Contextual Constraints: Dynamic Evolution of Snake Venom Phospholipase A2
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structural and Functional Underpinnings of Snake Venom PLA2s
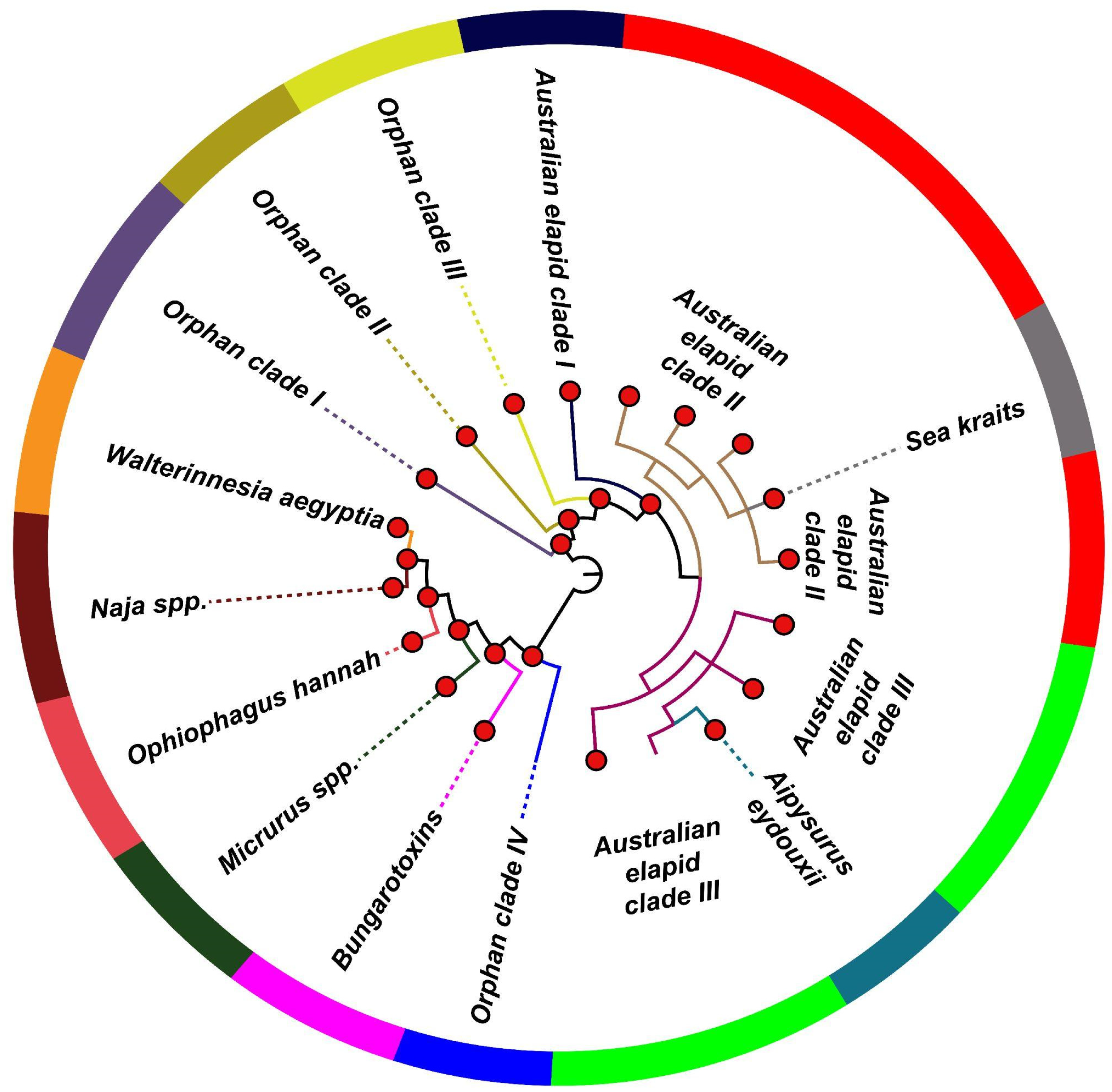
2.2. The Influence of Diverse Ecologies on the Evolution of Snake Venom PLA2s
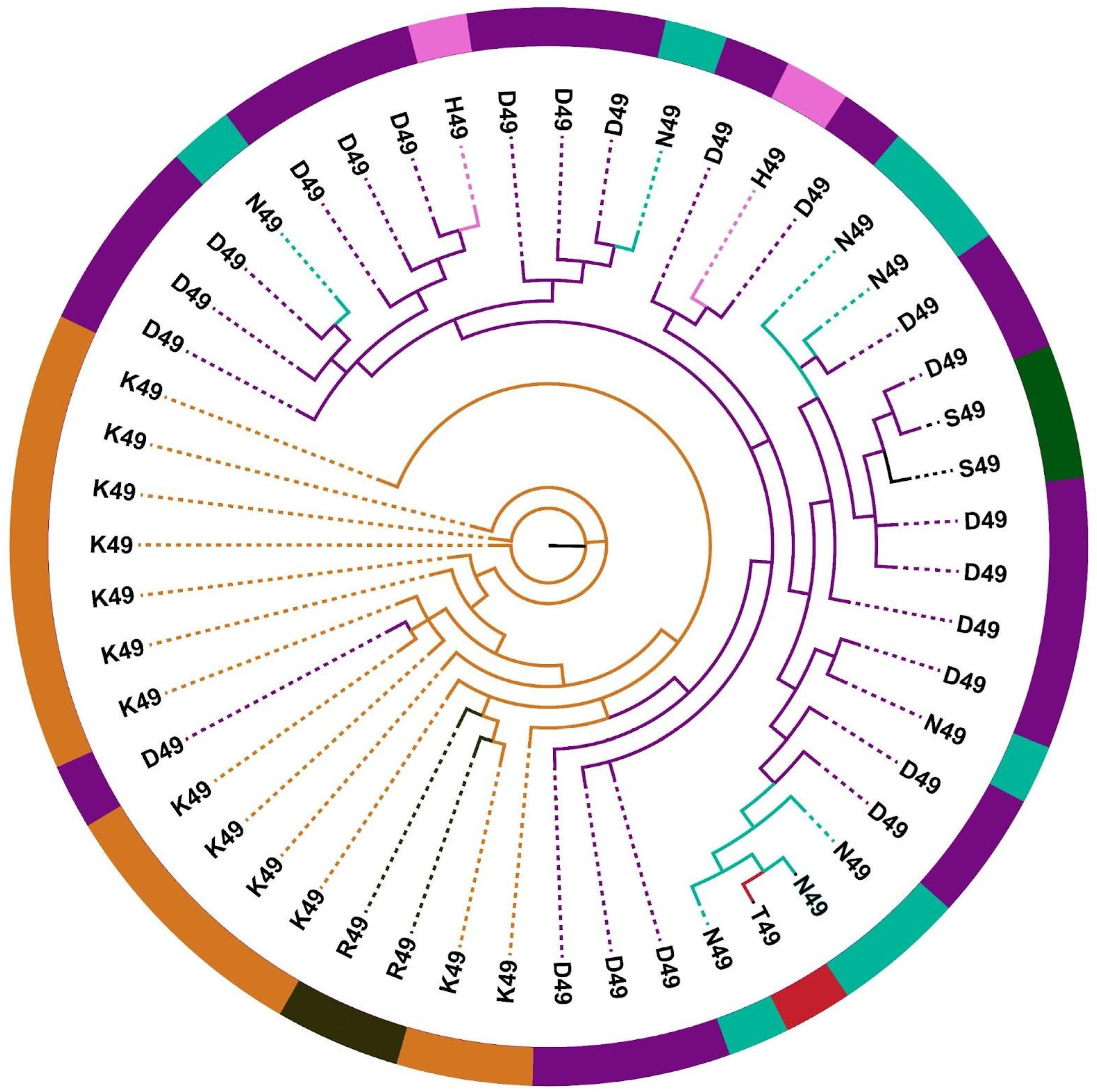
2.3. Distinct Phylogenetic Histories of Elapid and Viperid Venom PLA2s
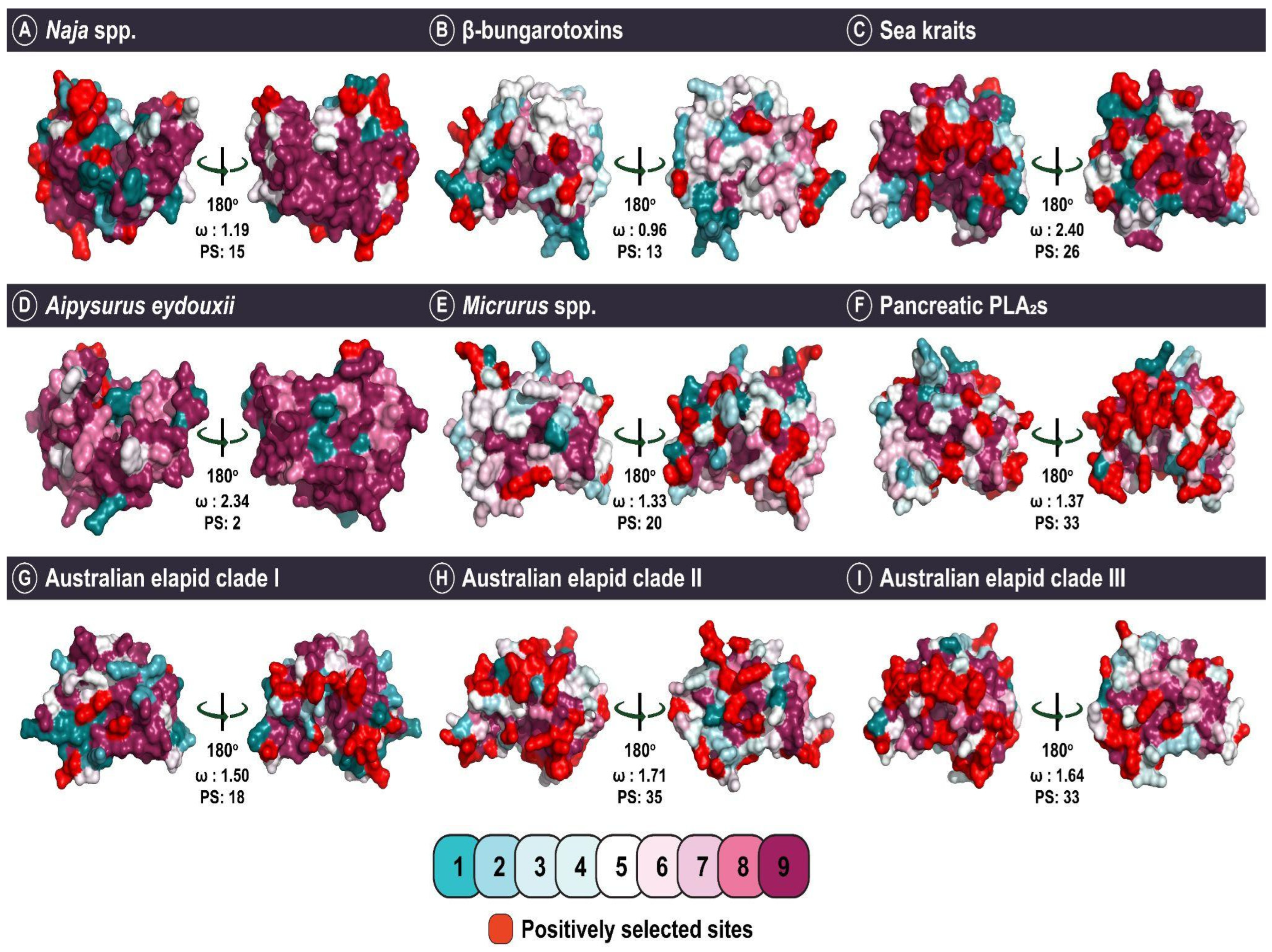
2.4. The Impact of Structure–Function, Ecology and Evolution on the Diversification of Snake Venom PLA2s
3. Conclusions
4. Material and Methods
4.1. Dataset Assembly and Phylogenetic Reconstructions
4.2. Selection Analyses
4.3. Structural Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jackson, T.N.; Jouanne, H.; Vidal, N. Snake venom in context: Neglected clades and concepts. Front. Ecol. Evol. 2019, 7, 332. [Google Scholar] [CrossRef] [Green Version]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casewell, N.R.; Jackson, T.N.; Laustsen, A.H.; Sunagar, K. Causes and consequences of snake venom variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [Green Version]
- Dennis, E.A. 9 Phospholipases. In The Enzymes; Elsevier: Amsterdam, The Netherlands, 1983; Volume 16, pp. 307–353. [Google Scholar]
- Dijkstra, B.W.; Kalk, K.H.; Hol, W.G.; Drenth, J. Structure of bovine pancreatic phospholipase A2 at 1.7 Å resolution. J. Mol. Biol. 1981, 147, 97–123. [Google Scholar] [CrossRef]
- Fry, M.; Ghosh, S.; East, J.; Franson, R. Role of human sperm phospholipase A2 in fertilization: Effects of a novel inhibitor of phospholipase A2 activity on membrane perturbations and oocyte penetration. Biol. Reprod. 1992, 47, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Arita, H.; Hanasaki, K.; Nakano, T.; Oka, S.; Teraoka, H.; Matsumoto, K. Novel proliferative effect of phospholipase A2 in Swiss 3T3 cells via specific binding site. J. Biol. Chem. 1991, 266, 19139–19141. [Google Scholar] [CrossRef]
- Murakami, M.; Sato, H.; Taketomi, Y. Updating phospholipase A2 biology. Biomolecules 2020, 10, 1457. [Google Scholar] [CrossRef]
- Dennis, E.A.; Rhee, S.G.; Billah, M.M.; Hannun, Y.A. Role of phospholipases in generating lipid second messengers in signal transduction 1. FASEB J. 1991, 5, 2068–2077. [Google Scholar] [CrossRef]
- Gutiérrez, J.; Lomonte, B. Venom Phospholipase A2 Enzymes: Structure, Function and Mechanism; Kini, R.M., Ed.; Wiley & Sons: Hoboken, NJ, USA, 1997. [Google Scholar]
- Župunski, V.; Kordiš, D. Strong and widespread action of site-specific positive selection in the snake venom Kunitz/BPTI protein family. Sci. Rep. 2016, 6, 37054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juárez, P.; Comas, I.; González-Candelas, F.; Calvete, J.J. Evolution of Snake Venom Disintegrins by Positive Darwinian Selection. Mol. Biol. Evol. 2008, 25, 2391–2407. [Google Scholar] [CrossRef] [Green Version]
- Sunagar, K.; Jackson, T.N.; Undheim, E.A.; Ali, S.; Antunes, A.; Fry, B.G. Three-fingered RAVERs: Rapid Accumulation of Variations in Exposed Residues of snake venom toxins. Toxins 2013, 5, 2172–2208. [Google Scholar] [CrossRef] [Green Version]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.; Kerkkamp, H.M.; Vos, R.A.; Guerreiro, I.; Calvete, J.J. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef] [Green Version]
- Sunagar, K.; Johnson, W.E.; O’Brien, S.J.; Vasconcelos, V.; Antunes, A. Evolution of CRISPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol. Biol. Evol. 2012, 29, 1807–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, V.J. Inventing an arsenal: Adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol. Biol. 2007, 7, 2. [Google Scholar] [CrossRef] [Green Version]
- Kini, R.M.; Chan, Y.M. Accelerated evolution and molecular surface of venom phospholipase A2 enzymes. J. Mol. Evol. 1999, 48, 125–132. [Google Scholar] [CrossRef]
- Oliveira, A.; Bleicher, L.; Schrago, C.G.; Junior, F.P.S. Conservation analysis and decomposition of residue correlation networks in the phospholipase A2 superfamily (PLA2s): Insights into the structure-function relationships of snake venom toxins. Toxicon 2018, 146, 50–60. [Google Scholar] [CrossRef]
- DUFTON, M.J.; HIDER, R.C. Classification of phospholipases A2 according to sequence. Evolutionary and pharmacological implications. Eur. J. Biochem. 1983, 137, 545–551. [Google Scholar] [CrossRef]
- Sunagar, K.; Jackson, T.; Reeks, T.; Fry, B. Group I Phospholipase A2 Enzymes; Oxford University Press: New York, NY, USA, 2015. [Google Scholar]
- Sunagar, K.; Tsai, I.; Lomonte, B.; Jackson, T.; Fry, B. Group II Phospholipase A2 Enzymes; Oxford University Press: New York, NY, USA, 2015. [Google Scholar]
- Jackson, T.N.; Koludarov, I. How the toxin got its toxicity. Front. Pharmacol. 2020, 11, 1893. [Google Scholar] [CrossRef]
- Dowell, N.; Giorgianni, M.; Kassner, V.; Selegue, J.; Sanchez, E.; Carroll, S. The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr. Biol. 2016, 26, 2434–2445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs, H.L.; Rossiter, W. Rapid evolution by positive selection and gene gain and loss: PLA2 venom genes in closely related Sistrurus rattlesnakes with divergent diets. J. Mol. Evol. 2008, 66, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Slowinski, J.B.; Knight, A.; Rooney, A.P. Inferring species trees from gene trees: A phylogenetic analysis of the Elapidae (Serpentes) based on the amino acid sequences of venom proteins. Mol. Phylogenet. Evol. 1997, 8, 349–362. [Google Scholar] [CrossRef] [Green Version]
- Kondo, K.; TODA, H.; NARITA, K.; LEE, C.-Y. Amino acid sequence of β2-bungarotoxin from Bungarus multicinctus venom. The amino acid substitutions in the B chains. J. Biochem. 1982, 91, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Kwong, P.D.; McDonald, N.Q.; Sigler, P.B.; Hendrickson, W.A. Structure of β2-bungarotoxin: Potassium channel binding by Kunitz modules and targeted phospholipase action. Structure 1995, 3, 1109–1119. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.Z.; Gopalakrishnakone, P.; Chung, M.C.; Kini, R.M. Complete Amino Acid Sequence of an Acidic, Cardiotoxic Phospholipase A2from the Venom ofOphiophagus hannah (King Cobra): A Novel Cobra Venom Enzyme with “Pancreatic Loop”. Arch. Biochem. Biophys. 1997, 338, 150–156. [Google Scholar] [CrossRef]
- Maraganore, J.M.; Heinrikson, R. The lysine-49 phospholipase A2 from the venom of Agkistrodon piscivorus piscivorus. Relation of structure and function to other phospholipases A2. J. Biol. Chem. 1986, 261, 4797–4804. [Google Scholar] [CrossRef]
- Van Den Bergh, C.J.; Slotboom, A.J.; Verheij, H.M.; De Haas, G.H. The role of aspartic acid-49 in the active site of phospholipase A2: A site-specific mutagenesis study of porcine pancreatic phospholipase A2 and the rationale of the enzymatic activity of [Iysine49] phospholipase A2 from Agkistrodon piscivorus piscivorus venom. Eur. J. Biochem. 1988, 176, 353–357. [Google Scholar] [PubMed]
- Polgár, J.; Magnenat, E.M.; Peitsch, M.C.; Wells, T.N.; Clemetson, K.J. Asp-49 is not an absolute prerequisite for the enzymic activity of low-Mr phospholipases A2: Purification, characterization and computer modelling of an enzymically active Ser-49 phospholipase A2, ecarpholin S, from the venom of Echis carinatus sochureki (saw-scaled viper). Biochem. J. 1996, 319, 961–968. [Google Scholar]
- Tsai, I.-H.; Wang, Y.-M.; Chen, Y.-H.; Tsai, T.-S.; Tu, M.-C. Venom phospholipases A2 of bamboo viper (Trimeresurus stejnegeri): Molecular characterization, geographic variations and evidence of multiple ancestries. Biochem. J. 2004, 377, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Chijiwa, T.; Tokunaga, E.; Ikeda, R.; Terada, K.; Ogawa, T.; Oda-Ueda, N.; Hattori, S.; Nozaki, M.; Ohno, M. Discovery of novel [Arg49]phospholipase A2 isozymes from Protobothrops elegans venom and regional evolution of Crotalinae snake venom phospholipase A2 isozymes in the southwestern islands of Japan and Taiwan. Toxicon 2006, 48, 672–682. [Google Scholar] [CrossRef]
- Gopalakrishnakone, P.; Kochva, E. Venom glands and some associated muscles in sea snakes. J. Morphol. 1990, 205, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Fry, B.; Kini, R.M. Eggs-only diet: Its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii). J. Mol. Evol. 2005, 60, 81–89. [Google Scholar] [CrossRef]
- Li, M.; Fry, B.G.; Kini, R.M. Putting the brakes on snake venom evolution: The unique molecular evolutionary patterns of Aipysurus eydouxii (Marbled sea snake) phospholipase A2 toxins. Mol. Biol. Evol. 2005, 22, 934–941. [Google Scholar] [CrossRef] [Green Version]
- da Silva Jr, N.J.; Aird, S.D. Prey specificity, comparative lethality and compositional differences of coral snake venoms. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2001, 128, 425–456. [Google Scholar] [CrossRef]
- Martins, M.; Oliveira, M.E. Natural history of snakes in forests of the Manaus region, Central Amazonia, Brazil. Herpetol. Nat. Hist. 1998, 6, 78–150. [Google Scholar]
- Campbell, J.A.; Lamar, W.W. The venomous reptiles of Latin America. In The Venomous Reptiles of Latin America; Cornell University Press: Ithaca, NY, USA, 1989. [Google Scholar]
- Souza, S.M.; Junqueira, A.B.; Jakovac, A.C.C.; Assunção, P.A.; Correia, J.A. Feeding behavior and ophiophagous habits of two poorly known Amazonian coral snakes, Micrurus albicinctus Amaral 1926 and Micrurus paraensis Cunha and Nascimento 1973 (Squamata, Elapidae). Herpetol. Notes 2011, 4, 369–372. [Google Scholar]
- Vitt, L.J.; Hulse, A.C. Observations on feeding habits and tail display of the Sonoran coral snake, Micruroides euryxanthus. Herpetologica 1973, 29, 302–304. [Google Scholar]
- Marques, O.; Sazima, I. Diet and feeding behavior of the coral snake, Micrurus corallinus, from the Atlantic forest in Brazil. Herpetol. Nat. Hist. 1997, 5, 88–93. [Google Scholar]
- Shine, R.; Shetty, S. Moving in two worlds: Aquatic and terrestrial locomotion in sea snakes (Laticauda colubrina, Laticaudidae). J. Evol. Biol. 2001, 14, 338–346. [Google Scholar] [CrossRef]
- Greene, H.W. Snakes: The Evolution of Mystery in Nature; University of California Press: Berkeley, CA, USA, 1997. [Google Scholar]
- Heatwole, H. Sea Snakes; University of New South Wales Press: Sydney, Australia, 1999. [Google Scholar]
- Sunagar, K.; Moran, Y. The rise and fall of an evolutionary innovation: Contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015, 11, e1005596. [Google Scholar] [CrossRef] [Green Version]
- Scanlon, J.D.; Lee, M.S. Phylogeny of Australasian venomous snakes (Colubroidea, Elapidae, Hydrophiinae) based on phenotypic and molecular evidence. Zool. Scr. 2004, 33, 335–366. [Google Scholar] [CrossRef]
- Wilson, S.K.; Swan, G. A Complete Guide to Reptiles of Australia; New Holland Publishers: Wahroonga, Australia, 2013. [Google Scholar]
- Jackson, T.N.; Sunagar, K.; Undheim, E.A.; Koludarov, I.; Chan, A.H.; Sanders, K.; Ali, S.A.; Hendrikx, I.; Dunstan, N.; Fry, B.G. Venom down under: Dynamic evolution of Australian elapid snake toxins. Toxins 2013, 5, 2621–2655. [Google Scholar] [CrossRef] [Green Version]
- Jackson, T.N.; Koludarov, I.; Ali, S.A.; Dobson, J.; Zdenek, C.N.; Dashevsky, D.; Op den Brouw, B.; Masci, P.P.; Nouwens, A.; Josh, P. Rapid radiations and the race to redundancy: An investigation of the evolution of Australian elapid snake venoms. Toxins 2016, 8, 309. [Google Scholar] [CrossRef]
- Malhotra, A.; Creer, S.; Harris, J.B.; Thorpe, R.S. The importance of being genomic: Non-coding and coding sequences suggest different models of toxin multi-gene family evolution. Toxicon 2015, 107, 344–358. [Google Scholar] [CrossRef]
- Koludarov, I.; Jackson, T.N.; Suranse, V.; Pozzi, A.; Sunagar, K.; Mikheyev, A.S. Reconstructing the evolutionary history of a functionally diverse gene family reveals complexity at the genetic origins of novelty. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 2005, 39, 197–218. [Google Scholar] [CrossRef] [Green Version]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar]
- Nielsen, R.; Yang, Z. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 2003, 20, 1231–1239. [Google Scholar] [CrossRef]
- Fry, B.; Undheim, E.; Jackson, T. Research methods. In Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery; Oxford University Press: New York, NY, USA, 2015; pp. 153–214. [Google Scholar]
- Lee, M.S.; Sanders, K.L.; King, B.; Palci, A. Diversification rates and phenotypic evolution in venomous snakes (Elapidae). R. Soc. Open Sci. 2016, 3, 150277. [Google Scholar] [CrossRef] [Green Version]
- Alencar, L.R.; Quental, T.B.; Grazziotin, F.G.; Alfaro, M.L.; Martins, M.; Venzon, M.; Zaher, H. Diversification in vipers: Phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenet. Evol. 2016, 105, 50–62. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree v1. 4. Molecular Evolution, Phylogenetics and Epidemiology; University of Edinburgh, Institute of Evolutionary Biology: Edinburgh, UK, 2012. [Google Scholar]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. FUBAR: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef] [Green Version]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [Green Version]
- Fraczkiewicz, R.; Braun, W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998, 19, 319–333. [Google Scholar] [CrossRef]
- Connolly, M.L. Analytical molecular surface calculation. J. Appl. Crystallogr. 1983, 16, 548–558. [Google Scholar] [CrossRef]
- Richmond, T.J. Solvent accessible surface area and excluded volume in proteins: Analytical equations for overlapping spheres and implications for the hydrophobic effect. J. Mol. Biol. 1984, 178, 63–89. [Google Scholar] [CrossRef]


| Feature | |
|---|---|
| Group I (Elapidae) | |
| Naja spp. | Phylogenetic clustering |
| Ophiophagus Hannah * | Phylogenetic clustering, ecological constraint |
| Walterinnesia aegyptia * | Phylogenetic clustering, ecological constraint |
| β-bungarotoxins | Phylogenetic clustering, structural constraint |
| Australian elapid clades I, II and III | Phylogenetic clustering, ecological constraint |
| Sea kraits | Phylogenetic clustering, ecological constraint |
| Aipysurus eydouxii | Phylogenetic clustering, dietary specialisation |
| Micrurus spp. | Phylogenetic clustering, dietary specialisation |
| Orphan clades I, II, III and IV * | Phylogenetic clustering |
| Pancreatic | Structural constraint |
| Group II (Viperidae) | |
| D49 | Catalysis |
| H49 * | Catalysis |
| K49 | Catalysis |
| N49 | Catalysis |
| R49 * | Catalysis |
| S49 | Catalysis |
| T49 * | Catalysis |
| Group | FUBAR a | MEME b | PAML c | Solvent Accessibility (Number of Residues) | |
|---|---|---|---|---|---|
| Exposed | Buried | ||||
| Elapidae | |||||
| Naja spp. | ω > 1 d: 3 ω < 1 e: 4 | 1 | 15 | 12 | 0 |
| ω **: 1.19 | |||||
| β-bungarotoxin | ω > 1 d: 14 ω < 1 e: 11 | 23 | 13 | 9 | 0 |
| ω **: 0.96 | |||||
| Australian elapid clade I | ω > 1 d: 6 ω < 1 e: 4 | 1 | 18 | 12 | 0 |
| ω **: 1.50 | |||||
| Australian elapid clade II | ω > 1 d: 24 ω < 1 e: 10 | 17 | 35 | 27 | 2 |
| ω **: 1.71 | |||||
| Australian elapid clade III | ω > 1 d: 24 ω < 1 e: 9 | 21 | 33 | 21 | 3 |
| ω **: 1.64 | |||||
| Sea kraits | ω > 1 d: 5 ω < 1 e: 1 | 1 | 26 | 16 | 3 |
| ω **: 2.40 | |||||
| A. eydouxii | ω > 1 d: 1 ω < 1 e: 0 | 0 | 2 | 1 | 1 |
| ω: 2.34 | |||||
| Micrurus spp. | ω > 1 d: 25 ω < 1 e: 6 | 33 | 24 | 15 | 1 |
| ω **: 1.34 | |||||
| Pancreatic PLA2s | ω > 1 d: 32 ω < 1 e: 13 | 21 | 33 | 21 | 1 |
| ω **: 1.37 | |||||
| Viperidae | |||||
| D49 | ω > 1 d: 25 ω < 1 e: 50 | 42 | 21 | 16 | 0 |
| ω **: 0.83 | |||||
| K49 | ω > 1 d: 13 ω < 1 e: 10 | 12 | 13 | 9 | 1 |
| ω **: 0.92 | |||||
| N49 | ω > 1 d: 17 ω < 1 e: 8 | 6 | 17 | 11 | 1 |
| ω **: 1.34 | |||||
| S49 | ω > 1 d: 1 ω < 1 e: 0 | 0 | 7 | 5 | 0 |
| ω **: 1.7044 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suranse, V.; Jackson, T.N.W.; Sunagar, K. Contextual Constraints: Dynamic Evolution of Snake Venom Phospholipase A2. Toxins 2022, 14, 420. https://doi.org/10.3390/toxins14060420
Suranse V, Jackson TNW, Sunagar K. Contextual Constraints: Dynamic Evolution of Snake Venom Phospholipase A2. Toxins. 2022; 14(6):420. https://doi.org/10.3390/toxins14060420
Chicago/Turabian StyleSuranse, Vivek, Timothy N. W. Jackson, and Kartik Sunagar. 2022. "Contextual Constraints: Dynamic Evolution of Snake Venom Phospholipase A2" Toxins 14, no. 6: 420. https://doi.org/10.3390/toxins14060420
APA StyleSuranse, V., Jackson, T. N. W., & Sunagar, K. (2022). Contextual Constraints: Dynamic Evolution of Snake Venom Phospholipase A2. Toxins, 14(6), 420. https://doi.org/10.3390/toxins14060420







