In Vitro Toxicity Evaluation of Cyanotoxins Cylindrospermopsin and Microcystin-LR on Human Kidney HEK293 Cells
Abstract
:1. Introduction
2. Results
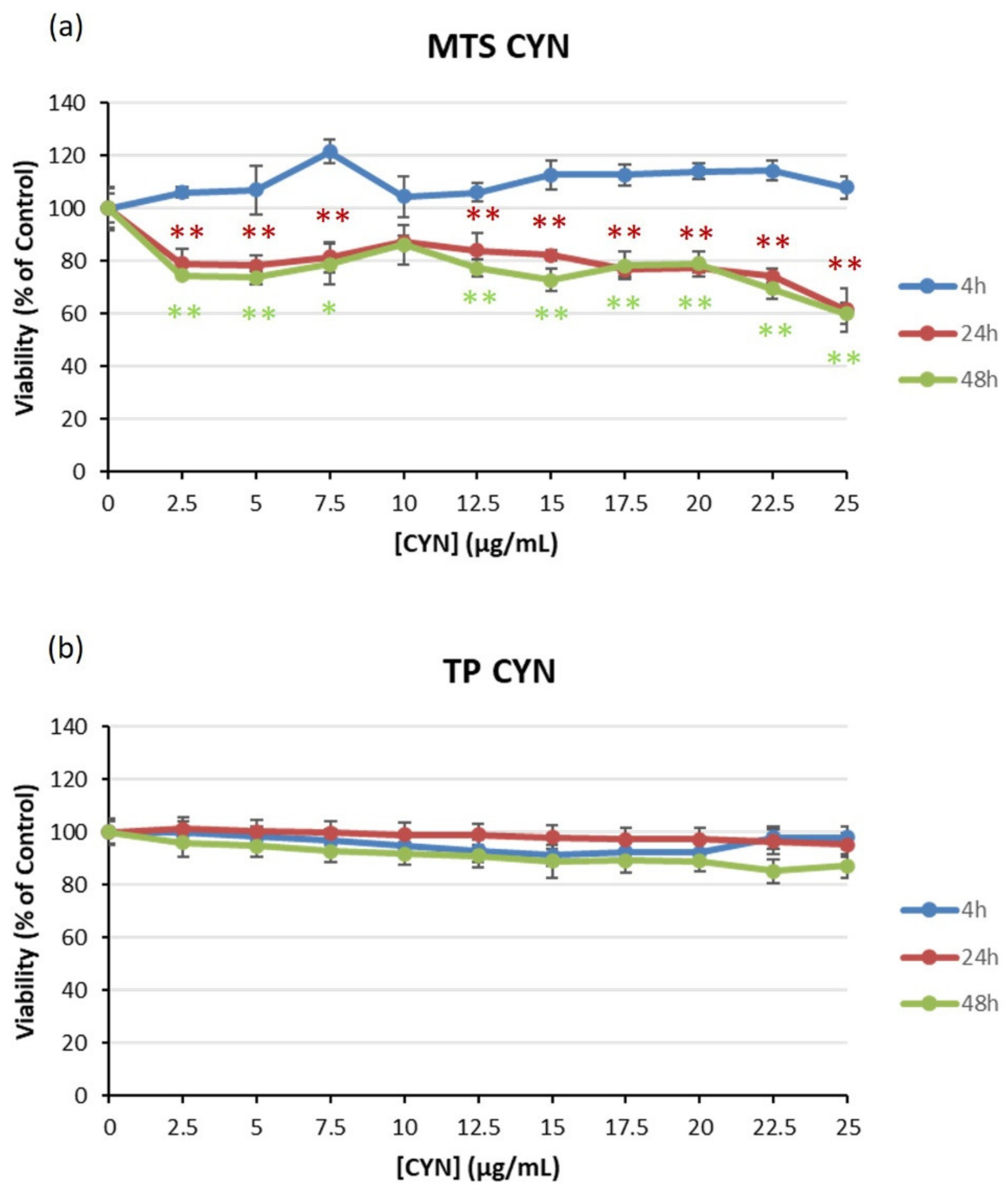
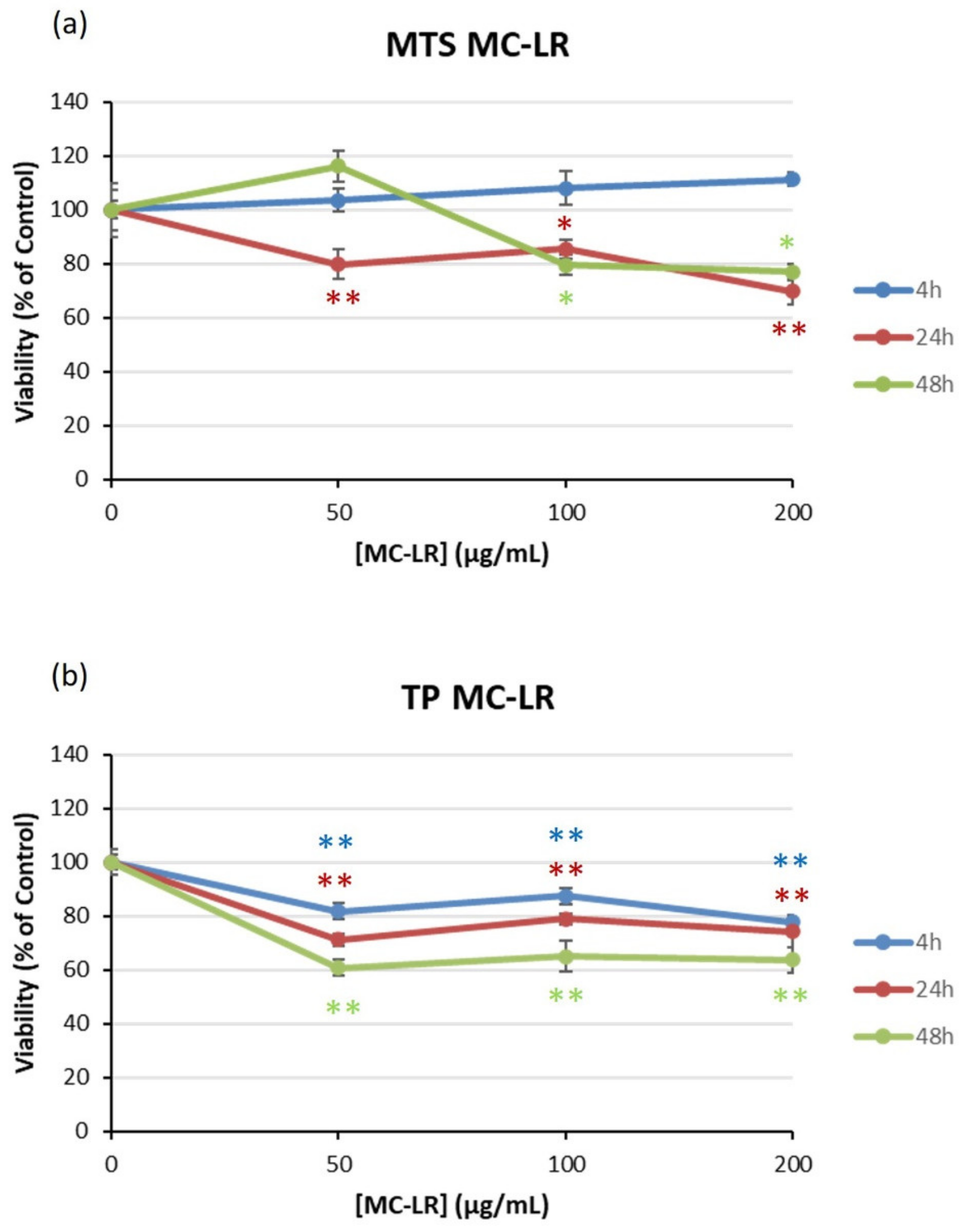
2.1. Viability of HEK293 Cells Exposed to Cyanotoxins
2.2. Effect of CYN on mRNA Expression
2.3. Proteomics Analysis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Supplies and Chemicals
5.2. Cell Culture and Treatment
5.3. Cytotoxicity Assays
5.4. Real-Time Quantitative PCR (qRT-PCR) Analysis after CYN Exposure
5.5. Proteomic Analysis
5.5.1. Sample Preparation
5.5.2. LC-MS/MS
5.5.3. Protein Identification
5.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Funari, E.; Testai, E. Human health risk assessment related to cyanotoxins exposure. Crit. Rev. Toxicol. 2008, 38, 97–125. [Google Scholar] [CrossRef] [PubMed]
- Catherine, A.; Bernard, C.; Spoof, L.; Bruno, M. Mycrocistins and nodularins. In Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis, 1st ed.; Meriluoto, J., Spoof, L., Cood, G.A., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 109–126. [Google Scholar] [CrossRef]
- Bouaïcha, N.; Miles, C.O.; Beach, D.G.; Labidi, Z.; Djabri, A.; Benayache, N.Y.; Nguyen-Quang, T. Structural diversity, characterization and toxicology of microcystins. Toxins 2019, 11, 714. [Google Scholar] [CrossRef] [Green Version]
- Diez-Quijada, L.; Puerto, M.; Gutiérrez-Praena, D.; Llana-Ruiz-Cabello, M.; Jos, Á.; Cameán, A.M. Microcystin-RR: Occurrence, content in water and food and toxicological studies. A review. Environ. Res. 2019, 168, 467–489. [Google Scholar] [CrossRef] [PubMed]
- Diez-Quijada, L.; Prieto, A.I.; Gumán-Guillé, R.; Jos, Á.; Cameán, A.M. Occurrence and toxicity of microcystin congeners other than MC-LR and MC-RR: A review. Food Chem. Toxicol. 2019, 125, 106–132. [Google Scholar] [CrossRef] [PubMed]
- Buratti, F.M.; Manganelli, M.; Vichi, S.; Stefanelli, M.; Scardala, S.; Testai, M.; Funari, E. Cyanotoxins: Producing organisms, occurrence, toxicity, mechanism of action and human health toxicological risk evaluation. Arch. Toxicol. 2017, 91, 1049–1130. [Google Scholar] [CrossRef]
- Mackintosh, C.; Beattie, K.A.; Klumpp, S.; Cohen, P.; Codd, G.A. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2 A from both mammals and higher plants. FEBS Lett. 1990, 264, 187–192. [Google Scholar] [CrossRef] [Green Version]
- Ingested Nitrate and Nitrite, and Cyanobacterial Peptide Toxins. Available online: https://www.ncbi.nlm.nih.gov/books/NBK326544/pdf/Bookshelf_NBK326544.pdf (accessed on 19 May 2022).
- Ohtani, I.; Moore, R.E.; Runnegar, M.T.C. Cylindrospermopsin—A potent hepatotoxin from the blue green alga Cylindrospermopsis raciborskii. J. Am. Chem. Soc. 1992, 114, 7941–7942. [Google Scholar] [CrossRef]
- Oliveira, F.; Diez-Quijada, L.; Turkina, M.V.; Morais, J.; Felpeto, A.B.; Azevedo, J.; Jos, A.; Camean, A.M.; Vasconcelos, V.; Martins, J.C.; et al. Physiological and metabolic responses of marine mussels exposed to toxic cyanobacteria Microcystis aeruginosa and Chrysosporum ovalisporum. Toxins 2020, 12, 196. [Google Scholar] [CrossRef] [Green Version]
- Terao, K.; Ohmori, S.; Igarashi, K.; Ohtani, I.; Watanabe, M.F.; Harada, K.I.; Ito, E.; Watanabe, M. Electron microscopic studies on experimental poisoning in mice induced by cylindrospermopsin isolated from blue-green alga Umezakia natans. Toxicon 1994, 32, 833–843. [Google Scholar] [CrossRef]
- Runnegar, M.T.; Kong, S.-M.; Zhong, Y.-Z.; Lu, S.C. Inhibition of reduced glutathione synthesis by cyanobacterial alkaloid cylindrospermopsin in cultured rat hepatocytes. Biochem. Pharmacol. 1995, 49, 219–225. [Google Scholar] [CrossRef]
- Froscio, S.M.; Humpage, A.R.; Burcham, P.C.; Falconer, I.R. Cylindrospermopsin-induced protein synthesis inhibition and its dissociation from acute toxicity in mouse hepatocytes. Environ. Toxicol. Int. J. 2003, 18, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.R.; Runnegar, M.T.; Jackson, A.R.; Falconer, I.R. Severe hepatotoxicity caused by the tropical cyanobacterium (blue-green alga) Cylindrospermopsis raciborskii (Woloszynska) Seenaya and Subba Raju isolated from a domestic water supply reservoir. Appl. Environ. Microbiol. 1985, 50, 1292–1295. [Google Scholar] [CrossRef] [Green Version]
- Žegura, B. An overview of the mechanisms of microcystin-LR genotoxicity and potential carcinogenicity. Mini Rev. Med. Chem. 2016, 16, 1042–1062. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa, M.G.; Gutiérrez-Praena, D.; Prieto, A.I.; Guzmán-Guillén, R.; Jos, Á.; Cameán, A.M. Neurotoxicity induced by microcystins and cylindrospermopsin: A review. Sci. Total Environ. 2019, 668, 547–565. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Yi, X.; Liu, W.; Zhang, C.; Massey, I.Y.; Yang, F.; Tian, L. A review of nephrotoxicity of microcystins. Toxins 2020, 12, 693. [Google Scholar] [CrossRef] [PubMed]
- Piyathilaka, M.A.P.C.; Pathmalal, M.M.; Tennekoon, K.H.; de Silva, B.G.D.N.K.; Samarakoon, S.R.; Chanthirika, S. Microcystin-LR-induced cytotoxicity and apoptosis in human embryonic kidney and human kidney adenocarcinoma cell lines. Microbiology 2015, 161, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Prieto, A.I.; Pichardo, S.; Jos, Á.; Moreno, I.; Cameán, A.M. Time-dependent oxidative stress responses after acute exposure to toxic cyanobacterial cells containing microcystins in Tilapia fish (Oreochromis niloticus) under laboratory conditions. Aquat. Toxicol. 2007, 84, 337–345. [Google Scholar] [CrossRef]
- Atencio, L.; Moreno, I.; Prieto, A.I.; Moyano, R.; Molina, A.M.; Cameán, A.M. Acute effects of microcystins MC-LR and MC-RR on acid and alkaline phosphatase activities and pathological changes in intraperitoneally exposed Tilapia fish (Oreochromis sp.). Toxicol. Pathol. 2008, 36, 449–458. [Google Scholar] [CrossRef]
- Froscio, S.M.; Cannon, E.; Lau, H.M.; Humpage, A.R. Limited uptake of the cyanobacterial toxin cylindrospermopsin by Vero cells. Toxicon 2009, 54, 862–868. [Google Scholar] [CrossRef]
- Puerto, M.; Jos, Á.; Pichardo, S.; Gutiérrez-Praena, D.; Cameán, A.M. Acute effects of pure cylindrospermopsin on the activity and transcription of antioxidant enzymes in tilapia (Oreochromis niloticus) exposed by gavage. Ecotoxicology 2011, 20, 1852–1860. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Praena, D.; Jos, Á.; Pichardo, S.; Moyano, R.; Blanco, A.; Monterde, J.G.; Cameán, A.M. Time-dependent histopathological changes induced in Tilapia (Oreochromis niloticus) after acute exposure to pure cylindrospermopsin by oral and intraperitoneal route. Ecotoxicol. Environ. Saf. 2012, 76, 102–113. [Google Scholar] [CrossRef]
- Guzmán-Guillén, R.; Prieto, A.I.; Vasconcelos, V.M.; Cameán, A.M. Cyanobacterium producing cylindrospermopsin cause oxidative stress at environmentally relevant concentrations in sub-chronically exposed tilapia (Oreochromis niloticus). Chemosphere 2013, 90, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Testai, E.; Buratti, F.M.; Funari, E.; Manganelli, M.; Vichi, S.; Arnich, N.; Biré, R.; Fessard, V.; Sialehaamoa, A.A. Review and analysis of occurrence, exposure and toxicity of cyanobacteria toxins in food. EFSA Support. Publ. 2016, 13, 998E. [Google Scholar] [CrossRef]
- Minasyan, A.; Christophoridis, C.; Wilson, A.E.; Zervou, S.-K.; Kaloudis, T.; Hiskia, A. Diversity of cyanobacteria and the presence of cyanotoxins in the epilimnion of Lake Yerevan (Armenia). Toxicon 2018, 150, 28–38. [Google Scholar] [CrossRef] [PubMed]
- León, C.; Boix, C.; Beltrán, E.; Peñuela, G.; López, F.; Sancho, J.V.; Hernández, F. Study of cyanotoxin degradation and evaluation of their transformation products in surface waters by LC-QTOF MS. Chemosphere 2019, 229, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gu, X.; Chen, H.; Mao, Z.; Shen, R.; Zeng, Q.; Ge, Y. Co-occurrence of multiple cyanotoxins and taste-and-odor compounds in the large eutrophic Lake Taihu, China: Dynamics, driving factors, and challenges for risk assessment. Environ. Pollut. 2022, 294, 118594. [Google Scholar] [CrossRef]
- Gutiérrez-Praena, D.; Guzmán-Guillén, R.; Pichardo, S.; Moreno, F.J.; Vasconcelos, V.; Jos, Á.; Cameán, A.M. Cytotoxic and morphological effects of microcystin-LR, cylindrospermopsin, and their combinations on the human hepatic cell line HepG2. Environ. Toxicol. 2019, 34, 240–251. [Google Scholar] [CrossRef]
- Hinojosa, M.G.; Prieto, A.I.; Gutiérrez-Praena, D.; Moreno, F.J.; Cameán, A.M.; Jos, Á. Neurotoxic assessment of microcystin-LR, cylindrospermopsin and their combination on the human neuroblastoma SH-SY5Y cell line. Chemosphere 2019, 224, 751–764. [Google Scholar] [CrossRef]
- Hercog, K.; Maisanaba, S.; Filipic, M.; Jos, Á.; Cameán, A.M.; Žegura, B. Genotoxic potential of the binary mixture of cyanotoxins microcystin-LR and cylindrospermopsin. Chemosphere 2017, 189, 319–329. [Google Scholar] [CrossRef]
- Díez-Quijada, L.; Prieto, A.I.; Puerto, M.; Jos, Á.; Cameán, A.M. In vitro mutagenic and genotoxic assessment of a mixture of the cyanotoxins microcystin-LR and cylindrospermopsin. Toxins 2019, 11, 318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humpage, A.R.; Falconer, I.R. Oral toxicity of the cyanobacterial toxin cylindrospermopsin in male Swiss Albino mice: Determination of no observed adverse effect level for deriving a drinking water guideline value. Environ. Toxicol. 2003, 18, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Hoeger, S.J.; Stemmer, K.; Feurstein, D.J.; Knobeloch, D.; Nussler, A.; Dietrich, D.R. The role of organic anion transporting polypeptides (OATPs/SLCOs) in the toxicity of different microcystin congeners in vitro: A comparison of primary human hepatocytes and OATP-transfected HEK293 cells. Toxicol. Appl. Pharmacol. 2010, 245, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, H.; Cai, Y.; Xie, P.; Xiao, W.; Che, J.; Ji, W.; Zhao, S. Microcystin-LR stabilizes c-myc protein by inhibiting proteinphosphatase 2A in HEK293 cells. Toxicology 2014, 319, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Takumi, S.; Shimono, T.; Ikema, S.; Hotta, Y.; Chigwechokha, P.K.; Sugiyama, Y.; Hashimoto, M.; Furukawa, T.; Komatsu, M. Overexpression of carboxylesterase contributes to the attenuation of cyanotoxin microcystin-LR toxicity. Comp. Biochem. Physiol. C 2017, 194, 22–27. [Google Scholar] [CrossRef]
- Ray, S.D.; Farris, F.F.; Hartman, A.C. Hormesis. In Encyclopedia of Toxicology, 3rd ed.; Wexler, P., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 944–948. [Google Scholar] [CrossRef]
- Li, T.; Huang, P.; Liang, J.; Fu, W.; Guo, Z.; Xu, L. Microcystin-LR (MCLR) induces a compensation of PP2A activity mediated by α4 protein in HEK293 cells. Int. J. Biol. Sci. 2011, 7, 740–752. [Google Scholar] [CrossRef]
- Dias, E.; Andrade, M.; Alverca, E.; Pereira, P.; Batoréu, M.C.C.; Jordan, P.; Silva, M.J. Comparative study of the cytotoxic effect of microcistin-LR and purified extracts from Microcystis aeruginosa on a kidney cell line. Toxicon 2009, 53, 487–495. [Google Scholar] [CrossRef]
- Froscio, S.M.; Fanok, S.; Humpage, A.R. Cytotoxicity screening for the cyanobacterial toxin cylindrospermopsin. J. Toxicol. Environ. Health A 2009, 72, 345–349. [Google Scholar] [CrossRef]
- Moraes, A.C.N.; Freire, D.S.; Habibi, H.; Lowe, J.; Magalhães, V.F. Cylindrospermopsin impairs tubular transport function in kidney cells LLC-PK1. Toxicol Lett. 2021, 344, 26–33. [Google Scholar] [CrossRef]
- Žegura, B.; Gajski, G.; Štraser, A.; Garaj-Vrhovac, V. Cylindrospermopsin induced DNA damage and alteration in the expression of genes involved in the response to DNA damage, apoptosis and oxidative stress. Toxicon 2011, 58, 471–479. [Google Scholar] [CrossRef]
- Štraser, A.; Filipič, M.; Žegura, B. Genotoxic effects of the cyanobacterial hepatotoxin cylindrospermopsin in the HepG2 cell line. Arch. Toxicol. 2011, 85, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Štraser, A.; Filipič, M.; Žegura, B. Cylindrospermopsin induced transcriptional responses in human hepatoma HepG2 cells. Toxicol. Vitro 2013, 27, 1809–1819. [Google Scholar] [CrossRef] [PubMed]
- Hercog, K.; Štampar, M.; Štern, A.; Filipič, M.; Žegura, B. Application of advanced HepG2 3D cell model for studying genotoxic activity of cyanobacterial toxin cylindrospermopsin. Environ. Pollut. 2020, 265, 114965. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, L.; Chen, G.; Wang, M.; Hu, T. Cylindrospermopsin impairs vascular smooth muscle cells by P53-mediated apoptosis due to ROS overproduction. Toxicol. Lett. 2021, 353, 83–92. [Google Scholar] [CrossRef]
- Wang, L.; Chen, G.; Xiao, G.; Han, L.; Wang, Q.; Hu, T. Cylindrospermopsin induces abnormal vascular development through impairing cytoskeleton and promoting vascular endothelial cell apoptosis by the Rho/ROCK signaling pathway. Environ. Res. 2020, 183, 109236. [Google Scholar] [CrossRef]
- Díez-Quijada, L.; Hercog, K.; Štampar, M.; Filipič, M.; Cameán, A.M.; Jos, Á.; Žegura, B. Genotoxic effects of cylindrospermopsin, microcystin-LR and their binary mixture in human hepatocellular carcinoma (HepG2) cell line. Toxins 2020, 12, 778. [Google Scholar] [CrossRef]
- Androutsopoulos, V.P.; Tsatsakis, A.M.; Spandidos, D.A. Cytochrome P450 CYP1A1: Wider roles in cancer progression and prevention. BMC Cancer 2009, 9, 187. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.Y.W.; Zhou, X.; Or, P.M.Y.; Kwan, Y.W.; Yeung, J.H.K. Tanshinone I increases CYP1A2 protein expression and enzyme activity in primary rat hepatocytes. Phytomedicine 2012, 19, 169–176. [Google Scholar] [CrossRef]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef]
- Bain, P.; Shaw, G.; Patel, B. Induction of p53-regulated gene expression in human cell lines exposed to the cyanobacterial toxin cylindrospermopsin. J. Toxicol. Environ. Health A 2007, 70, 1687–1693. [Google Scholar] [CrossRef]
- Michael, D.; Oren, M. The p53 and Mdm2 families in cancer. Curr. Opin. Genet. Dev. 2002, 12, 53–59. [Google Scholar] [CrossRef]
- Pichardo, S.; Cameán, A.M.; Jos, A. In vitro toxicological assessment of cylindrospermopsin: A review. Toxins 2017, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fridovich, I. The biology of oxygen radicals. Science 1978, 201, 875–880. [Google Scholar] [CrossRef] [PubMed]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef] [Green Version]
- Galasso, M.; Gambino, S.; Romanelli, M.G.; Donadelli, M.; Scupoli, M.T. Browsing the oldest antioxidant enzyme: Catalase and its multiple regulation in cancer. Free Radic. Biol. Med. 2021, 172, 264–272. [Google Scholar] [CrossRef]
- Araujo Eleutherio, E.C.; Silva Magalhaes, R.S.; de Araújo Brasil, A.; Monteiro Neto, J.R.; de Holanda Paranhos, L. SOD1, more than just an antioxidant. Arch. Biochem. Biophys. 2021, 697, 108701. [Google Scholar] [CrossRef]
- Stolwijk, J.M.; Falls-Hubert, K.C.; Searby, C.C.; Wagner, B.A.; Buettner, G.R. Simultaneous detection of the enzyme activities of GPx1 and GPx4 guide optimization of selenium in cell biological experiments. Redox Biol. 2020, 32, 101518. [Google Scholar] [CrossRef]
- Araújo, M.J.; Sousa, M.L.; Felpeto, A.B.; Turkina, M.V.; Fonseca, E.; Martins, J.C.; Vasconcelos, V.; Campos, A. Comparison of sample preparation methods for shotgun proteomic studies in aquaculture species. Proteomes 2021, 9, 46. [Google Scholar] [CrossRef]
- Campos, A.; Tedesco, S.; Vasconcelos, V.; Cristobal, S. Proteomic research in bivalves: Towards the identification of molecular markers of aquatic pollution. J. Proteom. 2012, 75, 4346–4359. [Google Scholar] [CrossRef]
- UniProt. Available online: https://www.uniprot.org/uniprot/P16152 (accessed on 19 May 2022).
- Puerto, M.; Campos, A.; Prieto, A.; Cameán, A.; de Almeida, A.M.; Coelho, A.V.; Vasconcelos, V. Differential protein expression in two bivalve species; Mytilus galloprovincialis and Corbicula fluminea; exposed to Cylindrospermopsis raciborskii cells. Aquat. Toxicol. 2011, 101, 109–116. [Google Scholar] [CrossRef]
- Menezes, C.; Valerio, E.; Días, E. The kidney vero-E6 cell line: A suitable model to study the toxicity of microcystins. In New Insights into Toxicity and Drug Testing; Gowder, S., Ed.; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef] [Green Version]
- Fu, W.; Yu, Y.; Xu, L. Identification of temporal differentially expressed protein responses to microcystin in human amniotic epithelial cells. Chem. Res. Toxicol. 2009, 22, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Xie, P.; Chen, J.; Liu, L.; Fan, H. A proteomic study on liver impairment in rat pups induced by maternal microcystin-LR exposure. Environ. Pollut. 2016, 212, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Cai, F.; Yan, W.; Li, C.; Wang, J. A proteomic analysis of MCLR-induced neurotoxicity: Implications for Alzheimer’s disease. Toxicol. Sci. 2012, 127, 485–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Yan, W.; Qiao, Q.; Chen, J.; Cai, F.; He, Y.; Zhang, X. Global effects of subchronic treatment of microcystin-LR on rat splenetic protein levels. J. Proteom. 2012, 77, 383–393. [Google Scholar] [CrossRef]
- Chen, T.; Wang, Q.; Cui, J.; Yang, W.; Shi, Q.; Hua, Z.; Ji, J.; Shen, P. Induction of apoptosis in mouse liver by microcystin-LR: A combined transcriptomic, proteomic, and simulation strategy. Mol. Cell. Proteom. 2005, 4, 958–974. [Google Scholar] [CrossRef] [Green Version]
- Liebel, S.; Regina Grötzner, S.; Dietrich Moura Costa, D.; Antônio Ferreira Randi, M.; Alberto de Oliveira Ribeiro, C.; Filipak Neto, F. Cylindrospermopsin effects on protein profile of HepG2 cells. Toxicol. Mech. Methods 2016, 26, 554–563. [Google Scholar] [CrossRef]
- Pichardo, S.; Jos, Á.; Zurita, J.L.; Salguero, M.; Cameán, A.M.; Repetto, G. The use of the fish cell lines RTG-2 and PLHC-1 to compare the toxic effects produced by microcystins LR and RR. Toxicol. Vitro 2005, 19, 865–873. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pichardo, S.; Jos, A.; Zurita, J.L.; Salguero, M.; Cameán, A.M.; Repetto, G. Acute and subacute toxic effects produced by microcystin-YR on the fish cell lines RTG-2 and PLHC-1. Toxicol. Vitro 2007, 21, 1460–1467. [Google Scholar] [CrossRef]
- Baltrop, J.A.; Owen, T.C.; Cory, A.H.; Cory, J.G. 5-((3-carboxyphenyl)-3-(4,5-dimethylthiazolyl)-3-(4-sulfophenyl)) tetrazolium, inner salt (MTS) and related analogs of 2-(4,5-dimethylthiazolyl)-2,5-diphenylterazolium bromide (MTT) reducing to purple water soluble formazan as cell-viability indicators. Bioorg. Med. Chem. Lett. 1991, 1, 611–614. [Google Scholar] [CrossRef]
- Maisanaba, S.; Hercog, K.; Filipic, M.; Jos, A.; Zegura, B. Genotoxic potential of montmorillonite clay mineral and alteration in the expression of genes involved in toxicity mechanisms in the human hepatoma cell line HepG2. J. Hazard. Mater. 2016, 304, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Danielsson, G.; Farinha, A.P.; Kuruvilla, J.; Warholm, P.; Cristobal, S. Shotgun proteomics to unravel marine mussel (Mytilus edulis) response to long-term exposure to low salinity and propranolol in a Baltic Sea microcosm. J. Proteom. 2016, 137, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Pérez, D.; Rodríguez, A.A.; Osorio, H.; Azevedo, J.; Castañeda, O.; Vasconcelos, V.; Antunes, A. Microcystin-LR detected in a low molecular weight fraction from a crude extract of Zoanthus sociatus. Toxins 2017, 9, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- String. Available online: https://string-db.org (accessed on 19 May 2022).

| Mechanisms Involved | Gene Symbol | CYN (µg/mL) | 4 h | 24 h | Entrez Gene Name |
|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | ||||
| Xenobiotic Metabolism | CYP1A1 | C- | 1.13 ± 0.67 | 1.03 ± 0.33 | Cytochrome P450 family 1 Subfamily A member 1 |
| 0.5 | 1.11 ± 0.11 | 1.06 ± 0.35 | |||
| 5 | 1.99 ± 0.05 ** | 24.21 ± 11.92 *** | |||
| B(a)P 30 µM | 1.03 ± 0.05 | 2.16 ± 0.33 | |||
| CYP1A2 | C- | 1.01 ± 0.19 | 1.02 ± 0.25 | Cytochrome P450 family 1 Subfamily A member 2 | |
| 0.5 | 0.90 ± 0.50 | 0.68 ± 0.16 | |||
| 5 | 0.87 ± 0.06 | 11.33 ± 0.17 * | |||
| B(a)P 30 µM | 1.23 ± 0.40 | 1.23 ± 0.18 | |||
| DNA damage responsive | TP53 | C- | 1.05 ± 0.19 | 1.00 ± 0.12 | Tumor protein P53 |
| 0.5 | 0.86 ± 0.21 | 1.14 ± 0.25 | |||
| 5 | 0.98 ± 0.23 | 3.38 ± 0.31 ** | |||
| B(a)P 30 µM | 0.9 ± 0.20 | 1.22 ± 0.21 | |||
| CDKN1A | C- | 1.03± 0.30 | 1.01 ± 0.17 | Cyclin Dependent Kinase Inhibitor 1A | |
| 0.5 | 0.72 ± 0.14 | 1.73 ± 0.40 | |||
| 5 | 1.01 ± 0.56 | 10.57 ± 1.82 * | |||
| B(a)P 30 µM | 0.88 ± 0.29 | 1.06 ± 0.68 | |||
| Oxidative stress | SOD1 | C- | 1.08 ± 0.47 | 1.01 ± 0.20 | Superoxide dismutase 1 |
| 0.5 | 1.08 ± 0.28 | 1.30 ± 0.25 | |||
| 5 | 2.04 ± 1.44 * | 1.10 ± 0.68 | |||
| B(a)P 30 µM | 1.04 ± 0.13 | 1.05 ± 0.21 | |||
| CAT | C- | 1.08 ± 0.48 | 1.00 ± 0.13 | Catalase | |
| 0.5 | 1.41 ± 0.52 | 1.45 ± 0.26 | |||
| 5 | 0.67 ± 0.83 | 2.96 ± 1.35 *** | |||
| B(a)P 30 µM | 1.21 ± 0.11 | 1.17 ± 0.10 | |||
| GPX1 | C- | 1.05 ± 0.43 | 1.05 ± 0.39 | Glutathione peroxidase 1 | |
| 0.5 | 1.04 ± 0.22 | 2.07 ± 0.46 | |||
| 5 | 0.89 ± 0.27 | 7.94 ± 2.35 *** | |||
| B(a)P 30 µM | 1.22 ± 0.18 | 1.36 ± 0.58 | |||
| Apoptosis/survival | BAX | C- | 1.02 ± 0.22 | 1.01 ± 0.14 | Apoptosis regulator BAX. |
| 0.5 | 0.74 ± 0.17 | 1.30 ± 0.25 | |||
| 5 | 0.99 ± 0.30 | 0.92 ± 0.22 | |||
| B(a)P 30 µM | 0.87 ± 0.10 | 1.14 ± 0.24 | |||
| BCL2 | C- | 1.01 ± 0.15 | 1.01 ± 0.16 | B-cell CLL/lymphoma 2 | |
| 0.5 | 0.78 ± 0.21 | 1.33 ± 0.26 * | |||
| 5 | 1.07 ± 0.02 | 3.85 ± 0.19 ** | |||
| B(a)P 30 µM | 0.79 ± 0.01 | 0.70 ± 0.13 * |
| AU | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Functional Category | Protein Name | Gene | C | 0.5 CYN | 1 CYN | 1 MC-LR | 0.5CYN + 1MCLR | 1CYN + 1MC-LR | ||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |||
| Cellular metabolism | Carbonyl reductase [NADPH] 1 | CBR1 | 2.3 × 10−4 | 2.6 × 10−4 | 4.7 × 10−4 | 3.7 × 10−4 | 9.0 × 10−4 | 5.3 × 10−4 | 9.0 × 10−4 | 3.0 × 10−4 | 8.7 × 10−4 | 1.7 × 10−4 | 1.6 × 10−3 ** | 4.5 × 10−4 |
| Phosphoglucomutase-2 | PGM2 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 9.0 × 10−5 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | 2.8 × 10−4 ** | 1.7 × 10−4 | 1.0 × 10−8 | 0 | |
| Lipid metabolism | Prosaposin | PSAP | 5.3 × 10−4 | 1.4 × 10−4 | 4.3 × 10−4 | 2.6 × 10−4 | 1.0 × 10−8 ** | 1.0 × 10−4 | 2.4 × 10−4 | 8.7 × 10−5 | 3.1 × 10−4 | 1.4 × 10−4 | 2.4 × 10−4 * | 1.4 × 10−4 |
| 3-ketoacyl-CoA thiolase, mitochondrial | ACAA2 | 6.4 × 10−4 | 2.7 × 10−4 | 5.7 × 10−4 | 3.9 × 10−4 | 1.0 × 10−8 * | 1.3 × 10−4 | 4.3 × 10−4 | 2.7 × 10−4 | 1.4 × 10−4 | 2.7 × 10−4 | 1.0 × 10−8 * | 0 | |
| Protein | Moesin | MSN | 9.9 × 10−4 | 2.9 × 10−4 | 7.8 × 10−4 | 2.8 × 10−4 | 4.3 × 10−4 * | 2.6 × 10−4 | 5.9 × 10−4 | 8.0 × 10−5 | 4.1 × 10−4 * | 1.5 × 10−4 | 1.5 × 10−4 ** | 2.1 × 10−4 |
| Cell adhesion | Integrin beta-1 | ITGB1 | 2.5 × 10−4 | 8.1 × 10−5 | 1.0 × 10−8 * | 7.9 × 10−5 | 1.0 × 10−8 * | 6.6 × 10−5 | 1.0 × 10−8 * | 7.4 × 10−5 | 1.0 × 10−8 ** | 0 | 1.0 × 10−8 ** | 0 |
| Fermitin family homolog 2 | FERMT2 | 2.9 × 10−4 | 1.0 × 10−4 | 3.3 × 10−4 | 1.7 × 10−4 | 1.0 × 10−8 | 7.7 × 10−5 | 8.6 × 10−5 | 1.0 × 10−4 | 1.7 × 10−4 | 1.3 × 10−4 | 1.0 × 10−8 * | 0 | |
| Protein Metabolism | Bleomycin hydrolase | BLMH | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 2.4 × 10−4 | 4.8 × 10−4 * | 2.4 × 10−4 | 1.0 × 10−8 | 0 |
| Protein regulation | E3 SUMO-protein ligase RanBP2 | RANBP2 | 4.1 × 10−5 | 3.2 × 10−5 | 3.7 × 10−5 | 2.1 × 10−5 | 1.2 × 10−4 * | 6.8 × 10−5 | 8.8 × 10−5 | 3.7 × 10−5 | 6.9 × 10−5 | 4.5 × 10−5 | 1.4 × 10−4 | 3.8 × 10−4 |
| E3 ubiquitin-protein ligase CHIP | STUB1 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | 1.7 × 10−4 | 2.5 × 10−4 | 1.0 × 10−8 | 1.8 × 10−4 | 4.0 × 10−4 ** | 1.7 × 10−4 | 4.1 × 10−4 | 2.4 × 10−4 | |
| Protein SGT1 homolog | SUGT1 | 4.3 × 10−4 | 1.8 × 10−4 | 3.9 × 10−4 | 3.1 × 10−4 | 1.0 × 10−8 * | 0 | 1.0 × 10−8 | 1.8 × 10−4 | 3.6 × 10−4 | 2.7 × 10−4 | 1.0 × 10−8 * | 0 | |
| ATP-dependent Clp protease proteolytic subunit, mitochondrial | CLPP | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | 4.3 × 10−4 ** | 3.6 × 10−4 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | |
| Protein synthesis | Translation initiation factor eIF-2B subunit alpha | EIF2B1 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | 1.0 × 10−8 | 0 | 3.6 × 10−4 * | 1.8 × 10−4 | 1.0 × 10−8 | 2.0 × 10−4 |
| Treacle protein | TCOF1 | 4.0 × 10−5 | 8.0 × 10−5 | 1.0 × 10−4 | 1.0 × 10−4 | 1.0 × 10−4 | 5.0 × 10−5 | 2.0 × 10−4 | 1.0 × 10−4 | 3.0 × 10−4 | 6.2 × 10−5 | 3.0 × 10−4 * | 9.3 × 10−5 | |
| 40S ribosomal protein S5 | RPS5 | 7.0 × 10−4 | 3.0 × 10−4 | 7.0 × 10−4 | 3.0 × 10−4 | 1.0 × 10−8 | 3.0 × 10−4 | 1.0 × 10−8 | 3.0 × 10−4 | 5.0 × 10−4 | 3.0 × 10−4 | 1.0 × 10−8 * | 0 | |
| Cisplatin resistance-associated overexpressed protein, isoform CRA_b | LUC7L3 | 1.0 × 10−8 | 1.3 × 10−4 | 1.5 × 10−4 | 2.5 × 10−4 | 4.1 × 10−4 ** | 1.2 × 10−4 | 2.5 × 10−4 | 2.0 × 10−5 | 2.5 × 10−4 | 1.0 × 10−4 | 3.0 × 10−4 | 5.2 × 10−5 | |
| Protein transport | Coatomer subunit gamma-1 | COPG1 | 7.0 × 10−4 | 5.1 × 10−5 | 3.2 × 10−4 * | 1.9 × 10−4 | 1.8 × 10−4 * | 2.5 × 10−4 | 2.2 × 10−4 * | 1.8 × 10−4 | 6.3 × 10−4 | 1.6 × 10−4 | 1.7 × 10−4 ** | 1.8 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diez-Quijada, L.; Puerto, M.; Gutiérrez-Praena, D.; Turkina, M.V.; Campos, A.; Vasconcelos, V.; Cameán, A.M.; Jos, Á. In Vitro Toxicity Evaluation of Cyanotoxins Cylindrospermopsin and Microcystin-LR on Human Kidney HEK293 Cells. Toxins 2022, 14, 429. https://doi.org/10.3390/toxins14070429
Diez-Quijada L, Puerto M, Gutiérrez-Praena D, Turkina MV, Campos A, Vasconcelos V, Cameán AM, Jos Á. In Vitro Toxicity Evaluation of Cyanotoxins Cylindrospermopsin and Microcystin-LR on Human Kidney HEK293 Cells. Toxins. 2022; 14(7):429. https://doi.org/10.3390/toxins14070429
Chicago/Turabian StyleDiez-Quijada, Leticia, María Puerto, Daniel Gutiérrez-Praena, Maria V. Turkina, Alexandre Campos, Vitor Vasconcelos, Ana M. Cameán, and Ángeles Jos. 2022. "In Vitro Toxicity Evaluation of Cyanotoxins Cylindrospermopsin and Microcystin-LR on Human Kidney HEK293 Cells" Toxins 14, no. 7: 429. https://doi.org/10.3390/toxins14070429





