Temporal Exposure to Bt Insecticide Causes Oxidative Stress in Larval Midgut Tissue
Abstract
:1. Introduction
2. Results
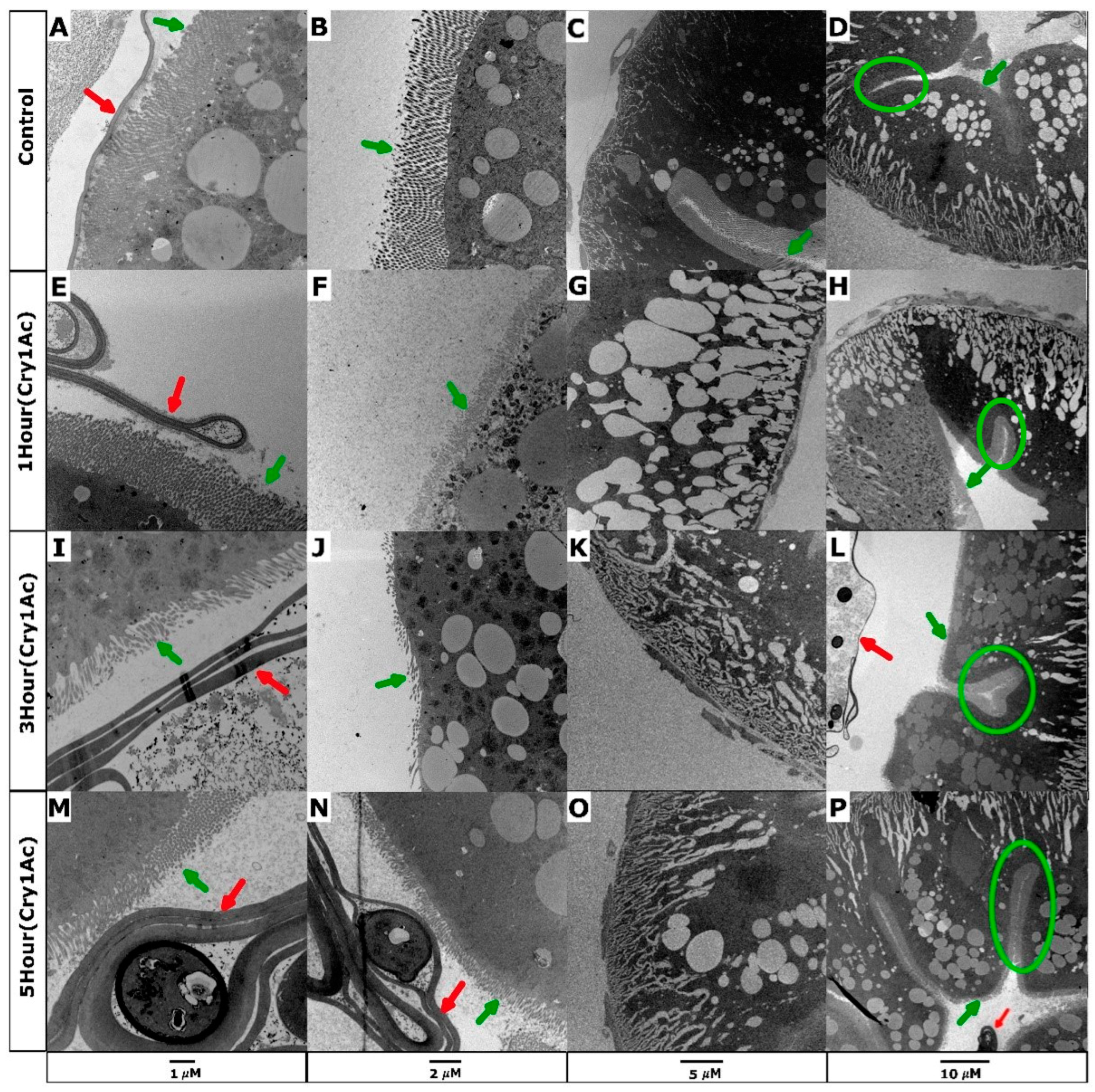
2.1. Cry1Ac Damages D. melanogaster Larval Midgut Epithelial Cells Expressing PxABCC2
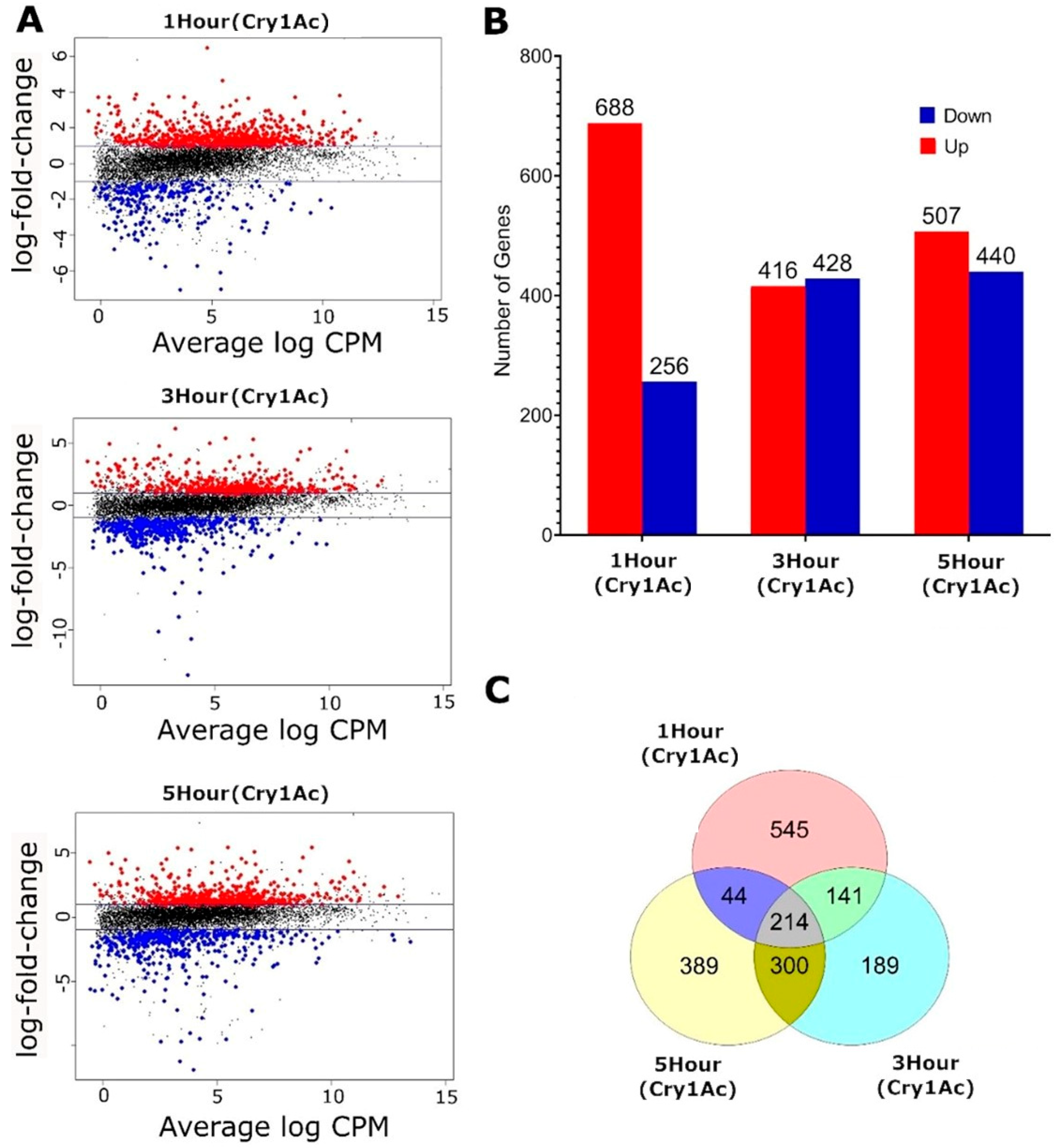
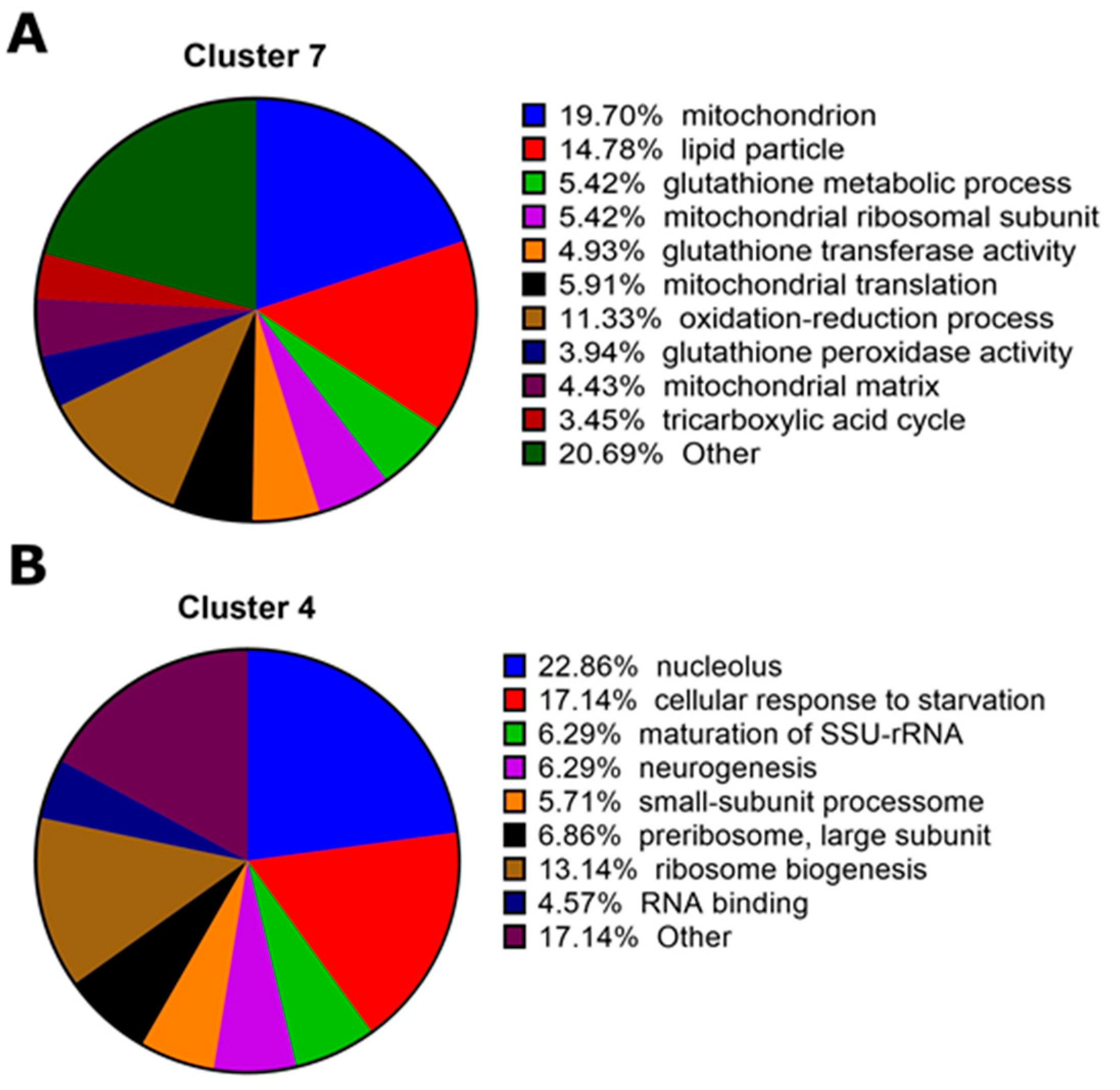
2.2. Transcriptomic Responses to Cry1Ac in the D. melanogaster Larval Midgut
2.3. qPCR Validation of 10 D. melanogaster Genes
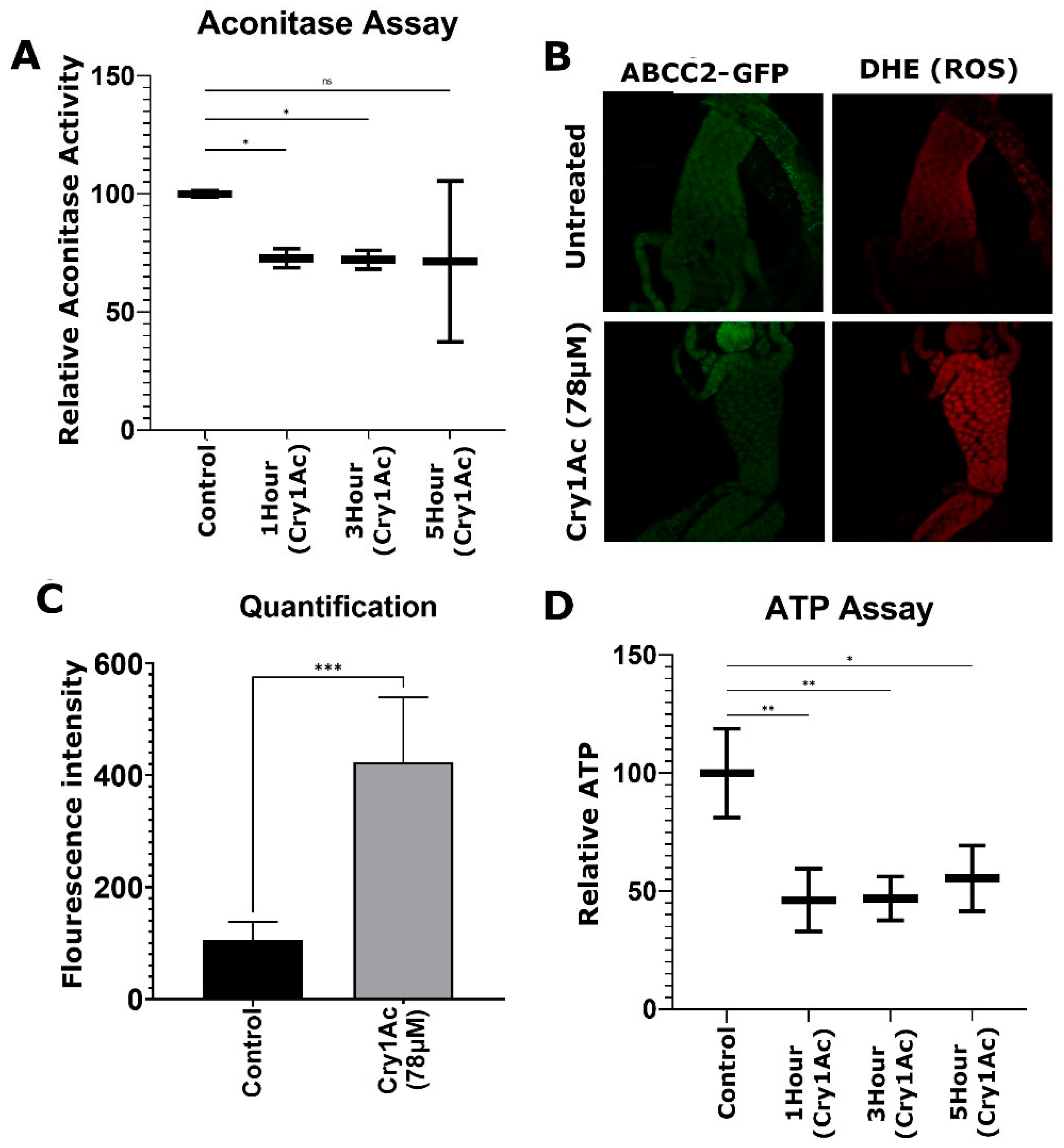
2.4. Cry1Ac Increased ROS Production in D. melanogaster Midgut Tissue
3. Discussion
4. Materials and Methods
4.1. Insect Strains and Rearing
4.2. Cry1Ac Production and Purification
4.3. Diet Preparation
4.4. Midgut Dissection
4.5. RNA Extraction
4.6. Transmission Electron Microscopy
4.7. ImageJ Quantification
4.8. RNA-Seq Library Preparation and Sequencing
4.9. Sequence Read Alignment
4.10. Differential Gene Expression Analysis
4.11. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
4.12. Analysis of Gene Co-Expression
4.13. cDNA Synthesis and qPCR
4.14. ROS Staining
4.15. ATP and Aconitase Activity Assays
4.16. Statistical Analysis and Imaging
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caccia, S.; Casartelli, M.; Tettamanti, G. The amazing complexity of insect midgut cells: Types, peculiarities, and functions. Cell Tissue Res. 2019, 377, 505–525. [Google Scholar] [CrossRef]
- Cognigni, P.; Bailey, A.P.; Miguel-Aliaga, I. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 2011, 13, 92–104. [Google Scholar] [CrossRef]
- Shanbhag, S.; Tripathi, S. Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J. Exp. Biol. 2009, 212, 1731–1744. [Google Scholar] [CrossRef]
- Terra, W.R. Evolution of digestive systems of insects. Annu. Rev. Entomol. 1990, 35, 181–200. [Google Scholar] [CrossRef]
- Buchon, N.; Osman, D.; David, F.P.; Fang, H.Y.; Boquete, J.P.; Deplancke, B.; Lemaitre, B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013, 3, 1725–1738. [Google Scholar] [CrossRef] [PubMed]
- Hofte, H.; Whiteley, H.R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 1989, 53, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Crickmore, N.; Berry, C.; Panneerselvam, S.; Mishra, R.; Connor, T.R.; Bonning, B.C. A structure-based nomenclature for Bacillus thuringiensis and other bacteria-derived pesticidal proteins. J. Invertebr. Pathol. 2021, 186, 107438. [Google Scholar] [CrossRef] [PubMed]
- Shelton, A.M.; Hossain, M.J.; Paranjape, V.; Azad, A.K.; Rahman, M.L.; Khan, A.; Prodhan, M.Z.H.; Rashid, M.A.; Majumder, R.; Hossain, M.A.; et al. Bt Eggplant Project in Bangladesh: History, Present Status, and Future Direction. Front. Bioeng. Biotechnol. 2018, 6, 106. [Google Scholar] [CrossRef]
- Hautea, D.M.; Taylo, L.D.; Masanga, A.P.; Sison, M.L.; Narciso, J.O.; Quilloy, R.B.; Hautea, R.A.; Shotkoski, F.A.; Shelton, A.M. Field Performance of Bt Eggplants (Solanum melongena L.) in the Philippines: Cry1Ac Expression and Control of the Eggplant Fruit and Shoot Borer (Leucinodes orbonalis Guenee). PLoS ONE 2016, 11, e0157498. [Google Scholar] [CrossRef]
- Gassmann, A.J.; Petzold-Maxwell, J.L.; Clifton, E.H.; Dunbar, M.W.; Hoffmann, A.M.; Ingber, D.A.; Keweshan, R.S. Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proc. Natl. Acad. Sci. USA 2014, 111, 5141–5146. [Google Scholar] [CrossRef]
- Tabashnik, B.E.; Carriere, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 2017, 35, 926–935. [Google Scholar] [CrossRef]
- Knight, P.J.; Knowles, B.H.; Ellar, D.J. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J. Biol. Chem. 1995, 270, 17765–17770. [Google Scholar] [CrossRef]
- Nagamatsu, Y.; Koike, T.; Sasaki, K.; Yoshimoto, A.; Furukawa, Y. The cadherin-like protein is essential to specificity determination and cytotoxic action of the Bacillus thuringiensis insecticidal CryIAa toxin. FEBS Lett. 1999, 460, 385–390. [Google Scholar] [CrossRef]
- Perera, O.P.; Willis, J.D.; Adang, M.J.; Jurat-Fuentes, J.L. Cloning and characterization of the Cry1Ac-binding alkaline phosphatase (HvALP) from Heliothis virescens. Insect Biochem. Mol. Biol. 2009, 39, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Gahan, L.J.; Pauchet, Y.; Vogel, H.; Heckel, D.G. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010, 6, e1001248. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Wang, J.; Guan, F.; Zhang, J.; Yu, S.; Liu, S.; Xue, Y.; Li, L.; Wu, S.; Wang, X.; et al. Dominant point mutation in a tetraspanin gene associated with field-evolved resistance of cotton bollworm to transgenic Bt cotton. Proc. Natl. Acad. Sci. USA 2018, 115, 11760–11765. [Google Scholar] [CrossRef]
- Guo, Z.; Sun, D.; Kang, S.; Zhou, J.; Gong, L.; Qin, J.; Guo, L.; Zhu, L.; Bai, Y.; Luo, L.; et al. CRISPR/Cas9-mediated knockout of both the PxABCC2 and PxABCC3 genes confers high-level resistance to Bacillus thuringiensis Cry1Ac toxin in the diamondback moth, Plutella xylostella (L.). Insect Biochem. Mol. Biol. 2019, 107, 31–38. [Google Scholar] [CrossRef]
- Liu, Z.; Fu, S.; Ma, X.; Baxter, S.W.; Vasseur, L.; Xiong, L.; Huang, Y.; Yang, G.; You, S.; You, M. Resistance to Bacillus thuringiensis Cry1Ac toxin requires mutations in two Plutella xylostella ATP-binding cassette transporter paralogs. PLoS Pathog. 2020, 16, e1008697. [Google Scholar] [CrossRef] [PubMed]
- Heckel, D.G. How do toxins from Bacillus thuringiensis kill insects? An evolutionary perspective. Arch. Insect Biochem. Physiol. 2020, 104, e21673. [Google Scholar] [CrossRef]
- Bretschneider, A.; Heckel, D.G.; Pauchet, Y. Three toxins, two receptors, one mechanism: Mode of action of Cry1A toxins from Bacillus thuringiensis in Heliothis virescens. Insect Biochem. Mol. Biol. 2016, 76, 109–117. [Google Scholar] [CrossRef]
- Tanaka, S.; Miyamoto, K.; Noda, H.; Jurat-Fuentes, J.L.; Yoshizawa, Y.; Endo, H.; Sato, R. The ATP-binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis. FEBS J. 2013, 280, 1782–1794. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhu, L.; Guo, L.; Wang, S.; Wu, Q.; Crickmore, N.; Zhou, X.; Bravo, A.; Soberon, M.; Guo, Z.; et al. A versatile contribution of both aminopeptidases N and ABC transporters to Bt Cry1Ac toxicity in the diamondback moth. BMC Biol. 2022, 20, 33. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Gomez, I.; Conde, J.; Munoz-Garay, C.; Sanchez, J.; Miranda, R.; Zhuang, M.; Gill, S.S.; Soberon, M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim. Biophys. Acta 2004, 1667, 38–46. [Google Scholar] [CrossRef]
- Guihard, G.; Vachon, V.; Laprade, R.; Schwartz, J.L. Kinetic properties of the channels formed by the Bacillus thuringiensis insecticidal crystal protein Cry1C in the plasma membrane of Sf9 cells. J. Membr. Biol. 2000, 175, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Vachon, V.; Paradis, M.J.; Marsolais, M.; Schwartz, J.L.; Laprade, R. Ionic permeabilities induced by Bacillus thuringiensis in Sf9 cells. J. Membr. Biol. 1995, 148, 57–63. [Google Scholar] [CrossRef]
- Kwa, M.S.; de Maagd, R.A.; Stiekema, W.J.; Vlak, J.M.; Bosch, D. Toxicity and binding properties of the Bacillus thuringiensis delta-endotoxin Cry1C to cultured insect cells. J. Invertebr. Pathol. 1998, 71, 121–127. [Google Scholar] [CrossRef]
- Endo, H.; Adegawa, S.; Kikuta, S.; Sato, R. The intracellular region of silkworm cadherin-like protein is not necessary to mediate the toxicity of Bacillus thuringiensis Cry1Aa and Cry1Ab toxins. Insect Biochem. Mol. Biol. 2018, 94, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; James, B.; Chakroun, M.; Ferre, J.; Downes, S. Isolating, characterising and identifying a Cry1Ac resistance mutation in field populations of Helicoverpa punctigera. Sci. Rep. 2018, 8, 2626. [Google Scholar] [CrossRef]
- Stevens, T.; Song, S.; Bruning, J.B.; Choo, A.; Baxter, S.W. Expressing a moth abcc2 gene in transgenic Drosophila causes susceptibility to Bt Cry1Ac without requiring a cadherin-like protein receptor. Insect Biochem. Mol. Biol. 2017, 80, 61–70. [Google Scholar] [CrossRef]
- Zhang, X.; Candas, M.; Griko, N.B.; Taussig, R.; Bulla, L.A., Jr. A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc. Natl. Acad. Sci. USA 2006, 103, 9897–9902. [Google Scholar] [CrossRef]
- Furlong, M.J.; Wright, D.J.; Dosdall, L.M. Diamondback moth ecology and management: Problems, progress, and prospects. Annu. Rev. Entomol. 2013, 58, 517–541. [Google Scholar] [CrossRef]
- Baxter, S.W.; Badenes-Perez, F.R.; Morrison, A.; Vogel, H.; Crickmore, N.; Kain, W.; Wang, P.; Heckel, D.G.; Jiggins, C.D. Parallel evolution of Bacillus thuringiensis toxin resistance in lepidoptera. Genetics 2011, 189, 675–679. [Google Scholar] [CrossRef] [PubMed]
- Kuraishi, T.; Binggeli, O.; Opota, O.; Buchon, N.; Lemaitre, B. Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2011, 108, 15966–15971. [Google Scholar] [CrossRef]
- Kleino, A.; Silverman, N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 2014, 42, 25–35. [Google Scholar] [CrossRef]
- Abu-Jamous, B.; Kelly, S. Clust: Automatic extraction of optimal co-expressed gene clusters from gene expression data. Genome Biol. 2018, 19, 172. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef]
- Schwartz, J.-L.; Laprade, R. Membrane permeabilisation by Bacillus thuringiensis toxins: Protein insertion and pore formation. In Entomopathogenic Bacteria: From Laboratory to Field Application; Charles, J.-F., Delécluse, A., Roux, C.N.-L., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 199–217. [Google Scholar]
- Schwartz, J.L.; Garneau, L.; Masson, L.; Brousseau, R. Early response of cultured lepidopteran cells to exposure to delta-endotoxin from Bacillus thuringiensis: Involvement of calcium and anionic channels. Biochim. Biophys. Acta 1991, 1065, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Contreras, L.; Drago, I.; Zampese, E.; Pozzan, T. Mitochondria: The calcium connection. Biochim. Biophys. Acta 2010, 1797, 607–618. [Google Scholar] [CrossRef]
- Martelli, F.; Zhongyuan, Z.; Wang, J.; Wong, C.O.; Karagas, N.E.; Roessner, U.; Rupasinghe, T.; Venkatachalam, K.; Perry, T.; Bellen, H.J.; et al. Low doses of the neonicotinoid insecticide imidacloprid induce ROS triggering neurological and metabolic impairments in Drosophila. Proc. Natl. Acad. Sci. USA 2020, 117, 25840–25850. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.J.; Levine, R.L.; Sohal, R.S. Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. Sci. USA 1997, 94, 11168–11172. [Google Scholar] [CrossRef] [PubMed]
- Markow, T.A. The secret lives of Drosophila flies. eLife 2015, 4, e06793. [Google Scholar] [CrossRef]
- Thomas, W.E.; Ellar, D.J. Bacillus thuringiensis var israelensis crystal delta-endotoxin: Effects on insect and mammalian cells in vitro and in vivo. J. Cell Sci. 1983, 60, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Gringorten, J.L.; Sohi, S.S.; Masson, L. Activity spectra of Bacillus thuringiensis delta-endotoxins against eight insect cell lines. In Vitro Cell Dev. Biol. Anim. 1999, 35, 299–303. [Google Scholar] [CrossRef]
- Nawrot-Esposito, M.P.; Babin, A.; Pasco, M.; Poirie, M.; Gatti, J.L.; Gallet, A. Bacillus thuringiensis Bioinsecticides Induce Developmental Defects in Non-Target Drosophila melanogaster Larvae. Insects 2020, 11, 697. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.D.; Tadei, W.P.; Abdalla, F.C. Occurrence of apocrine secretion in the larval gut epithelial cells of Aedes aegypti L., Anopheles albitarsis Lynch-Arribalzaga and Culex quinquefasciatus say (Diptera: Culicidae): A defense strategy against infection by Bacillus sphaericus Neide? Neotrop. Entomol. 2009, 38, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Vodovar, N.; Vinals, M.; Liehl, P.; Basset, A.; Degrouard, J.; Spellman, P.; Boccard, F.; Lemaitre, B. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. USA 2005, 102, 11414–11419. [Google Scholar] [CrossRef]
- Opota, O.; Vallet-Gely, I.; Vincentelli, R.; Kellenberger, C.; Iacovache, I.; Gonzalez, M.R.; Roussel, A.; van der Goot, F.G.; Lemaitre, B. Monalysin, a novel ss-pore-forming toxin from the Drosophila pathogen Pseudomonas entomophila, contributes to host intestinal damage and lethality. PLoS Pathog. 2011, 7, e1002259. [Google Scholar] [CrossRef]
- Lee, K.Z.; Lestradet, M.; Socha, C.; Schirmeier, S.; Schmitz, A.; Spenle, C.; Lefebvre, O.; Keime, C.; Yamba, W.M.; Bou Aoun, R.; et al. Enterocyte Purge and Rapid Recovery Is a Resilience Reaction of the Gut Epithelium to Pore-Forming Toxin Attack. Cell Host Microbe 2016, 20, 716–730. [Google Scholar] [CrossRef]
- Oppert, B.; Martynov, A.G.; Elpidina, E.N. Bacillus thuringiensis Cry3Aa protoxin intoxication of Tenebrio molitor induces widespread changes in the expression of serine peptidase transcripts. Comp. Biochem. Physiol. Part D Genom. Proteom. 2012, 7, 233–242. [Google Scholar] [CrossRef]
- Li, S.; Xu, X.; Shakeel, M.; Xu, J.; Zheng, Z.; Zheng, J.; Yu, X.; Zhao, Q.; Jin, F. Bacillus thuringiensis Suppresses the Humoral Immune System to Overcome Defense Mechanism of Plutella xylostella. Front. Physiol. 2018, 9, 1478. [Google Scholar] [CrossRef]
- Tamez-Guerra, P.; Valadez-Lira, J.A.; Alcocer-Gonzalez, J.M.; Oppert, B.; Gomez-Flores, R.; Tamez-Guerra, R.; Rodriguez-Padilla, C. Detection of genes encoding antimicrobial peptides in Mexican strains of Trichoplusia ni (Hubner) exposed to Bacillus thuringiensis. J. Invertebr. Pathol. 2008, 98, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.; Sanko, T.J.; Bezuidenhout, C.C.; van den Berg, J. Transcriptome and differentially expressed genes of Busseola fusca (Lepidoptera: Noctuidae) larvae challenged with Cry1Ab toxin. Gene 2019, 710, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Sparks, M.E.; Blackburn, M.B.; Kuhar, D.; Gundersen-Rindal, D.E. Transcriptome of the Lymantria dispar (gypsy moth) larval midgut in response to infection by Bacillus thuringiensis. PLoS ONE 2013, 8, e61190. [Google Scholar] [CrossRef]
- Hillyer, J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016, 58, 102–118. [Google Scholar] [CrossRef] [PubMed]
- Obata, F.; Tanaka, S.; Kashio, S.; Tsujimura, H.; Sato, R.; Miura, M. Induction of rapid and selective cell necrosis in Drosophila using Bacillus thuringiensis Cry toxin and its silkworm receptor. BMC Biol. 2015, 13, 48. [Google Scholar] [CrossRef] [PubMed]
- Cancino-Rodezno, A.; Alexander, C.; Villasenor, R.; Pacheco, S.; Porta, H.; Pauchet, Y.; Soberon, M.; Gill, S.S.; Bravo, A. The mitogen-activated protein kinase p38 is involved in insect defense against Cry toxins from Bacillus thuringiensis. Insect Biochem. Mol. Biol. 2010, 40, 58–63. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Poidevin, M.; Lemaitre, B. The Drosophila MAPK p38c regulates oxidative stress and lipid homeostasis in the intestine. PLoS Genet. 2014, 10, e1004659. [Google Scholar] [CrossRef]
- Ferraboschi, P.; Ciceri, S.; Grisenti, P. Applications of Lysozyme, an Innate Immune Defense Factor, as an Alternative Antibiotic. Antibiotics 2021, 10, 1534. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; St Leger, R.J.; Wu, L.P. Fungal peptide Destruxin A plays a specific role in suppressing the innate immune response in Drosophila melanogaster. J. Biol. Chem. 2007, 282, 8969–8977. [Google Scholar] [CrossRef] [PubMed]
- Broderick, N.A.; Raffa, K.F.; Handelsman, J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc. Natl. Acad. Sci. USA 2006, 103, 15196–15199. [Google Scholar] [CrossRef] [PubMed]
- Raymond, B.; Johnston, P.R.; Wright, D.J.; Ellis, R.J.; Crickmore, N.; Bonsall, M.B. A mid-gut microbiota is not required for the pathogenicity of Bacillus thuringiensis to diamondback moth larvae. Environ. Microbiol. 2009, 11, 2556–2563. [Google Scholar] [CrossRef]
- Johnston, P.R.; Crickmore, N. Gut bacteria are not required for the insecticidal activity of Bacillus thuringiensis toward the tobacco hornworm, Manduca sexta. Appl. Environ. Microbiol. 2009, 75, 5094–5099. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, X.; De Mandal, S.; Shakeel, M.; Hua, Y.; Shoukat, R.F.; Fu, D.; Jin, F. Gut microbiota mediate Plutella xylostella susceptibility to Bt Cry1Ac protoxin is associated with host immune response. Environ. Pollut. 2021, 271, 116271. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, W.; Zhang, Q.; Zhang, X.; Zhang, Z. Microbiota and transcriptome changes of Culex pipiens pallens larvae exposed to Bacillus thuringiensis israelensis. Sci. Rep. 2021, 11, 20241. [Google Scholar] [CrossRef]
- Sutherland, P.W.; Harris, M.O.; Markwick, N.P. Effects of Starvation and the Bacillus thuringiensis Endotoxin Cry1Ac on the Midgut Cells, Feeding Behavior, and Growth of Lightbrown Apple Moth Larvae. Ann. Entomol. Soc. Am. 2003, 96, 250–264. [Google Scholar] [CrossRef]
- Ribeiro, L.M.S.; Wanderley-Teixeira, V.; Siqueira, H.Á.A.d.; Oliveira, A.; Lemos, A.; Teixeira, Á.A.C. Midgut histopathology of resistant and susceptible Plutella xylostella exposed to commercial formulations of Bacillus thuringiensis. Bull. Insectology 2013, 66, 161–171. [Google Scholar]
- Brookes, P.S.; Yoon, Y.; Robotham, J.L.; Anders, M.W.; Sheu, S.S. Calcium, ATP, and ROS: A mitochondrial love-hate triangle. Am. J. Physiol. Cell Physiol. 2004, 287, C817–C833. [Google Scholar] [CrossRef]
- Salminen, T.S.; Vale, P.F. Drosophila as a Model System to Investigate the Effects of Mitochondrial Variation on Innate Immunity. Front. Immunol. 2020, 11, 521. [Google Scholar] [CrossRef]
- Inoue, M.; Sato, E.F.; Nishikawa, M.; Park, A.M.; Kira, Y.; Imada, I.; Utsumi, K. Mitochondrial generation of reactive oxygen species and its role in aerobic life. Curr. Med. Chem. 2003, 10, 2495–2505. [Google Scholar] [CrossRef]
- Arai, R.; Ueda, H.; Kitayama, A.; Kamiya, N.; Nagamune, T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001, 14, 529–532. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]



| 1 H | 3 H | 5 H | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FlyBase ID Symbol | logFC | FDR | logFC | FDR | logFC | FDR | |||||
| IMD | FBgn0038928 | Fadd | ns | −0.049 | 1.000 | ns | 0.174 | 1.000 | ∧ | 1.433 | 0.004 |
| FBgn0014018 | Rel | ∧ | 2.377 | 0.001 | ∧ | 1.208 | 0.048 | ns | 0.703 | 0.443 | |
| FBgn0010303 | hep | ns | 0.029 | 1.000 | ns | 1.596 | 0.033 | ns | 0.959 | 0.240 | |
| FBgn0001297 | kay | ∧ | 2.154 | 0.001 | ns | 0.959 | 0.121 | ns | 0.519 | 0.766 | |
| Lys | FBgn0004427 | LysD | ns | −0.689 | 0.654 | ns | −1.311 | 0.208 | ∨ | −1.901 | 0.046 |
| FBgn0004431 | LysX | ∨ | −7.066 | 0.000 | ∨ | −6.186 | 0.000 | ∨ | −3.052 | 0.000 | |
| AMP | FBgn0038530 | AttD | ∧ | 3.649 | 0.004 | ∧ | 4.053 | 0.002 | ∧ | 5.054 | 0.001 |
| FBgn0000276 | CecA1 | ∧ | 2.722 | 0.005 | ∧ | 2.471 | 0.008 | ∧ | 1.731 | 0.049 | |
| FBgn0000277 | CecA2 | ∧ | 3.337 | 0.003 | ∧ | 2.933 | 0.004 | ∧ | 2.419 | 0.012 | |
| FBgn0004240 | DptA | ∨ | −3.313 | 0.003 | ∨ | −2.084 | 0.027 | ∨ | −2.056 | 0.025 | |
| FBgn0041581 | AttB | ∧ | 3.728 | 0.004 | ∧ | 4.030 | 0.002 | ns | 2.078 | 0.055 | |
| FBgn0262881 | CG43236 | ∨ | −3.635 | 0.020 | ∨ | −2.906 | 0.050 | ns | −1.919 | 0.160 | |
| Flybase ID Symbol | 1 H | 3 H | 5 H | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| logFC | FDR | logFC | FDR | logFC | FDR | ||||||
| Autophagy | FBgn0026404 | Dronc | ∧ | 2.416 | 0.004 | ∧ | 1.643 | 0.036 | ∧ | 1.832 | 0.017 |
| FBgn0010501 | Dcp-1 | ns | 0.827 | 0.153 | ∧ | 1.344 | 0.004 | ∧ | 2.427 | 0.000 | |
| FBgn0025624 | CG4025 | ns | 0.848 | 0.237 | ns | 0.699 | 0.508 | ∧ | 1.629 | 0.006 | |
| FBgn0265464 | Traf6 | ns | 0.105 | 1.000 | ns | 0.773 | 0.211 | ∧ | 1.204 | 0.005 | |
| FBgn0010213 | Sod2 | ∧ | 1.426 | 0.003 | ns | 0.987 | 0.040 | ns | 0.242 | 1.000 | |
| FBgn0010303 | hep | ns | 0.029 | 1.000 | ∧ | 1.596 | 0.033 | ns | 0.959 | 0.240 | |
| FBgn0002567 | Rab32 | ns | −0.018 | 1.000 | ∨ | −1.454 | 0.005 | ns | −0.363 | 1.000 | |
| FBgn0034897 | Sesn | ns | −0.583 | 0.651 | ∨ | −2.211 | 0.002 | ns | −1.124 | 0.077 | |
| FBgn0261108 | Atg13 | ∨ | −1.495 | 0.010 | ns | −0.404 | 1.000 | ns | 0.040 | 1.000 | |
| FBgn0003997 | hid | ∨ | −4.011 | 0.004 | ns | −2.077 | 0.052 | ns | 0.022 | 1.000 | |
| FBgn0262656 | Myc | ∨ | −3.611 | 0.003 | ns | −1.549 | 0.142 | ns | −1.511 | 0.137 | |
| FBgn0039044 | p53 | ∨ | −1.857 | 0.011 | ns | −1.097 | 0.145 | ns | −0.429 | 0.875 | |
| FBgn0086357 | Sec61alpha | ∧ | 1.271 | 0.022 | ns | 0.176 | 1.000 | ns | −0.231 | 1.000 | |
| FBgn0010638 | Sec61beta | ∧ | 1.256 | 0.019 | ns | 0.490 | 0.907 | ns | −0.447 | 0.893 | |
| FBgn0003360 | sesB | ∧ | 1.499 | 0.004 | ns | 0.827 | 0.206 | ns | −0.219 | 1.000 | |
| Apoptosis | FBgn0026404 | Dronc | ∧ | 2.416 | 0.004 | ∧ | 1.643 | 0.036 | ∧ | 1.832 | 0.017 |
| FBgn0067102 | GlcT | ∧ | 2.137 | 0.001 | ns | 1.140 | 0.016 | ∧ | 1.247 | 0.007 | |
| FBgn0003892 | ptc | ∨ | −1.535 | 0.009 | ns | −1.166 | 0.046 | ∨ | −1.472 | 0.010 | |
| FBgn0010501 | Dcp−1 | ns | 0.827 | 0.153 | ∧ | 1.344 | 0.004 | ∧ | 2.427 | 0.000 | |
| FBgn0015245 | Hsp60A | ns | 0.322 | 1.000 | ∨ | −1.464 | 0.007 | ∨ | −1.620 | 0.004 | |
| FBgn0033784 | SCCRO3 | ns | 0.457 | 0.912 | ns | 1.072 | 0.028 | ∧ | 1.534 | 0.002 | |
| FBgn0022027 | Vps25 | ns | 0.335 | 1.000 | ns | 1.072 | 0.020 | ∧ | 1.345 | 0.003 | |
| FBgn0025878 | wrapper | ns | −0.884 | 0.608 | ns | 3.667 | 0.002 | ∧ | 2.580 | 0.012 | |
| FBgn0013762 | Cdk5 | ns | 0.791 | 0.316 | ns | 0.877 | 0.236 | ∧ | 1.246 | 0.034 | |
| FBgn0033783 | CG17019 | ns | 0.686 | 0.506 | ns | 1.049 | 0.147 | ∧ | 1.679 | 0.012 | |
| FBgn0053346 | CG33346 | ns | −1.041 | 0.273 | ns | −0.255 | 1.000 | ∧ | 3.068 | 0.002 | |
| FBgn0036831 | CG6839 | ns | −0.417 | 0.911 | ns | −1.251 | 0.183 | ∨ | −2.384 | 0.009 | |
| FBgn0036165 | chrb | ns | 0.291 | 1.000 | ns | 0.540 | 0.801 | ∧ | 1.675 | 0.011 | |
| FBgn0038928 | Fadd | ns | −0.049 | 1.000 | ns | 0.174 | 1.000 | ∧ | 1.433 | 0.004 | |
| FBgn0259108 | futsch | ns | −1.558 | 0.114 | ns | −1.085 | 0.362 | ∨ | −1.922 | 0.048 | |
| FBgn0013726 | pnut | ns | 0.478 | 0.881 | ns | 0.941 | 0.075 | ∧ | 1.361 | 0.004 | |
| FBgn0034279 | CG18635 | ns | 1.028 | 0.142 | ∧ | 1.407 | 0.032 | ns | 0.913 | 0.219 | |
| FBgn0045035 | tefu | ns | −0.949 | 0.222 | ∨ | −1.619 | 0.021 | ns | −1.068 | 0.132 | |
| FBgn0004569 | aos | ∨ | −1.380 | 0.014 | ns | −0.623 | 0.636 | ns | −0.724 | 0.370 | |
| FBgn0261108 | Atg13 | ∨ | −1.495 | 0.010 | ns | −0.404 | 1.000 | ns | 0.040 | 1.000 | |
| FBgn0264291 | Det | ∨ | −1.847 | 0.030 | ns | −1.130 | 0.216 | ns | −0.875 | 0.372 | |
| FBgn0024732 | Drep1 | ∨ | −1.382 | 0.047 | ns | −1.366 | 0.053 | ns | −0.824 | 0.335 | |
| FBgn0003997 | hid | ∨ | −4.011 | 0.004 | ns | −2.077 | 0.052 | ns | 0.022 | 1.000 | |
| FBgn0262656 | Myc | ∨ | −3.611 | 0.003 | ns | −1.549 | 0.142 | ns | −1.511 | 0.137 | |
| FBgn0039044 | p53 | ∨ | −1.857 | 0.011 | ns | −1.097 | 0.145 | ns | −0.429 | 0.875 | |
| FBgn0038519 | Prx3 | ∧ | 1.442 | 0.005 | ns | 0.944 | 0.090 | ns | −0.452 | 0.911 | |
| FBgn0003360 | sesB | ∧ | 1.499 | 0.004 | ns | 0.827 | 0.206 | ns | −0.219 | 1.000 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muita, B.K.; Baxter, S.W. Temporal Exposure to Bt Insecticide Causes Oxidative Stress in Larval Midgut Tissue. Toxins 2023, 15, 323. https://doi.org/10.3390/toxins15050323
Muita BK, Baxter SW. Temporal Exposure to Bt Insecticide Causes Oxidative Stress in Larval Midgut Tissue. Toxins. 2023; 15(5):323. https://doi.org/10.3390/toxins15050323
Chicago/Turabian StyleMuita, Biko K., and Simon W. Baxter. 2023. "Temporal Exposure to Bt Insecticide Causes Oxidative Stress in Larval Midgut Tissue" Toxins 15, no. 5: 323. https://doi.org/10.3390/toxins15050323






