A Novel SELEX Based on Immobilizing Libraries Enables Screening of Saxitoxin Aptamers for BLI Aptasensor Applications
Abstract
:1. Introduction
2. Results and Discussion
2.1. Selection of Aptamers by IMC-SELEX
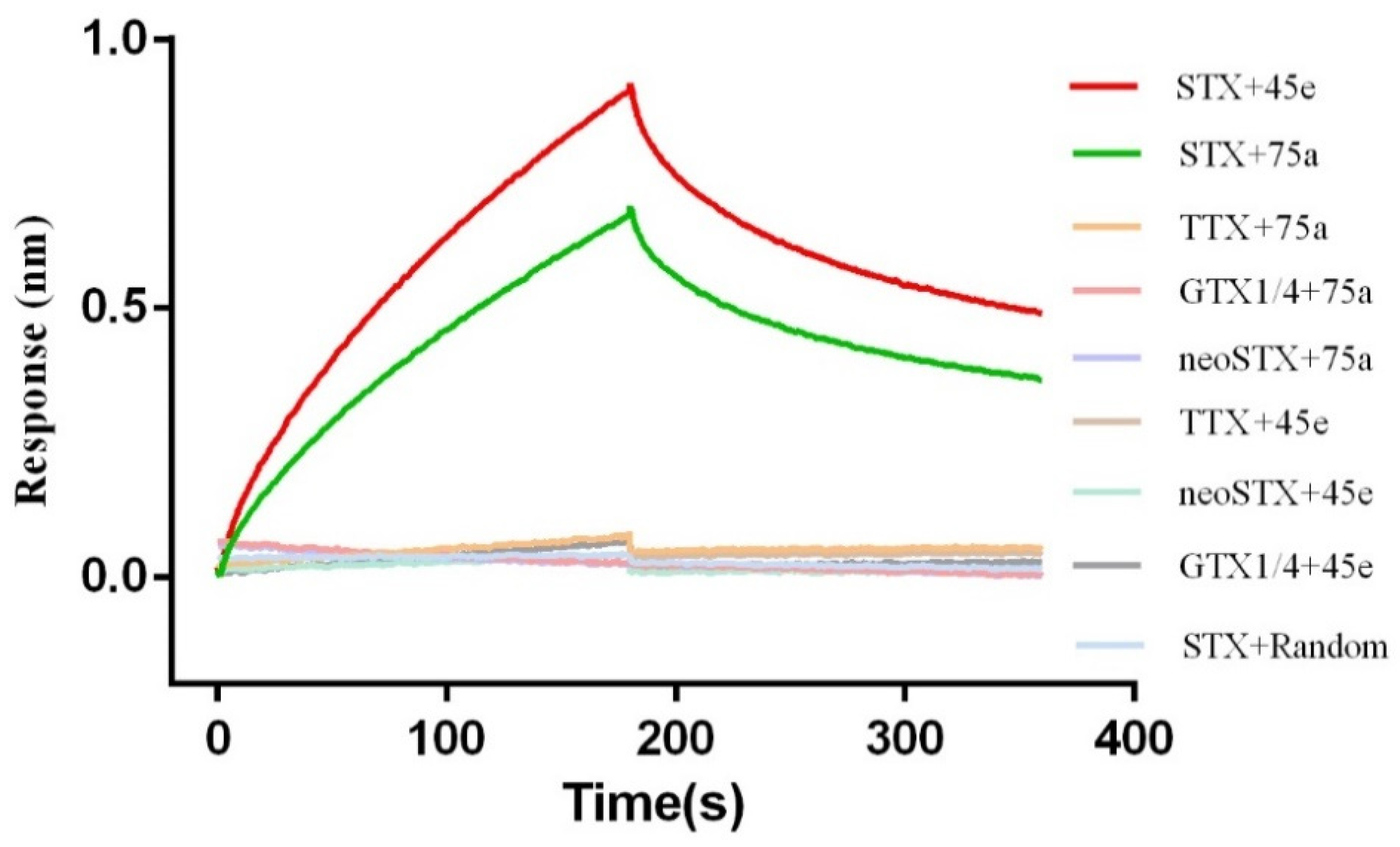
2.2. Identification of Affinity and Specificity of STX and Aptamer
2.3. Truncation and Optimization of Aptamers
2.4. Molecular Docking and Molecular Dynamics Simulations (MDS)
2.4.1. 3-D Structure Modeling of Aptamer 45e
2.4.2. Docking and Molecular Dynamics Simulation of Aptamer 45e with Saxitoxin
2.4.3. Further Optimization of Aptamer 45e
2.5. Microscale Thermophoresis (MST)
2.6. BLI Aptasensor for STX Detection
3. Conclusions
4. Material and Methods
4.1. Materials and Reagents
4.2. Selection of Aptamers In Vitro
4.2.1. Design of Screening Libraries
4.2.2. Selection of Aptamers by Ni-NTA Column
4.2.3. Clonal Sequencing to Select the Target Aptamer
4.3. Identification of Affinity and Specificity of STX and Aptamer
4.4. Molecular Docking and Molecular Dynamics Simulations (MDS)
4.4.1. 3-D Structure Modeling of G-Quadruplex 45e
4.4.2. Docking of Aptamer 45e with Saxitoxin
4.4.3. Molecular Dynamics (MD) Simulation
4.5. Microscale Thermophoresis (MST)
4.6. Preparation of Aptasensor
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deeds, J.R.; Landsberg, J.H.; Etheridge, S.M.; Pitcher, G.C.; Longan, S.W. Non-traditional vectors for paralytic shellfish poisoning. Mar. Drugs 2008, 6, 308–348. [Google Scholar] [CrossRef] [PubMed]
- Morabito, S.; Silvestro, S.; Faggio, C. How the marine biotoxins affect human health. Nat. Prod. Res. 2018, 32, 621–631. [Google Scholar] [CrossRef]
- Cusick, K.D.; Sayler, G.S. An overview on the marine neurotoxin, saxitoxin: Genetics, molecular targets, methods of detection and ecological functions. Mar. Drugs 2013, 11, 991–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeDoux, M.; Hall, S. Proficiency testing of eight French laboratories in using the AOAC mouse bioassay for paralytic shellfish poisoning: Interlaboratory collaborative study. J. AOAC Int. 2000, 83, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Garthwaite, I.; Ross, K.M.; Miles, C.O.; Briggs, L.R.; Towers, N.R.; Borrell, T.; Busby, P. Integrated enzyme-linked immunosorbent assay screening system for amnesic, neurotoxic, diarrhetic, and paralytic shellfish poisoning toxins found in New Zealand. J. AOAC Int. 2001, 84, 1643–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wharton, R.E.; Feyereisen, M.C.; Gonzalez, A.L.; Abbott, N.L.; Hamelin, E.I.; Johnson, R.C. Quantification of saxitoxin in human blood by ELISA. Toxicon 2017, 133, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.M.; Beach, D.G.; Reeves, K.L.; Gibbs, R.S.; Kerrin, E.S.; Mccarron, P.; Quilliam, M.A. Hydrophilic interaction liquid chromatography-tandem mass spectrometry for quantitation of paralytic shellfish toxins: Validation and application to reference materials. Anal. Bioanal. Chem. 2017, 409, 5675–5687. [Google Scholar] [CrossRef]
- Zhuo, L.; Yin, Y.; Fu, W.; Qiu, B.; Lin, Z.; Yang, Y.; Zheng, L.; Li, J.; Chen, G. Determination of paralytic shellfish poisoning toxins by HILIC–MS/MS coupled with dispersive solid phase extraction. Food Chem. 2013, 137, 115–121. [Google Scholar] [CrossRef]
- Cianca, R.C.C.; Pallares, M.A.; Barbosa, R.D.; Adan, L.V.; Martins, J.M.L.; Gago-Martínez, A. Application of precolumn oxidation HPLC method with fluorescence detection to evaluate saxitoxin levels in discrete brain regions of rats. Toxicon 2007, 49, 89–99. [Google Scholar] [CrossRef]
- Turner, A.D.; Stubbs, B.; Coates, L.; Dhanji-Rapkova, M.; Hatfield, R.G.; Lewis, A.M.; Rowland-Pilgrim, S.; O’Neil, A.; Stubbs, P.; Ross, S.; et al. Variability of paralytic shellfish toxin occurrence and profiles in bivalve molluscs from Great Britain from official control monitoring as determined by pre-column oxidation liquid chromatography and implications for applying immunochemical tests. Harmful Algae 2014, 31, 87–99. [Google Scholar] [CrossRef]
- Veronica, R.; Ana, B.; Mercedes, A.; Alvaro, A.; Luis, B. Liquid Chromatography with a Fluorimetric Detection Method for Analysis of Paralytic Shellfish Toxins and Tetrodotoxin Based on a Porous Graphitic Carbon Column. Toxins 2016, 8, 196. [Google Scholar]
- Jeffrey, V.D.R.; Gibbs, R.S.; Muggah, P.M.; Rourke, W.A.; Macneil, J.D.; Quilliam, M.A.; Collaborators. Liquid chromatography post-column oxidation (PCOX) method for the determination of paralytic shellfish toxins in mussels, clams, oysters, and scallops: Collaborative study. J. Aoac. Int. 2011, 94, 1154–1175. [Google Scholar]
- Famulok, M.; Mayer, G.; Blind, M. Nucleic acid aptamers from selection in vitro to applications in vivo. Acc. Chem. Res. 2000, 33, 591–599. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yin, Q.; Chang, Y.; Zhang, Q.; Brennan, J.D.; Li, Y. In vitro selection of circular DNA aptamers for biosensing applications. Angew. Chem. Int. Ed. 2019, 58, 8013–8017. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; He, L.; Hu, Y.; Luo, Z.; Zhang, J. A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem. Sci. 2020, 11, 12157–12164. [Google Scholar] [CrossRef]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef]
- Ning, Y.; Hu, J.; Lu, F. Aptamers used for biosensors and targeted therapy. Biomed. Pharmacother. 2020, 132, 110902. [Google Scholar] [CrossRef]
- Qi, X.; Yan, X.; Zhao, Y.; Li, L.; Wang, S. Highly sensitive and specific detection of small molecules using advanced aptasensors based on split aptamers: A review. TrAC Trends Anal. Chem. 2020, 133, 116069. [Google Scholar] [CrossRef]
- Gao, S.; Hu, B.; Zheng, X.; Cao, Y.; Liu, D.; Sun, M.; Jiao, B.; Wang, L. Gonyautoxin 1/4 aptamers with high-affinity and high-specificity: From efficient selection to aptasensor application. Biosens. Bioelectron. 2016, 79, 938–944. [Google Scholar] [CrossRef]
- Gao, S.; Zheng, X.; Hu, B.; Sun, M.; Wu, J.; Jiao, B.; Wang, L. Enzyme-linked, aptamer-based, competitive biolayer interferometry biosensor for palytoxin. Biosens. Bioelectron. 2017, 89, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Hu, B.; Zhou, R.; Liu, D.; Peng, D.; Li, Z.; Li, Z.; Jiao, B.; Wang, L. Rapid and sensitive detection of nodularin-R in water by a label-free BLI aptasensor. Analyst 2018, 143, 4316–4322. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hu, B.; Zhou, R.; Zhang, X.; Wang, R.; Gao, Y.; Sun, M.; Jiao, B.; Wang, L. Selection and application of aptamers with high-affinity and high-specificity against dinophysistoxin-1. RSC Adv. 2020, 10, 8181–8189. [Google Scholar] [CrossRef]
- Handy, S.M.; Yakes, B.J.; DeGrasse, J.A.; Campbell, K.; Elliott, C.T.; Kanyuck, K.M.; DeGrasse, S.L. First report of the use of a saxitoxin–protein conjugate to develop a DNA aptamer to a small molecule toxin. Toxicon 2013, 61, 30–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Hu, B.; Gao, S.; Liu, D.; Sun, M.; Jiao, B.; Wang, L. A saxitoxin-binding aptamer with higher affinity and inhibitory activity optimized by rational site-directed mutagenesis and truncation. Toxicon 2015, 101, 41–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, L.; Jiang, L.; Song, Y.; Ding, Y.; Zhang, J.; Wu, X.; Tang, D. Amperometric aptasensor for saxitoxin using a gold electrode modified with carbon nanotubes on a self-assembled monolayer, and methylene blue as an electrochemical indicator probe. Microchim. Acta 2016, 183, 1971–1980. [Google Scholar] [CrossRef]
- Gao, S.; Zheng, X.; Wu, J. A biolayer interferometry-based competitive biosensor for rapid and sensitive detection of saxitoxin. Sens. Actuators B Chem. 2017, 246, 169–174. [Google Scholar] [CrossRef]
- Cheng, S.; Zheng, B.; Yao, D.; Wang, Y.; Tian, J.; Liu, L.; Liang, H.; Ding, Y. Determination of Saxitoxin by Aptamer-Based Surface-Enhanced Raman Scattering. Anal. Lett. 2019, 52, 902–918. [Google Scholar] [CrossRef]
- Caglayan, M.O.; Ustundag, Z. Saxitoxin aptasensor based on attenuated internal reflection ellipsometry for seafood. Toxicon 2020, 187, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, B.; Kalyani, N.; Anand, A.; Khan, E.; Das, S.; Bansal, V.; Kumar, A.; Sharma, T.K. GOLD SELEX: A novel SELEX approach for the development of high-affinity aptamers against small molecules without residual activity. Microchim. Acta 2020, 187, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Diao, D.L.; Qiao, N.; Wu, X.; Li, J.Y.; Lou, X.H. An efficient method to evaluate experimental factor influence on in vitro binding of aptamers. Anal. Biochem. 2018, 556, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Duan, N.; Xia, Y.; Hun, X.; Wang, H.; Wang, Z. Magnetic Separation-Based Multiple SELEX for Effectively Selecting Aptamers against Saxitoxin, Domoic Acid, and Tetrodotoxin. J. Agric. Food. Chem. 2018, 66, 9801–9809. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.-J.; Park, J.-H.; Lee, B.; Kim, M.-G. Label-free direct detection of saxitoxin based on a localized surface plasmon resonance aptasensor. Toxins 2019, 11, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kökpinar, Ö.; Walter, J.G.; Shoham, Y.; Stahl, F.; Scheper, T. Aptamer-based downstream processing of his-tagged proteins utilizing magnetic beads. Biotechnol. Bioeng. 2011, 108, 2371–2379. [Google Scholar] [CrossRef]
- Yang, L.; Gao, T.; Li, W.; Luo, Y.; Ullah, S.; Fang, X.; Cao, Y.; Pei, R. Ni-Nitrilotriacetic Acid Affinity SELEX Method for Selection of DNA Aptamers Specific to the N-Cadherin Protein. ACS Comb. Sci. 2020, 22, 867–872. [Google Scholar] [CrossRef]
- Murphy, J.C.; Jewell, D.L.; White, K.I.; Fox, G.E.; Willson, R.C. Nucleic acid separations utilizing immobilized metal affinity chromatography. Biotechnol. Prog. 2003, 19, 982–986. [Google Scholar] [CrossRef]
- Kowalska, E.; Bartnicki, F.; Pels, K.; Strzalka, W. The impact of immobilized metal affinity chromatography (IMAC) resins on DNA aptamer selection. Anal. Bioanal. Chem. 2014, 406, 5495–5499. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.H.; Wang, W.; Jones, R.A.; Patel, D.J. Formation of an amino-acid-binding pocket through adaptive zippering-up of a large DNA hairpin loop. Chem. Biol. 1998, 5, 555–572. [Google Scholar] [CrossRef] [Green Version]
- Qiao, N.; Li, J.; Wu, X.; Diao, D.L.; Zhao, J.X.; Li, J.Y.; Ren, X.J.; Ding, X.F.; Shangguan, D.H.; Lou, X.H. Speeding up in Vitro Discovery of Structure-Switching Aptamers via Magnetic Cross-Linking Precipitation. Anal. Chem. 2019, 91, 13383–13389. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.M.; Zou, R.X.; Lou, X.H. Label-Free Fluorescent Detection of Ions, Proteins, and Small Molecules Using Structure-Switching Aptamers, SYBR Gold, and Exonuclease I. Anal. Chem. 2012, 84, 3554–3560. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.M.; Zhang, F.Y.; Goldys, E.M.; Gao, F.; Liu, G.Z. Advances in structure-switching aptasensing towards real time detection of cytokines. Trac.-Trend Anal. Chem. 2018, 102, 379–396. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Nikolaus, N.; Strehlitz, B. Capture-SELEX: Selection of DNA Aptamers for Aminoglycoside Antibiotics. J. Anal. Methods Chem. 2012, 2012, 415697. [Google Scholar] [CrossRef]
- Paniel, N.; Istamboulie, G.; Triki, A.; Lozano, C.; Barthelmebs, L.; Noguer, T. Selection of DNA aptamers against penicillin G using Capture-SELEX for the development of an impedimetric sensor. Talanta 2017, 162, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, R.; Karimzadeh, F.; Kermanpur, A.; Kharaziha, M. The novel immobilization of G-quadruplex aptamer on Cu deposited surface using electrochemical method. Mater. Lett. 2021, 282, 128703. [Google Scholar] [CrossRef]
- Liu, Y.; Le, C.; Tyrrell, D.L.; Le, X.C.; Li, X.-F. Aptamer binding assay for the E antigen of hepatitis B using modified aptamers with G-quadruplex structures. Anal. Chem. 2020, 92, 6495–6501. [Google Scholar] [CrossRef] [Green Version]
- Xi, H.; Juhas, M.; Zhang, Y. G-quadruplex based biosensor: A potential tool for SARS-CoV-2 detection. Biosens. Bioelectron. 2020, 167, 112494. [Google Scholar] [CrossRef]
- Masoud, S.S.; Nagasawa, K. i-Motif-Binding Ligands and Their Effects on the Structure and Biological Functions of i-Motif. Chem. Pharm. Bull. 2018, 66, 1091–1103. [Google Scholar] [CrossRef] [Green Version]
- Kumaraswamy, S.; Tobias, R. Label-free kinetic analysis of an antibody–antigen interaction using biolayer interferometry. In Protein-Protein Interactions; Springer: Berlin/Heidelberg, Germany, 2015; pp. 165–182. [Google Scholar]
- Shechtman, O. The coefficient of variation as an index of measurement reliability. In Methods of Clinical Epidemiology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 39–49. [Google Scholar]
- Ullah, N.; Chen, W.; Noureen, B.; Tian, Y.; Du, L.; Wu, C.; Ma, J. An Electrochemical Ti3C2Tx Aptasensor for Sensitive and Label-Free Detection of Marine Biological Toxins. Sensors 2021, 21, 4938. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, L.; Ma, R.; Wang, L.; Yan, X.; Qi, X.; Wang, S.; Mao, X. A competitive colorimetric aptasensor transduced by hybridization chain reaction-facilitated catalysis of AuNPs nanozyme for highly sensitive detection of saxitoxin. Anal. Chim. Acta 2021, 1173, 338710. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, Y.; Yan, X.; Qi, X.; Wang, L.; Ma, R.; Wang, S.; Mao, X. Development of a terminal-fixed aptamer and a label-free colorimetric aptasensor for highly sensitive detection of saxitoxin. Sens. Actuators B Chem. 2021, 344, 130320. [Google Scholar] [CrossRef]
- Qi, X.; Yan, X.; Zhao, L.; Huang, Y.; Wang, S.; Liang, X. A facile label-free electrochemical aptasensor constructed with nanotetrahedron and aptamer-triplex for sensitive detection of small molecule: Saxitoxin. J. Electroanal. Chem. 2020, 858, 113805. [Google Scholar] [CrossRef]
- Cheng, S.; Zheng, B.; Yao, D.; Kuai, S.; Tian, J.; Liang, H.; Ding, Y. Study of the binding way between saxitoxin and its aptamer and a fluorescent aptasensor for detection of saxitoxin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 204, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Concepcion, J.; Witte, K.; Wartchow, C.; Choo, S.; Yao, D.F.; Persson, H.; Wei, J.; Li, P.; Heidecker, B.; Ma, W.L.; et al. Label-Free Detection of Biomolecular Interactions Using BioLayer Interferometry for Kinetic Characterization. Comb. Chem. High Throughput Screen. 2009, 12, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Le Berre, M.; Kilcoyne, M.; Kane, M. Generation of a panel of high affinity antibodies and development of a biosensor-based immunoassay for the detection of okadaic acid in shellfish. Toxicon 2015, 103, 169–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS Mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34 (Suppl. S2), W676–W682. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Ponder, J.W. Polarizable atomic multipole water model for molecular mechanics simulation. J. Phys. Chem. B 2003, 107, 5933–5947. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Cieplak, P.; Kollman, P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000, 21, 1049–1074. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed] [Green Version]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, M.; Li, G.; Zhang, Q.; Liu, J.; Huang, Q. De novo post-SELEX optimization of a G-quadruplex DNA aptamer binding to marine toxin gonyautoxin 1/4. Comput. Struct. Biotechnol. J. 2020, 18, 3425–3433. [Google Scholar] [CrossRef] [PubMed]




| Contribution | ∆EVDW | ∆Eele | ∆Gpolar | ∆Gnonpolar | ∆Gtotal |
|---|---|---|---|---|---|
| Energy (kcal/mol) | −25.02 ± 0.41 | −381.64 ± 1.81 | 386.11 ± 1.59 | −2.83 ± 0.03 | −23.38 ± 0.53 |
| No | Concentration of STX (ng/mL) | Recovery (%) | CV (%) |
|---|---|---|---|
| 1 | 100 | 106 | 4.3 |
| 2 | 200 | 110 | 6.7 |
| 3 | 500 | 97 | 2.4 |
| Method | Sample | Selecting Method of Aptamer | Kd (nM) | Linear Range | Limit of Detection | Recovery (%) | References | |
|---|---|---|---|---|---|---|---|---|
| HILIC-MS/MS | bivalve aquatic products, Standard addition | — | — | — | 18–25 nmol/kg tissue | 92–96 | Thomas et al. 2017 [7] | |
| Scomberomorus niphonius, oyster, blood clam | — | — | — | 2.9–4.3μg/kg | 76.8–93.6 | Zhuo, et al. 2013 [8] | ||
| Precolumn oxidation HPLC | rats’ brain homogenate | — | — | 0.05–20 ng/mL | 11.19 ± 0.11pg/20μL injection | 59.5 ± 1.5 for total brain | Cianca et al. 2007 [9] | |
| Postcolumn oxidation HPLC-FLD | mussel, clam, scallop, oyster | — | — | 0.007–3.812mg STX.diHCl/kg | 0.0038–0.0044mg STX.diHCl/kg | 78.8–83.4 | Veronica et al. 2016 [11] | |
| — | — | 101–123 | Jeffrey et al. 2011 [12] | |||||
| Direct ELISA | human whole blood | — | — | 0.010–1.0 ng/mL | 0.02 ng/mL | — | Wharton et al. 2017 [6] | |
| dried human blood | 0.06 ng/mL | |||||||
| Aptasensor | 1.Electrochemical aptasensor | mussels | Fixing saxitoxin | APTSTX1 (3840) | 0.9–30 nM | 0.38 nM | 63–120.5 | Hou et al. 2016 [28] |
| seawater | Fixing saxitoxin | APTSTX1 (3840) | 1–400 nM | 0.92 nM | 94.4–111 | Qi et al. 2020 [56] | ||
| 2.Spectroscopic ellipsometry (SE) aptasensor | fish and shrimp | Fixing saxitoxin | APTSTX1 (3840) | 0.1–1000 ng/mL | 0.11 ng/mL | — | Caglayan et al. 2020 [31] | |
| 3.Competitive biosensor | shellfish, ribbon fish and water | Site-directed mutation and truncation of APTSTX1 | M30f (133) | 100–800 ng/mL | 0.50 ng/mL | 101.4–107.3 | Gao et al. 2017 [29] | |
| 4.Fluorescenceswitch sensor | Shellfish | 0–24 ng/mL | 1.80 ng/mL | 105.7–111.2 | Cheng et al. 2018 [57] | |||
| 5.Colorimetric aptasensor | shellfish | Site-directed mutation and truncation of APTSTX1 | M30f (133) | 78.13–2500 pM | 42.46 pM | 106.2–113.5 | Zhao et al. 2021 [54] | |
| seawater and scallop | M30f Engineered by terminal hybridization | TF-M-30f (0.917) | 0.1457− 37.30 nM | 142.3 pM | 98.21–114.1 | Li et al. 2021 [55] | ||
| 6.LSPR aptasensor | mussels | GO-SELEX | 50.75 ± 14.97 | 5–10,000 μg/L | 2.46 μg/L | 96.13–116.05 | Ha et al. 2019 [35] | |
| 7.Fluorescence assay | clam | MRGO-SELEX | STX-41(61.44 ± 23.18) | — | 0.39 ng/mL | 84.59–96.13 | Gu et al. 2018 [34] | |
| 8.Biosensor | seawater | IMC-SELEX | 75a (136) | — | — | — | This work | |
| seawater | 45e-1(19) | 50–800 ng/ml | 0.50 ng/mL | 97–106 | ||||
| 9.Electrochemical Ti3C2Tx | mussel tissues | SELEX | — | 1.0–200 nM | 0.03 nM | 103 | Ullah et al. 2021 [53] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, R.; Gao, Y.; Yang, C.; Zhang, X.; Hu, B.; Zhao, L.; Guo, H.; Sun, M.; Wang, L.; Jiao, B. A Novel SELEX Based on Immobilizing Libraries Enables Screening of Saxitoxin Aptamers for BLI Aptasensor Applications. Toxins 2022, 14, 228. https://doi.org/10.3390/toxins14030228
Zhou R, Gao Y, Yang C, Zhang X, Hu B, Zhao L, Guo H, Sun M, Wang L, Jiao B. A Novel SELEX Based on Immobilizing Libraries Enables Screening of Saxitoxin Aptamers for BLI Aptasensor Applications. Toxins. 2022; 14(3):228. https://doi.org/10.3390/toxins14030228
Chicago/Turabian StyleZhou, Rong, Yun Gao, Chengfang Yang, Xiaojuan Zhang, Bo Hu, Luming Zhao, Han Guo, Mingjuan Sun, Lianghua Wang, and Binghua Jiao. 2022. "A Novel SELEX Based on Immobilizing Libraries Enables Screening of Saxitoxin Aptamers for BLI Aptasensor Applications" Toxins 14, no. 3: 228. https://doi.org/10.3390/toxins14030228







