A Computational Understanding of Inter-Individual Variability in CYP2D6 Activity to Investigate the Impact of Missense Mutations on Ochratoxin A Metabolism
Abstract
:1. Introduction
2. Results and Discussion
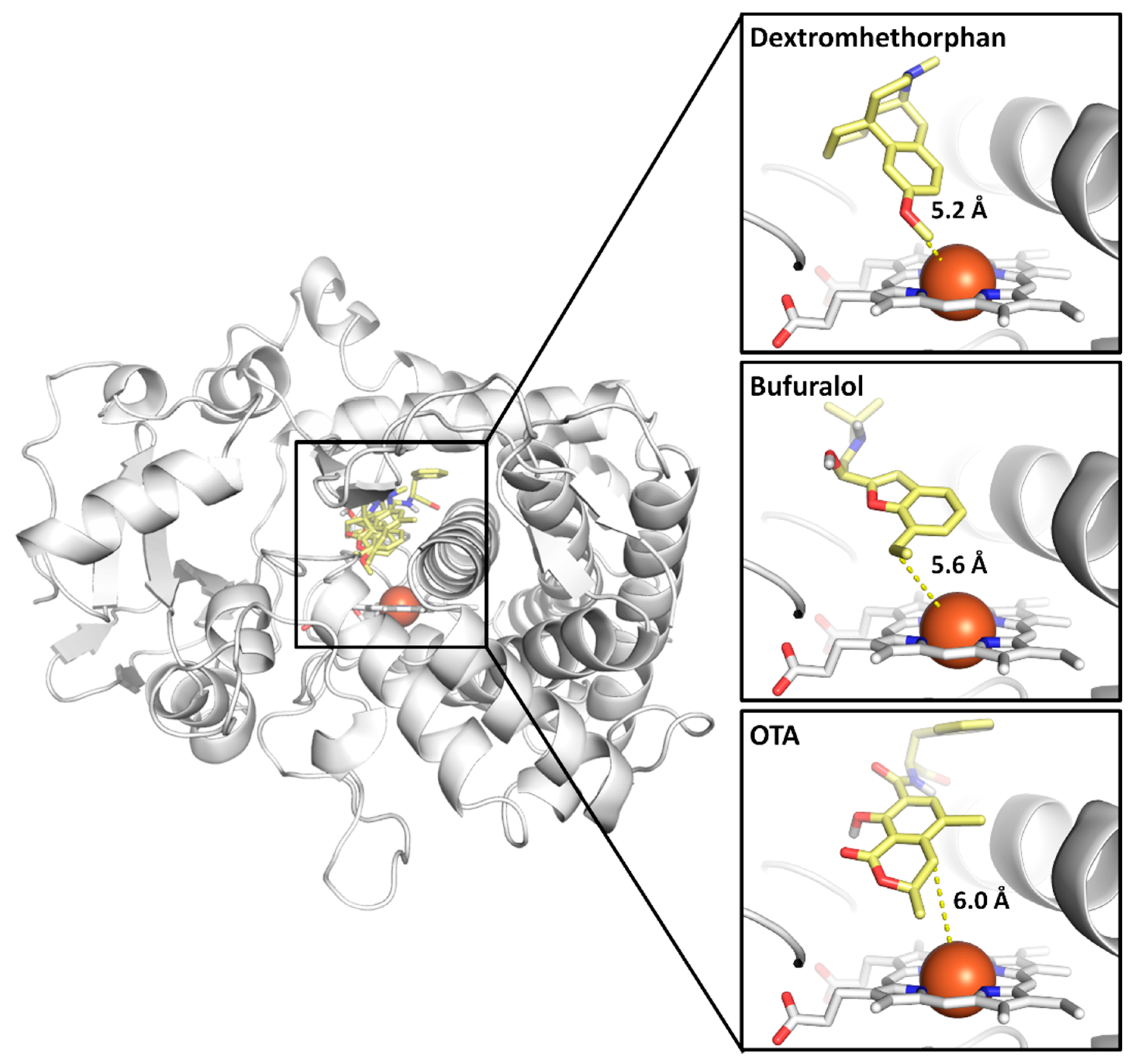
2.1. Assessing Model Efficacy
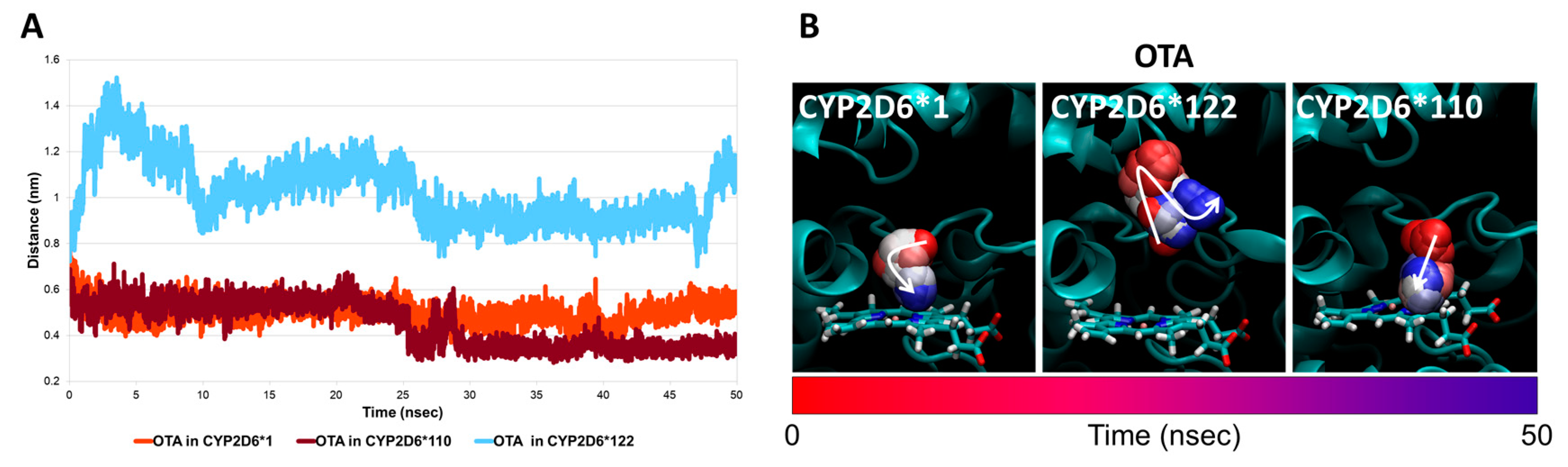
2.2. Analysis of Uncharacterised CYP2D6 Variants
3. Conclusions
4. Materials and Methods
4.1. Data Source
4.2. Model and Ligands Preparation
4.3. Docking Analysis
4.4. Molecular Dynamics
4.5. Statistical Analysis
4.6. Multiple-Sequence Alignment Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kasteel, E.E.J.; Darney, K.; Kramer, N.I.; Dorne, J.; Lautz, L.S. Human variability in isoform-specific UDP-glucuronosyltransferases: Markers of acute and chronic exposure, polymorphisms and uncertainty factors. Arch. Toxicol. 2020, 94, 2637–2661. [Google Scholar] [CrossRef] [PubMed]
- Machalz, D.; Pach, S.; Bermudez, M.; Bureik, M.; Wolber, G. Structural insights into understudied human cytochrome P450 enzymes. Drug Discov. Today 2021, 26, 2456–2464. [Google Scholar] [CrossRef] [PubMed]
- Dorne, J. Metabolism, variability and risk assessment. Toxicology 2010, 268, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.; Crosby, I.; Yip, V.; Maguire, P.; Pirmohamed, M.; Turner, R.M. A Review of the Important Role of CYP2D6 in Pharmacogenomics. Genes 2020, 11, 1295. [Google Scholar] [CrossRef]
- Nofziger, C.; Turner, A.J.; Sangkuhl, K.; Whirl-Carrillo, M.; Agundez, J.A.G.; Black, J.L.; Dunnenberger, H.M.; Ruano, G.; Kennedy, M.A.; Phillips, M.S.; et al. PharmVar GeneFocus: CYP2D6. Clin. Pharmacol. Ther. 2020, 107, 154–170. [Google Scholar] [CrossRef] [Green Version]
- Darney, K.; Lautz, L.S.; Bechaux, C.; Wiecek, W.; Testai, E.; Amzal, B.; Dorne, J. Human variability in polymorphic CYP2D6 metabolism: Implications for the risk assessment of chemicals in food and emerging designer drugs. Environ. Int. 2021, 156, 106760. [Google Scholar] [CrossRef]
- Magarbeh, L.; Gorbovskaya, I.; Le Foll, B.; Jhirad, R.; Muller, D.J. Reviewing pharmacogenetics to advance precision medicine for opioids. Biomed. Pharmacother. 2021, 142, 112060. [Google Scholar] [CrossRef]
- Friedrich, D.C.; Genro, J.P.; Sortica, V.A.; Suarez-Kurtz, G.; de Moraes, M.E.; Pena, S.D.J.; dos Santos, A.K.R.; Romano-Silva, M.A.; Hutz, M.H. Distribution of CYP2D6 Alleles and Phenotypes in the Brazilian Population. PLoS ONE 2014, 9, e110691. [Google Scholar] [CrossRef] [Green Version]
- Soria-Chacartegui, P.; Villapalos-Garcia, G.; Zubiaur, P.; Abad-Santos, F.; Koller, D. Genetic Polymorphisms Associated With the Pharmacokinetics, Pharmacodynamics and Adverse Effects of Olanzapine, Aripiprazole and Risperidone. Front. Pharmacol. 2021, 12, 1738. [Google Scholar] [CrossRef]
- Faponle, A.S.; Roy, A.; Adelegan, A.A.; Gauld, J.W. Molecular Dynamics Simulations of a Cytochrome P450 from Tepidiphilus thermophilus (P450-TT) Reveal How Its Substrate-Binding Channel Opens. Molecules 2021, 26, 3614. [Google Scholar] [CrossRef]
- Don, C.G.; Smiesko, M. Microsecond MD simulations of human CYP2D6 wild-type and five allelic variants reveal mechanistic insights on the function. PLoS ONE 2018, 13, e0202534. [Google Scholar] [CrossRef] [PubMed]
- De Waal, P.W.; Sunden, K.F.; Furge, L.L. Molecular Dynamics of CYP2D6 Polymorphisms in the Absence and Presence of a Mechanism-Based Inactivator Reveals Changes in Local Flexibility and Dominant Substrate Access Channels. PLoS ONE 2014, 9, e108607. [Google Scholar] [CrossRef]
- Fink-Gremmels, J. Conclusions from the workshops on Ochratoxin A in food: Recent developments and significance, organized by ILSI Europe in Baden (Austria), 29 June–1 July 2005. Food Addit. Contam. 2005, 22, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Ma, W.; Ma, Z.Y.; Zhang, Q.H.; Li, H.M. The Occurrence and Contamination Level of Ochratoxin A in Plant and Animal-Derived Food Commodities. Molecules 2021, 26, 6928. [Google Scholar] [CrossRef]
- EFSA. Risk assessment of ochratoxin A in food. EFSA J. 2020, 18, e06113. [Google Scholar]
- Tao, Y.F.; Xie, S.Y.; Xu, F.F.; Liu, A.M.; Wang, Y.X.; Chen, D.M.; Pan, Y.H.; Huang, L.L.; Peng, D.P.; Wang, X.; et al. Ochratoxin A: Toxicity, oxidative stress and metabolism. Food Chem. Toxicol. 2018, 112, 320–331. [Google Scholar] [CrossRef]
- Heussner, A.H.; Bingle, L.E.H. Comparative Ochratoxin Toxicity: A Review of the Available Data. Toxins 2015, 7, 4253–4282. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.H.; Dohnal, V.; Huang, L.L.; Kuca, K.; Wang, X.; Chen, G.Y.; Yuan, Z.H. Metabolic Pathways of Ochratoxin A. Curr. Drug Metab. 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Ali, N.; Munoz, K.; Degen, G.H. Ochratoxin A and its metabolites in urines of German adults-An assessment of variables in biomarker analysis. Toxicol. Lett. 2017, 275, 19–26. [Google Scholar] [CrossRef]
- Dellafiora, L.; Gonaus, C.; Streit, B.; Galaverna, G.; Moll, W.D.; Vogtentanz, G.; Schatzmayr, G.; Dall’Asta, C.; Prasad, S. An In Silico Target Fishing Approach to Identify Novel Ochratoxin A Hydrolyzing Enzyme. Toxins 2020, 12, 258. [Google Scholar] [CrossRef]
- Pfohl-Leszkowicz, A.; Manderville, R.A. Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 2007, 51, 61–99. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Yamazoe, Y. Unimolecular and Bimolecular Binding System for the Prediction of CYP2D6-Mediated Metabolism. Drug Metab. Dispos. 2012, 40, 486–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- IARC. Chapter 6—Mycotoxins and human health. In Improving Public HEalth Through Mycotoxin Control–IARC Scientific Publication N. 158; Pit, J., Wild, C., Baan, R., Gelderblom, W., Miller, J., Riley, R., Wu, F., Eds.; International Agency for Research on Cancer: Lyon, France, 2012. [Google Scholar]
- Malir, F.; Ostry, V.; Pfohl-Leszkowicz, A.; Malir, J.; Toman, J. Ochratoxin A: 50 Years of Research. Toxins 2016, 8, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfohl-Leszkowicz, A.; Hadjeba-Medjdoub, K.; Ballet, N.; Schrickx, J.; Fink-Gremmels, J. Assessment and characterisation of yeast-based products intended to mitigate ochratoxin exposure using in vitro and in vivo models. Food Addit. Contam. A 2015, 32, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.N.; Viktorova, J.; Ruml, T. Mycotoxins: Biotransformation and Bioavailability Assessment Using Caco-2 Cell Monolayer. Toxins 2020, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Sakuyama, K.; Sasaki, T.; Ujiie, S.; Obata, K.; Mizugaki, M.; Ishikawa, M.; Hiratsuka, M. Functional Characterization of 17 CYP2D6 Allelic Variants (CYP2D6.2, 10, 14A-B, 18, 27, 36, 39, 47-51, 53-55, and 57). Drug Metab. Dispos. 2008, 36, 2460–2467. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.F.; Neiner, A.; Kharasch, E.D. Stereoselective Bupropion Hydroxylation by Cytochrome P450 CYP2B6 and Cytochrome P450 Oxidoreductase Genetic Variants. Drug Metab. Dispos. 2020, 48, 438–445. [Google Scholar] [CrossRef] [Green Version]
- Vizeli, P.; Straumann, I.; Holze, F.; Schmid, Y.; Dolder, P.C.; Liechti, M.E. Genetic influence of CYP2D6 on pharmacokinetics and acute subjective effects of LSD in a pooled analysis. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Louisse, J.; Dorne, J.L.C.M.; Dellafiora, L. Investigating the interaction between organic anion transporter 1 and ochratoxin A: An in silico structural study to depict early molecular events of substrate recruitment and the impact of single point mutations. Toxicol. Lett. 2022, 355, 19–30. [Google Scholar] [CrossRef]
- Xu, L.; Chen, L.Y. Molecular determinant of substrate binding and specificity of cytochrome P450 2J2. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Wang, A.; Stout, C.D.; Zhang, Q.H.; Johnson, E.F. Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding. J. Biol. Chem. 2015, 290, 5092–5104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, K.; Woo, S.M.; Siu, T.; Cortopassi, W.A.; Duarte, F.; Paton, R.S. Cation-pi interactions in protein-ligand binding: Theory and data-mining reveal different roles for lysine and arginine. Chem. Sci. 2018, 9, 2655–2665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyson, H.J.; Wright, P.E.; Scheraga, H.A. The role of hydrophobic interactions in initiation and propagation of protein folding. Proc. Natl. Acad. Sci. USA 2006, 103, 13057–13061. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.T.; Lu, H.Y.; Zhang, N.; Zhu, Z.F.; Wang, S.Q.; Li, M.H. PremPS: Predicting the impact of missense mutations on protein stability. PLoS Comput. Biol. 2020, 16, e1008543. [Google Scholar] [CrossRef]
- Manoharan, A.; Shewade, D.G.; Ravindranath, P.A.; Rajkumar, R.P.; Ramprasad, V.L.; Adithan, S.; Damodaran, S.E. Resequencing CYP2D6 gene in Indian population: CYP2D6*41 identified as the major reduced function allele. Pharmacogenomics 2019, 20, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acid Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Li, T.; Bonkovsky, H.L.; Guo, J.T. Structural analysis of heme proteins: Implications for design and prediction. BMC Struct. Biol. 2011, 11, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, A.J.; Lambert, J.C.; Thayer, K.; Dorne, J. Sourcing data on chemical properties and hazard data from the US-EPA CompTox Chemicals Dashboard: A practical guide for human risk assessment. Environ. Int. 2021, 154, 106566. [Google Scholar] [CrossRef]
- Nikolov, I.G.; Chernozemsky, I.N.; Idle, J.R. Genetic Predisposition to Balkan Endemic Nephropathy: Ability to Hydroxylate Debrisoquine as a Host Risk Factor. IARC Sci. Publ. 1991, 289–296. [Google Scholar]
- Guengerich, F.P. Characterization of Human Cytochrome-P450 Enzymes. Faseb J. 1992, 6, 745–748. [Google Scholar] [CrossRef] [Green Version]
- Pfohl-Leszkowicz, A.; Pinelli, E.; Bartsch, H.; Mohr, U.; Castegnaro, M. Sex- and strain-specific expression of cytochrome P450s in ochratoxin A-induced genotoxicity and carcinogenicity in rats. Mol. Carcinog. 1998, 23, 76–85. [Google Scholar] [CrossRef]
- Manderville, R.A.; Pfohl-Leszkowicz, A. Bioactivation and DNA adduction as a rationale for ochratoxin A carcinogenesis. World Mycotoxin J. 2008, 1, 357–367. [Google Scholar] [CrossRef]
- El Adlouni, C.; Pinelli, E.; Azémar, B.; Zaoui, D.; Beaune, P.; Pfohl-Leszkowicz, A. Phenobarbital increases DNA adduct and metabolites formed by ochratoxin A: Role of CYP 2C9 and microsomal glutathione-S-transferase. Environ. Mol. Mutagen. 2000, 35, 123–131. [Google Scholar] [CrossRef]
- Atanasova, S.Y.; von Ahsen, N.; Toncheva, D.I.; Dimitrov, T.G.; Oellerich, M.; Armstrong, V.W. Genetic polymorphisms of cytochrome P450 among patients with Balkan endemic nephropathy (BEN). Clin. Biochem. 2005, 38, 223–228. [Google Scholar] [CrossRef]
- Pfohl-Leszkowicz, A. Ochratoxin A and aristolochic acid involvement in nephropathies and associated urothelial tract tumours. Arhiv Za Higijenu Rada I Toksikologiju-Arch. Ind. Hyg. Toxicol. 2009, 60, 465–483. [Google Scholar] [CrossRef] [Green Version]
- Don, C.G.; Smiesko, M. Out-compute drug side effects: Focus on cytochrome P450 2D6 modeling. Wiley Interdiscip. Rev.-Comput. Mol. Sci. 2018, 8, e1366. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.J.; Gindulyte, A.; He, J.; He, S.Q.; Li, Q.L.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acid Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acid Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using Modeller. Curr. Protoc. Bioinform. 2006, 15, 5.6.1–5.6.37. [Google Scholar]
- Dellafiora, L.; Paolella, S.; Dall’Asta, C.; Dossena, A.; Cozzini, P.; Galaverna, G. Hybrid in Silico/in Vitro Approach for the Identification of Angiotensin I Converting Enzyme Inhibitory Peptides from Parma Dry-Cured Ham. J. Agr. Food Chem. 2015, 22, 6366–6375. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Rojas, W.; Olivero-Verbel, J. Potential interaction of natural dietary bioactive compounds with COX-2. J. Mol. Graph. Model. 2011, 30, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Mena, P.; Cozzini, P.; Brighenti, F.; Del Rio, D. Modelling the possible bioactivity of ellagitannin-derived metabolites. In silico tools to evaluate their potential xenoestrogenic behavior. Food Funct. 2013, 4, 1442–1451. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.; Mittal, J.; Feig, M.; Mackerell, A.D.J. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Silva, D.A.; Yan, Y.J.; Huang, X.H. Force field development for cofactors in the photosystem II. J. Comput. Chem. 2012, 33, 1969–1980. [Google Scholar] [CrossRef] [PubMed]
- Panneerselvam, S.; Yesudhas, D.; Durai, P.; Anwar, M.A.; Gosu, V.; Choi, S. A Combined Molecular Docking/Dynamics Approach to Probe the Binding Mode of Cancer Drugs with Cytochrome P450 3A4. Molecules 2015, 20, 14915–14935. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acid Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorne, J.L.C.M.; Cirlini, M.; Louisse, J.; Pedroni, L.; Galaverna, G.; Dellafiora, L. A Computational Understanding of Inter-Individual Variability in CYP2D6 Activity to Investigate the Impact of Missense Mutations on Ochratoxin A Metabolism. Toxins 2022, 14, 207. https://doi.org/10.3390/toxins14030207
Dorne JLCM, Cirlini M, Louisse J, Pedroni L, Galaverna G, Dellafiora L. A Computational Understanding of Inter-Individual Variability in CYP2D6 Activity to Investigate the Impact of Missense Mutations on Ochratoxin A Metabolism. Toxins. 2022; 14(3):207. https://doi.org/10.3390/toxins14030207
Chicago/Turabian StyleDorne, Jean Lou C. M., Martina Cirlini, Jochem Louisse, Lorenzo Pedroni, Gianni Galaverna, and Luca Dellafiora. 2022. "A Computational Understanding of Inter-Individual Variability in CYP2D6 Activity to Investigate the Impact of Missense Mutations on Ochratoxin A Metabolism" Toxins 14, no. 3: 207. https://doi.org/10.3390/toxins14030207
APA StyleDorne, J. L. C. M., Cirlini, M., Louisse, J., Pedroni, L., Galaverna, G., & Dellafiora, L. (2022). A Computational Understanding of Inter-Individual Variability in CYP2D6 Activity to Investigate the Impact of Missense Mutations on Ochratoxin A Metabolism. Toxins, 14(3), 207. https://doi.org/10.3390/toxins14030207






