes, A.D.; Selwood, A.I.; Jensen, D.J.; Munday, R.; Cooney, J.M.; Beuzenberg, V. Polyhydroxylated amide analogs of yessotoxin from Protoceratium reticulatum. Toxicon 2005, 45, 61–71. [Google Scholar] [CrossRef] [PubMed]




| Species | LM | NGS |
|---|---|---|
| Pseudo-nitzschia calliantha | - | + |
| Pseudo-nitzschia delicatissima | - | + |
| Pseudo-nitzschia pungens | - | + |
| Alexandrium andersonii | - | + |
| Alexandrium minutum | - | + |
| Alexandrium ostenfeldii | - | + |
| Amphidoma languida | - | + |
| Dinophysis acuminata | + | + |
| Dinophysis acuta | + | + |
| Dinophysis caudata | + | - |
| Dinophysis sacculus | + | - |
| Gonyaulax spinifera | + | + |
| Gymnodinium catenatum | - | + |
| Karlodinium veneficum | - | + |
| Lingulodinium polyedra | + | + |
| Phalacroma rotundatum | + | + |
| Polykrikos hartmannii | - | + |
| Prorocentrum cordatum | + | + |
| Protoceratium reticulatum | + | + |
| Aureococcus anophagefferens | - | + |
| RDA Model Cell Abundance vs. Environmental Data | CCA Model Toxin Variant Abundance vs. Environmental Data | |
|---|---|---|
| Proportion of the total variance (inertia) explained | ||
| Constrained | 55.68% | 36% |
| Model significance (ANOVA) | ||
| p-values | p = 0.001 | p = 0.001 |
| R2 | 0.57 | n/a |
| R2adj | 0.49 | n/a |
| Axes significance (ANOVA) and contribution to total variance explained by the model | ||
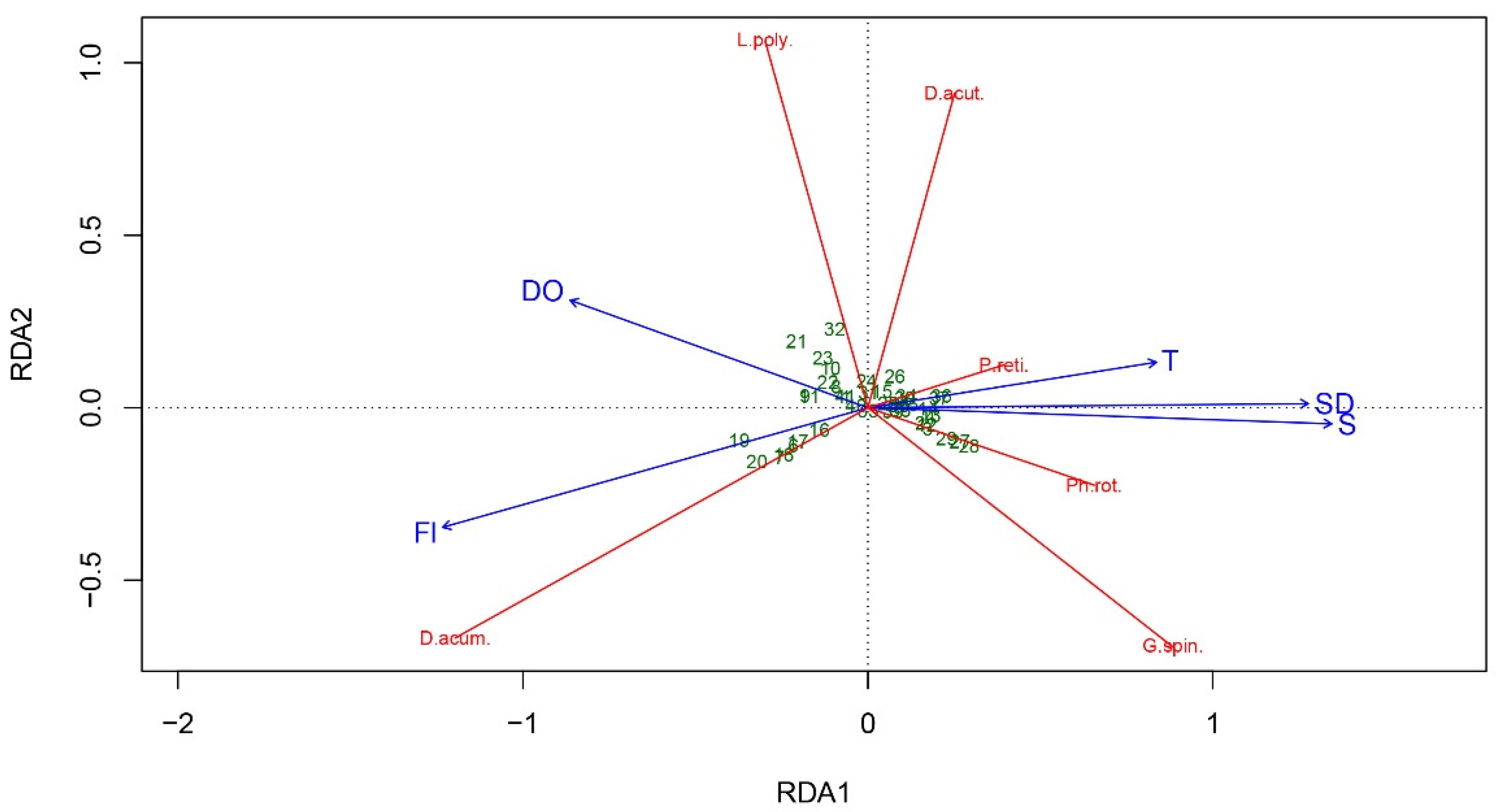
| p-values | RDA1 axis p = 0.001; 36.04% RDA2 axis p = 0.001; 8.94% Contribution of variables to the axis explains the biggest portion of variance in data: RDA1 – T, S, DO, and SD | CCA1 axis p = 0.001; 22.92% CCA2 axis p = 0.048; 9.75%. Contribution of variables to the axis explains the biggest portion of variance in data: CCA1 – T, S, Fl, and SD |
| Model term significance (ANOVA) and percentage contribution to total variance explained by the model | ||
| p-values and percent contribution | T: p = 0.001; 11.05% S: p = 0.001; 20.65% DO: p = 0.015; 4.11% Fl: p = 0.015; 5.64% SD: p = 0.064; n/a. | T: p = 0.008; 3.58% S: p = 0.001; 5.01% DO: p = 0.760; n/a Fl: p = 0.034; 2.61% SD: p = 0.001; 8.01% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dzhembekova, N.; Moncheva, S.; Slabakova, N.; Zlateva, I.; Nagai, S.; Wietkamp, S.; Wellkamp, M.; Tillmann, U.; Krock, B. New Knowledge on Distribution and Abundance of Toxic Microalgal Species and Related Toxins in the Northwestern Black Sea. Toxins 2022, 14, 685. https://doi.org/10.3390/toxins14100685
Dzhembekova N, Moncheva S, Slabakova N, Zlateva I, Nagai S, Wietkamp S, Wellkamp M, Tillmann U, Krock B. New Knowledge on Distribution and Abundance of Toxic Microalgal Species and Related Toxins in the Northwestern Black Sea. Toxins. 2022; 14(10):685. https://doi.org/10.3390/toxins14100685
Chicago/Turabian StyleDzhembekova, Nina, Snejana Moncheva, Nataliya Slabakova, Ivelina Zlateva, Satoshi Nagai, Stephan Wietkamp, Marvin Wellkamp, Urban Tillmann, and Bernd Krock. 2022. "New Knowledge on Distribution and Abundance of Toxic Microalgal Species and Related Toxins in the Northwestern Black Sea" Toxins 14, no. 10: 685. https://doi.org/10.3390/toxins14100685
APA StyleDzhembekova, N., Moncheva, S., Slabakova, N., Zlateva, I., Nagai, S., Wietkamp, S., Wellkamp, M., Tillmann, U., & Krock, B. (2022). New Knowledge on Distribution and Abundance of Toxic Microalgal Species and Related Toxins in the Northwestern Black Sea. Toxins, 14(10), 685. https://doi.org/10.3390/toxins14100685






