Machine Learning with Alpha Toxin Phenotype to Predict Clinical Outcome in Patients with Staphylococcus aureus Bloodstream Infection
Abstract
:1. Introduction
2. Results
2.1. Bacterial Isolates and Patient Characteristics
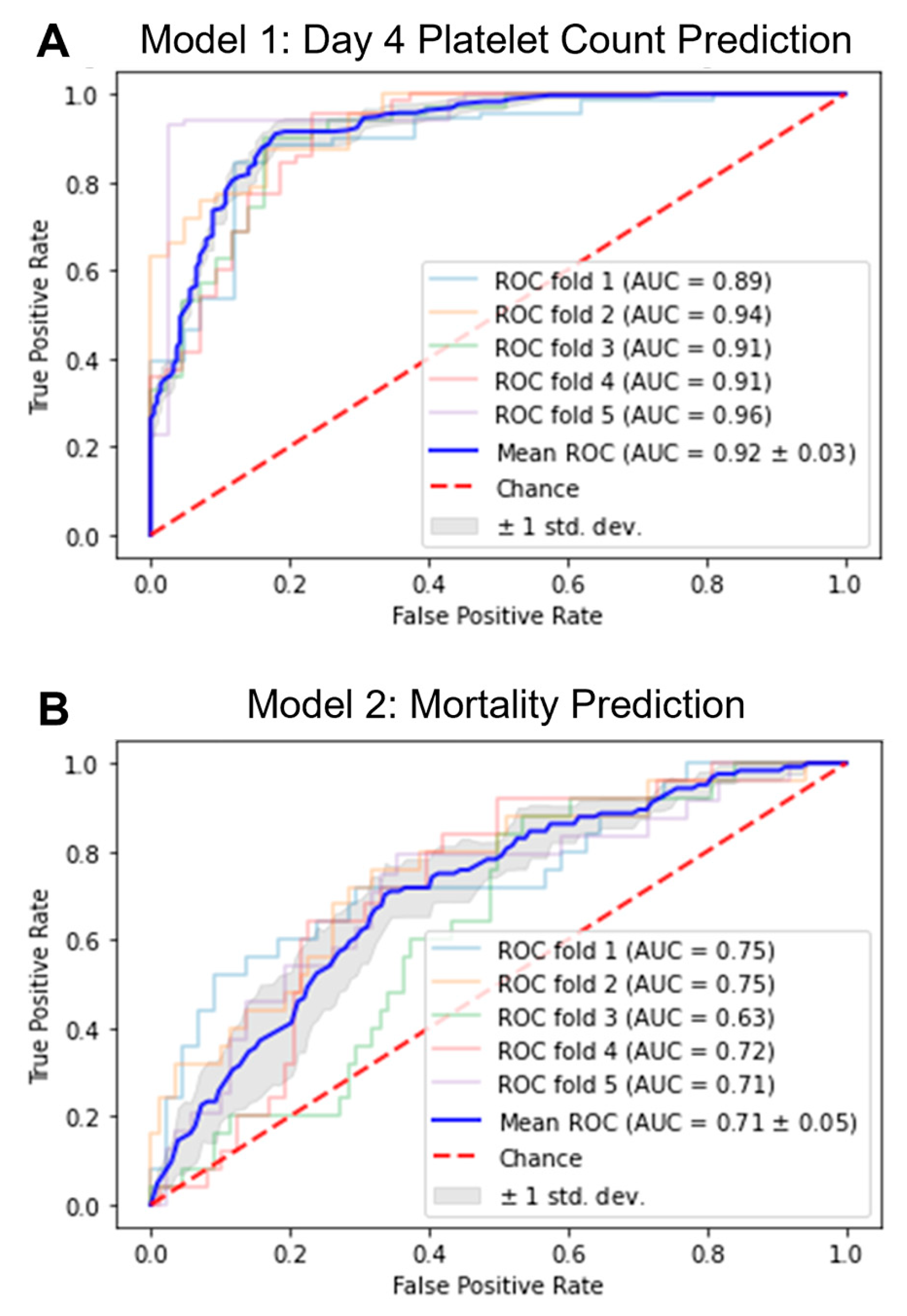
2.2. Development of ML Models
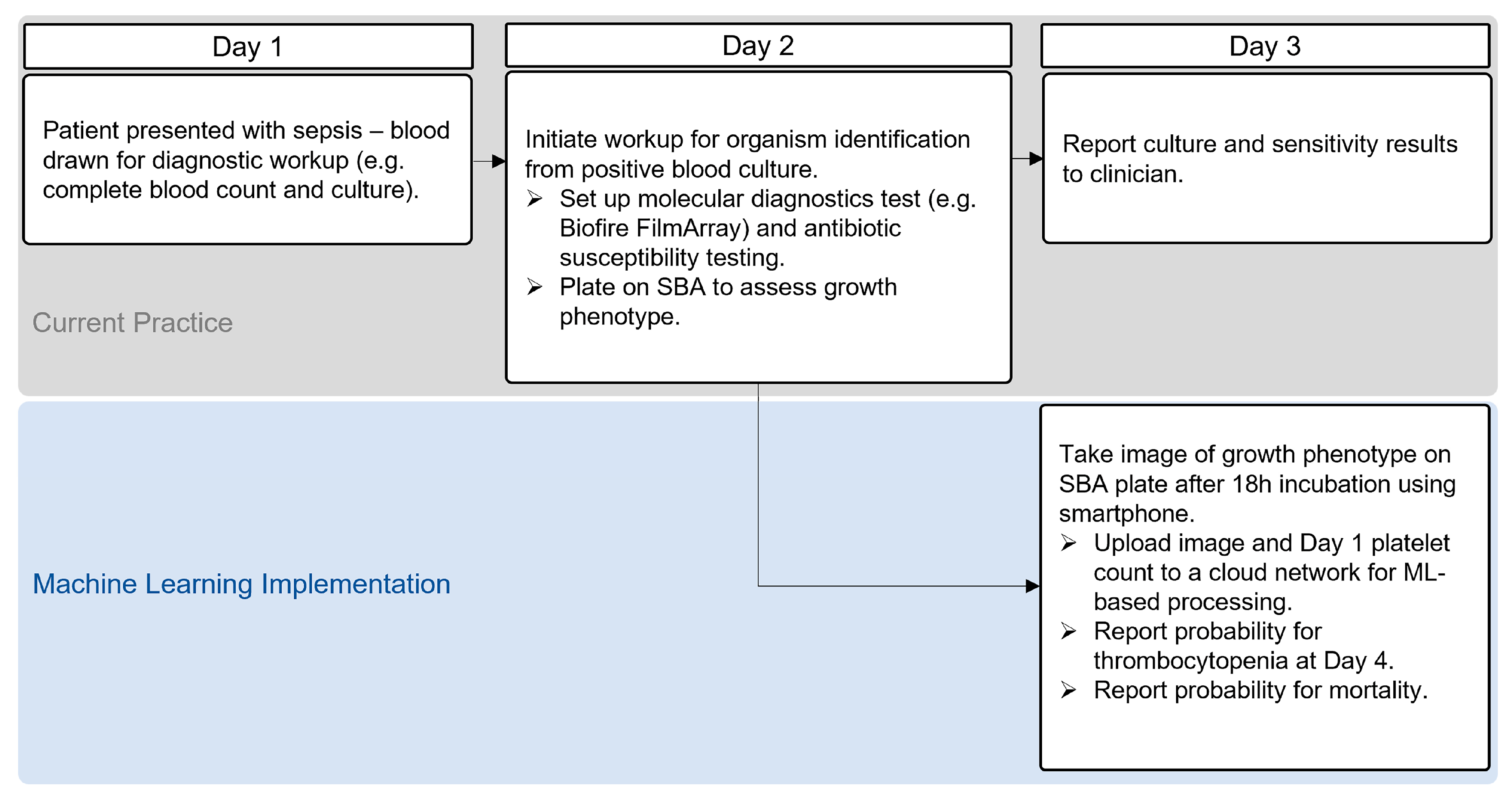
2.3. Application of ML Models
3. Discussion
4. Materials and Methods
4.1. Patient Cohort and Bacterial Isolate Selection
4.2. Bacterial Growth Conditions
4.3. Rabbit Erythrocyte Lysis Assay
4.4. Machine Learning Algorithm Development
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Magill, S.S.; O’Leary, E.; Janelle, S.J.; Thompson, D.L.; Dumyati, G.; Nadle, J.; Wilson, L.E.; Kainer, M.A.; Lynfield, R.; Greissman, S.; et al. Changes in prevalence of health care–associated infections in U.S. hospitals. N. Engl. J. Med. 2018, 379, 1732–1744. [Google Scholar] [CrossRef]
- Thammavongsa, V.; Kim, H.K.; Missiakas, D.; Schneewind, O. Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 2015, 13, 529–543. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.K.; Thammavongsa, V.; Schneewind, O.; Missiakas, D. Recurrent infections and immune evasion strategies of Staphylococcus aureus. Curr. Opin. Microbiol. 2012, 15, 92–99. [Google Scholar] [CrossRef] [Green Version]
- Minejima, E.; Mai, N.; Bui, N.; Mert, M.; Mack, W.J.; She, R.C.; Nieberg, P.; Spellberg, B.; Wong-Beringer, A. Defining the breakpoint duration of Staphylococcus aureus bacteremia predictive of poor outcomes. Clin. Infect Dis. 2020, 70, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Bubeck Wardenburg, J.; Bae, T.; Otto, M.; DeLeo, F.R.; Schneewind, O. Poring over pores: α-hemolysin and panton-valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 2007, 13, 1405–1406. [Google Scholar] [CrossRef] [PubMed]
- Surewaard, B.G.J.; Thanabalasuriar, A.; Zeng, Z.; Tkaczyk, C.; Cohen, T.S.; Bardoel, B.W.; Jorch, S.K.; Deppermann, C.; Bubeck Wardenburg, J.; Davis, R.P.; et al. α-Toxin induces platelet aggregation and liver injury during Staphylococcus aureus sepsis. Cell Host Microbe 2018, 24, 271–284.e3. [Google Scholar] [CrossRef] [PubMed]
- Inoshima, I.; Inoshima, N.; Wilke, G.A.; Powers, M.E.; Frank, K.M.; Wang, Y.; Wardenburg, J.B. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat. Med. 2011, 17, 1310–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, K.M.; Zhou, T.; Moreno-Vinasco, L.; Hollett, B.; Garcia, J.G.N.; Bubeck Wardenburg, J. Host response signature to Staphylococcus aureus alpha-hemolysin implicates pulmonary Th17 response. Infect. Immun. 2012, 80, 3161–3169. [Google Scholar] [CrossRef] [Green Version]
- Rauch, S.; DeDent, A.C.; Kim, H.K.; Bubeck Wardenburg, J.; Missiakas, D.M.; Schneewind, O. Abscess formation and alpha-hemolysin induced toxicity in a mouse model of Staphylococcus aureus peritoneal infection. Infect. Immun. 2012, 80, 3721–3732. [Google Scholar] [CrossRef] [Green Version]
- Ragle, B.E.; Wardenburg, J.B. Anti-alpha-hemolysin monoclonal antibodies mediate protection against Staphylococcus aureus pneumonia. Infect. Immun. 2009, 77, 2712–2718. [Google Scholar] [CrossRef] [Green Version]
- Alhurayri, F.; Porter, E.; Douglas-Louis, R.; Minejima, E.; Wardenburg, J.B.; Wong-Beringer, A. Increased risk of thrombocytopenia and death in patients with bacteremia caused by high alpha toxin-producing methicillin-resistant Staphylococcus aureus. Toxins 2021, 13, 726. [Google Scholar] [CrossRef] [PubMed]
- Powers, M.E.; Kim, H.K.; Wang, Y.; Bubeck Wardenburg, J. ADAM10 mediates vascular injury induced by Staphylococcus aureus α-hemolysin. J. Infect. Dis. 2012, 206, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Powers, M.E.; Becker, R.E.N.; Sailer, A.; Turner, J.R.; Wardenburg, J.B. Synergistic action of Staphylococcus aureus α-toxin on platelets and myeloid lineage cells contributes to lethal sepsis. Cell Host Microbe 2015, 17, 775–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaki, J.; Synold, T.; Wong-Beringer, A. Antivirulence potential of TR-700 and clindamycin on clinical isolates of Staphylococcus aureus producing phenol-soluble modulins. Antimicrob. Agents Chemother. 2011, 55, 4432–4435. [Google Scholar] [CrossRef] [Green Version]
- Stulik, L.; Malafa, S.; Hudcova, J.; Rouha, H.; Henics, B.Z.; Craven, D.E.; Sonnevend, A.M.; Nagy, E. α-Hemolysin activity of methicillin-susceptible Staphylococcus aureus predicts ventilator-associated pneumonia. Am. J. Respir. Crit. Care Med. 2014, 190, 1139–1148. [Google Scholar] [CrossRef] [Green Version]
- Shamout, F.; Zhu, T.; Clifton, D.A. Machine learning for clinical outcome prediction. IEEE Rev. Biomed. Eng. 2021, 14, 116–126. [Google Scholar] [CrossRef]
- Richens, J.G.; Lee, C.M.; Johri, S. Improving the accuracy of medical diagnosis with causal machine learning. Nat. Commun. 2020, 11, 3923. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar]
- Egli, A.; Schrenzel, J.; Greub, G. Digital microbiology. Clin. Microbiol. Infect. 2020, 26, 1324–1331. [Google Scholar] [CrossRef]
- Smith, K.P.; Kirby, J.E. Image analysis and artificial intelligence in infectious disease diagnostics. Clin. Microbiol. Infect. 2020, 26, 1318–1323. [Google Scholar] [CrossRef]
- Peiffer-Smadja, N.; Dellière, S.; Rodriguez, C.; Birgand, G.; Lescure, F.-X.; Fourati, S.; Ruppé, E. Machine learning in the clinical microbiology laboratory: Has the time come for routine practice? Clin. Microbiol. Infect. 2020, 26, 1300–1309. [Google Scholar] [CrossRef]
- Soriano, A.; Marco, F.; Martínez, J.A.; Pisos, E.; Almela, M.; Dimova, V.P.; Alamo, D.; Ortega, M.; Lopez, J.; Mensa, J. Influence of vancomycin minimum inhibitory concentration on the treatment of methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 2008, 46, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Douglas-Louis, R.; Lou, M.; Lee, B.; Minejima, E.; Bubeck-Wardenburg, J.; Wong-Beringer, A. Prognostic significance of early platelet dynamics in Staphylococcus aureus bacteremia. BMC Infect. Dis. 2023, 23, 82. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Uchiyama, S.; Olson, J.; Morodomi, Y.; Cornax, I.; Ando, N.; Kohno, Y.; Kyaw, M.M.T.; Aguilar, B.; Haste, N.M.; et al. Repurposed drugs block toxin-driven platelet clearance by the hepatic ashwell-morell receptor to clear Staphylococcus aureus bacteremia. Sci. Transl. Med. 2021, 13, eabd6737. [Google Scholar] [CrossRef] [PubMed]
- Moraveji, Z.; Tabatabaei, M.; Shirzad Aski, H.; Khoshbakht, R. Characterization of hemolysins of Staphylococcus strains isolated from human and bovine, Southern Iran. Iran. J. Vet. Res. 2014, 15, 326–330. [Google Scholar]
- Huseby, M.; Shi, K.; Brown, C.K.; Digre, J.; Mengistu, F.; Seo, K.S.; Bohach, G.A.; Schlievert, P.M.; Ohlendorf, D.H.; Earhart, C.A. Structure and biological activities of beta toxin from Staphylococcus aureus. J. Bacteriol. 2007, 189, 8719–8726. [Google Scholar] [CrossRef] [Green Version]
- Ulloa, E.R.; Uchiyama, S.; Gillespie, R.; Nizet, V.; Sakoulas, G. Ticagrelor Increases Platelet-Mediated Staphylococcus Aureus Killing, Resulting in Clearance of Bacteremia. J. Infect. Dis. 2021, 224, 1566–1569. [Google Scholar] [CrossRef]
- Caffrey, A.R.; Appaneal, H.J.; LaPlante, K.L.; Lopes, V.V.; Ulloa, E.R.; Nizet, V.; Sakoulas, G. Impact of Clopidogrel on Clinical Outcomes in Patients with Staphylococcus Aureus Bacteremia: A National Retrospective Cohort Study. Antimicrob. Agents Chemother. 2022, 66, e02117-21. [Google Scholar] [CrossRef]
- Trier, D.A.; Gank, K.D.; Kupferwasser, D.; Yount, N.Y.; French, W.J.; Michelson, A.D.; Kupferwasser, L.I.; Xiong, Y.Q.; Bayer, A.S.; Yeaman, M.R. Platelet Antistaphylococcal Responses Occur through P2X1 and P2Y12 Receptor-Induced Activation and Kinocidin Release. Infect. Immun. 2008, 76, 5706–5713. [Google Scholar] [CrossRef] [Green Version]
- Bayer, A.S.; Ramos, M.D.; Menzies, B.E.; Yeaman, M.R.; Shen, A.J.; Cheung, A.L. Hyperproduction of Alpha-Toxin by Staphylococcus Aureus Results in Paradoxically Reduced Virulence in Experimental Endocarditis: A Host Defense Role for Platelet Microbicidal Proteins. Infect. Immun. 1997, 65, 4652–4660. [Google Scholar] [CrossRef] [Green Version]
- Oury, C.; Meyers, S.; Jacques, N.; Leeten, K.; Jiang, Z.; Musumeci, L.; Lox, M.; Debuisson, M.; Goffin, E.; Pirotte, B.; et al. Protective Effect of Ticagrelor Against Infective Endocarditis Induced by Virulent Staphylococcus Aureus in Mice. JACC Basic Transl. Sci. 2023; in press. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)–A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Wada, K. Labelme: Image Polygonal Annotation with Python. Available online: https://github.com/wkentaro/labelme (accessed on 6 April 2023).
- Bradski, G. The openCV Library. Available online: https://opencv.org/ (accessed on 6 April 2023).
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Milton-Barker, A. Inception v3 Deep Convolutional Architecture for Classifying Acute Myeloid/Lymphoblastic Leukemia. Available online: https://www.intel.com/content/www/us/en/developer/articles/technical/inception-v3-deep-convolutional-architecture-for-classifying-acute-myeloidlymphoblastic.html (accessed on 10 October 2022).

| All Patients (n = 229) | Normal Platelet (n = 151) | D1 Thrombocytopenia (n = 78) | p Value | |
|---|---|---|---|---|
| Age, y, mean (SD) | 60 (16.4) | 60 (16.6) | 60 (16.0) | 0.96 |
| Sex (male) | 163 (71.2) | 111 (73.5) | 52 (66.7) | 0.29 |
| Comorbidities | ||||
| None | 24 (10.5) | 14 (9.3) | 10 (12.8) | 0.50 |
| Coronary artery disease | 30 (13.1) | 16 (10.6) | 14 (17.9) | 0.15 |
| Heart failure | 30 (13.1) | 19 (12.6) | 11 (14.1) | 0.69 |
| Diabetes | 98 (42.8) | 65 (43.0) | 33 (42.3) | >0.99 |
| Hypertension | 116 (50.7) | 82 (54.3) | 34 (43.6) | 0.13 |
| Intravenous drug use | 35 (15.3) | 23 (15.2) | 12 (15.4) | >0.99 |
| Liver disease | 41 (18.0) | 22 (14.6) | 19 (24.4) | 0.072 |
| Renal disease | 65 (28.4) | 41 (27.2) | 24 (30.8) | 0.64 |
| Dialysis | 47 (20.5) | 29 (19.2) | 18 (23.1) | 0.49 |
| Source risk category | ||||
| Low a | 49 (21.4) | 33 (21.9) | 16 (20.5) | 0.87 |
| Intermediate b | 129 (56.3) | 94 (62.2) | 35 (44.9) | 0.017 |
| High c | 51 (22.3) | 24 (15.9) | 27 (34.6) | 0.0023 |
| Microbiology | ||||
| MRSA | 84 (36.7) | 53 (35.1) | 31 (39.7) | 0.56 |
| Severity of illness | ||||
| Sepsis | 197 (86.0) | 126 (83.4) | 71 (91.0) | 0.16 |
| Severe sepsis | 122 (53.3) | 63 (41.7) | 59 (75.6) | <0.0001 |
| Septic shock | 35 (15.3) | 15 (9.9) | 20 (25.6) | 0.0032 |
| Concurrent antiplatelets | 61 (26.6) | 39 (25.8) | 22 (28.2) | 0.75 |
| Duration of SAB, days | 3 (1–5) | 2 (1–4) | 4 (1–6) | 0.0016 |
| Persistent SAB | 91 (39.7) | 48 (31.8) | 43 (55.1) | 0.001 |
| 30-day mortality | 43 (18.8) | 17 (11.3) | 26 (33.3) | 0.0001 |
| Metric | Model 1: Day 4 Platelet Count | Model 2: Mortality | ||
|---|---|---|---|---|
| Mean | Standard Deviation | Mean | Standard Deviation | |
| Accuracy | 0.823 | 0.032 | 0.691 | 0.064 |
| F1-Score | 0.806 | 0.049 | 0.511 | 0.025 |
| Precision | 0.782 | 0.102 | 0.392 | 0.045 |
| Sensitivity | 0.891 | 0.082 | 0.750 | 0.076 |
| Specificity | 0.724 | 0.187 | 0.672 | 0.101 |
| AUC | 0.920 | 0.027 | 0.711 | 0.048 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beadell, B.; Nehra, S.; Gusenov, E.; Huse, H.; Wong-Beringer, A. Machine Learning with Alpha Toxin Phenotype to Predict Clinical Outcome in Patients with Staphylococcus aureus Bloodstream Infection. Toxins 2023, 15, 417. https://doi.org/10.3390/toxins15070417
Beadell B, Nehra S, Gusenov E, Huse H, Wong-Beringer A. Machine Learning with Alpha Toxin Phenotype to Predict Clinical Outcome in Patients with Staphylococcus aureus Bloodstream Infection. Toxins. 2023; 15(7):417. https://doi.org/10.3390/toxins15070417
Chicago/Turabian StyleBeadell, Brent, Surya Nehra, Elizabeth Gusenov, Holly Huse, and Annie Wong-Beringer. 2023. "Machine Learning with Alpha Toxin Phenotype to Predict Clinical Outcome in Patients with Staphylococcus aureus Bloodstream Infection" Toxins 15, no. 7: 417. https://doi.org/10.3390/toxins15070417
APA StyleBeadell, B., Nehra, S., Gusenov, E., Huse, H., & Wong-Beringer, A. (2023). Machine Learning with Alpha Toxin Phenotype to Predict Clinical Outcome in Patients with Staphylococcus aureus Bloodstream Infection. Toxins, 15(7), 417. https://doi.org/10.3390/toxins15070417





