The Growing Importance of Three-Dimensional Models and Microphysiological Systems in the Assessment of Mycotoxin Toxicity
Abstract
:1. Introduction
2. Towards More Predictive Toxicology
3. 3D Spheroids
3.1. Considerations for Spheroid Handling
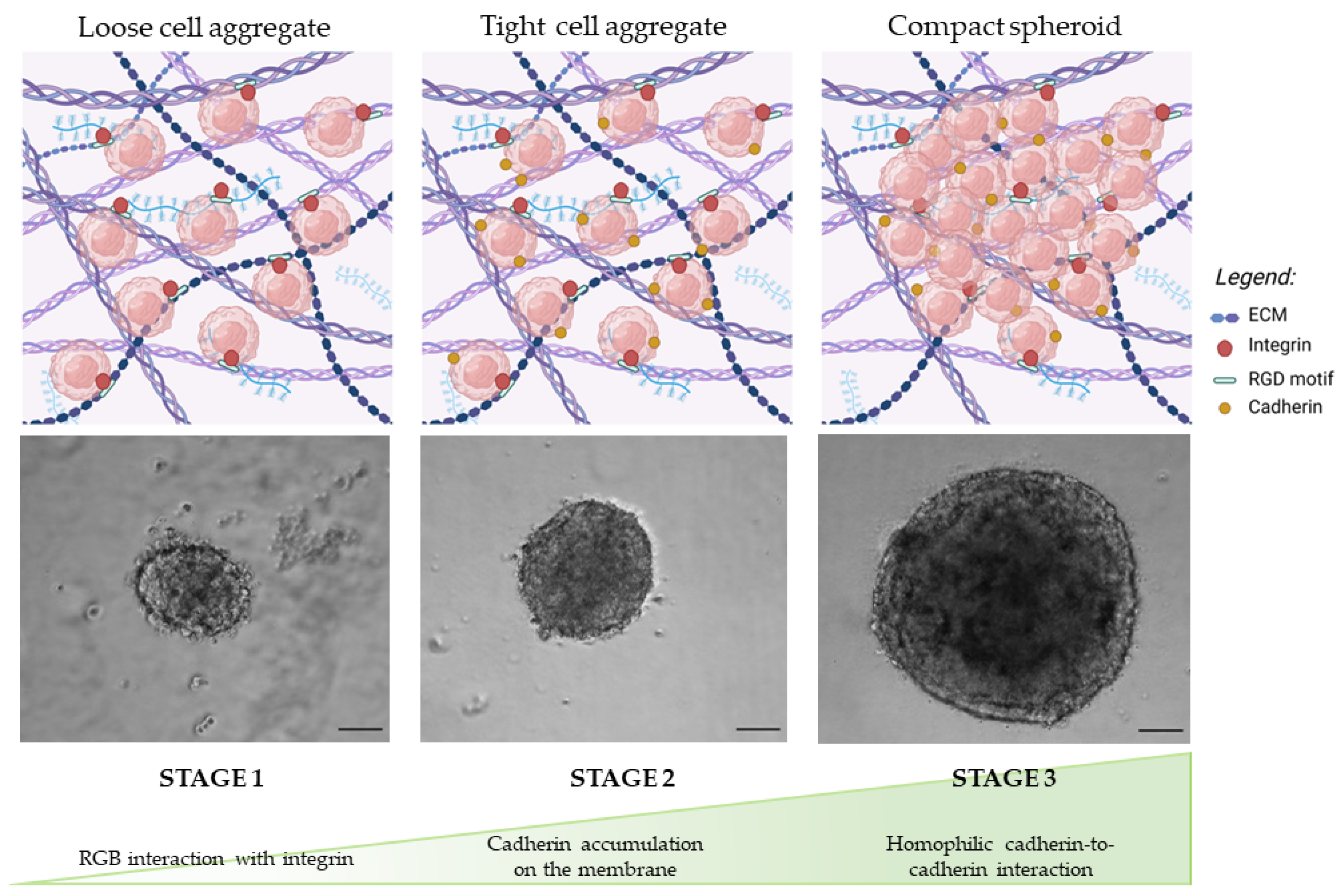
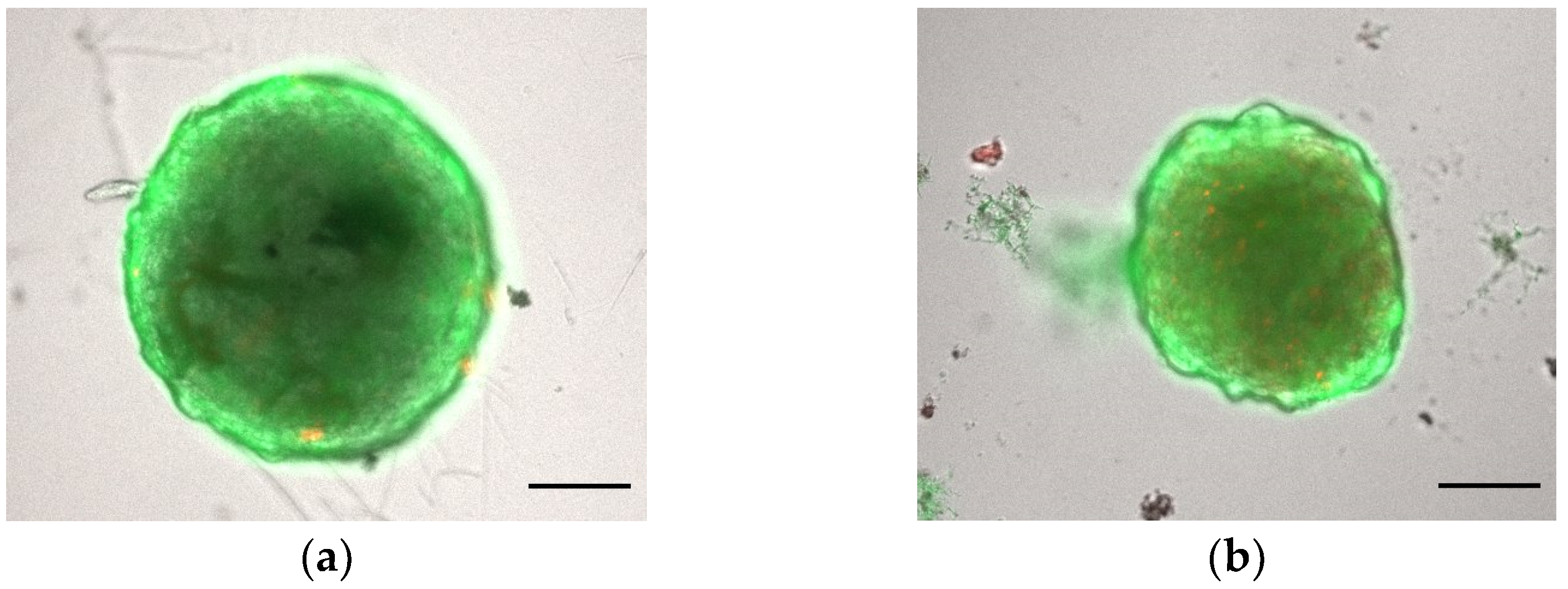
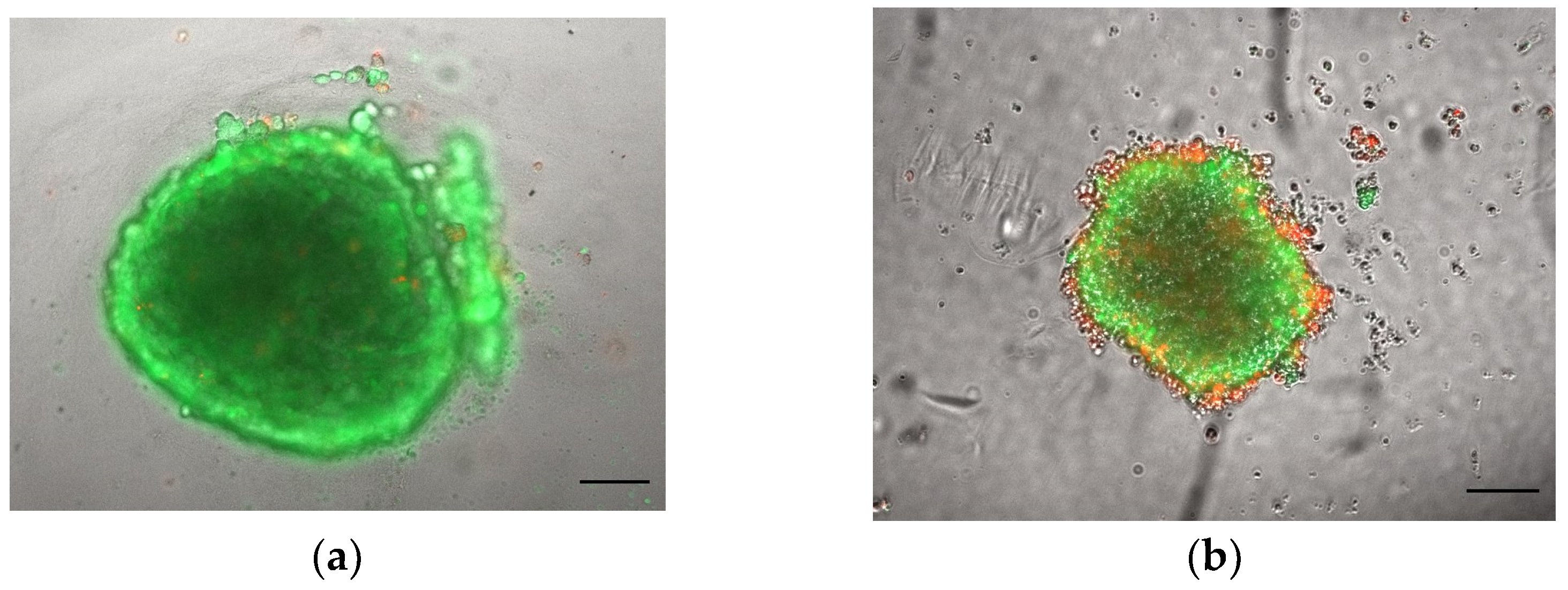
3.1.1. Morphology
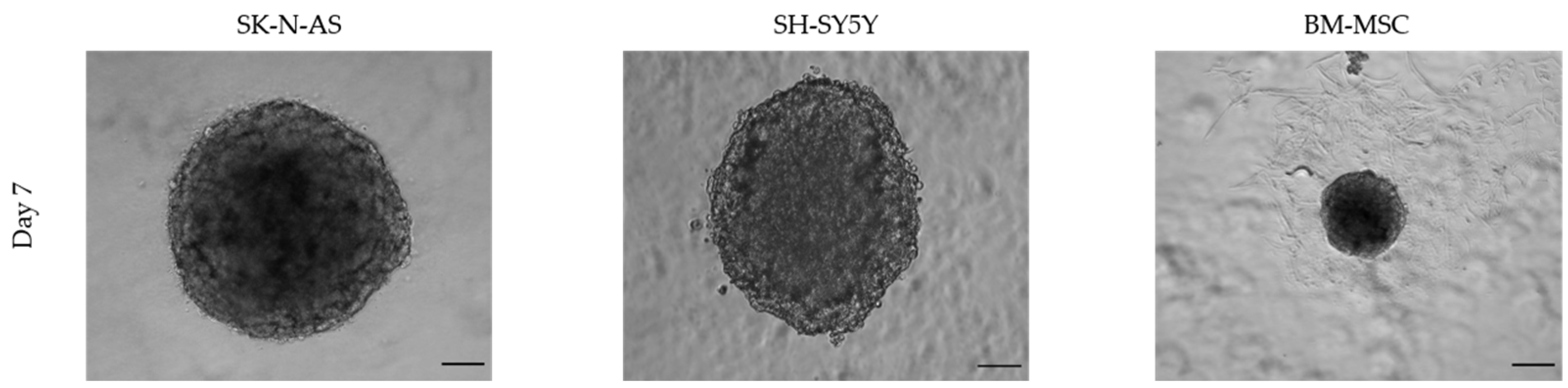
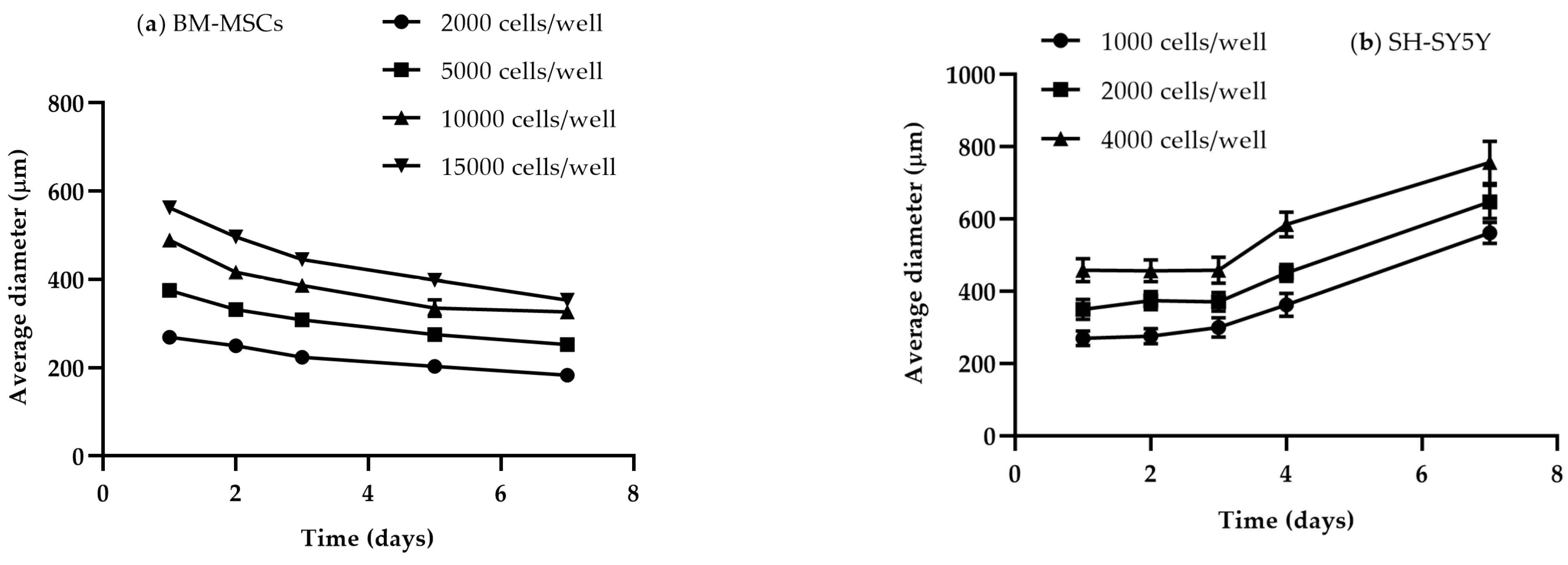
3.1.2. Size and Time
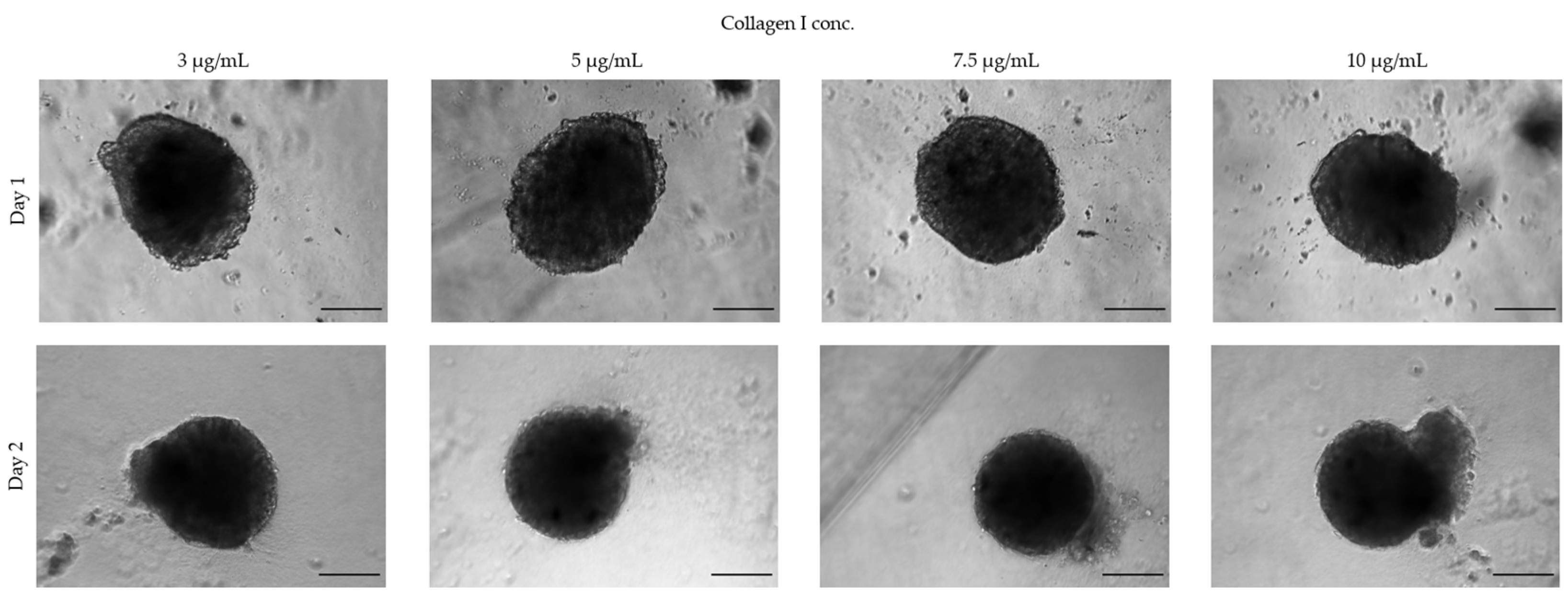
3.1.3. Extracellular Matrices
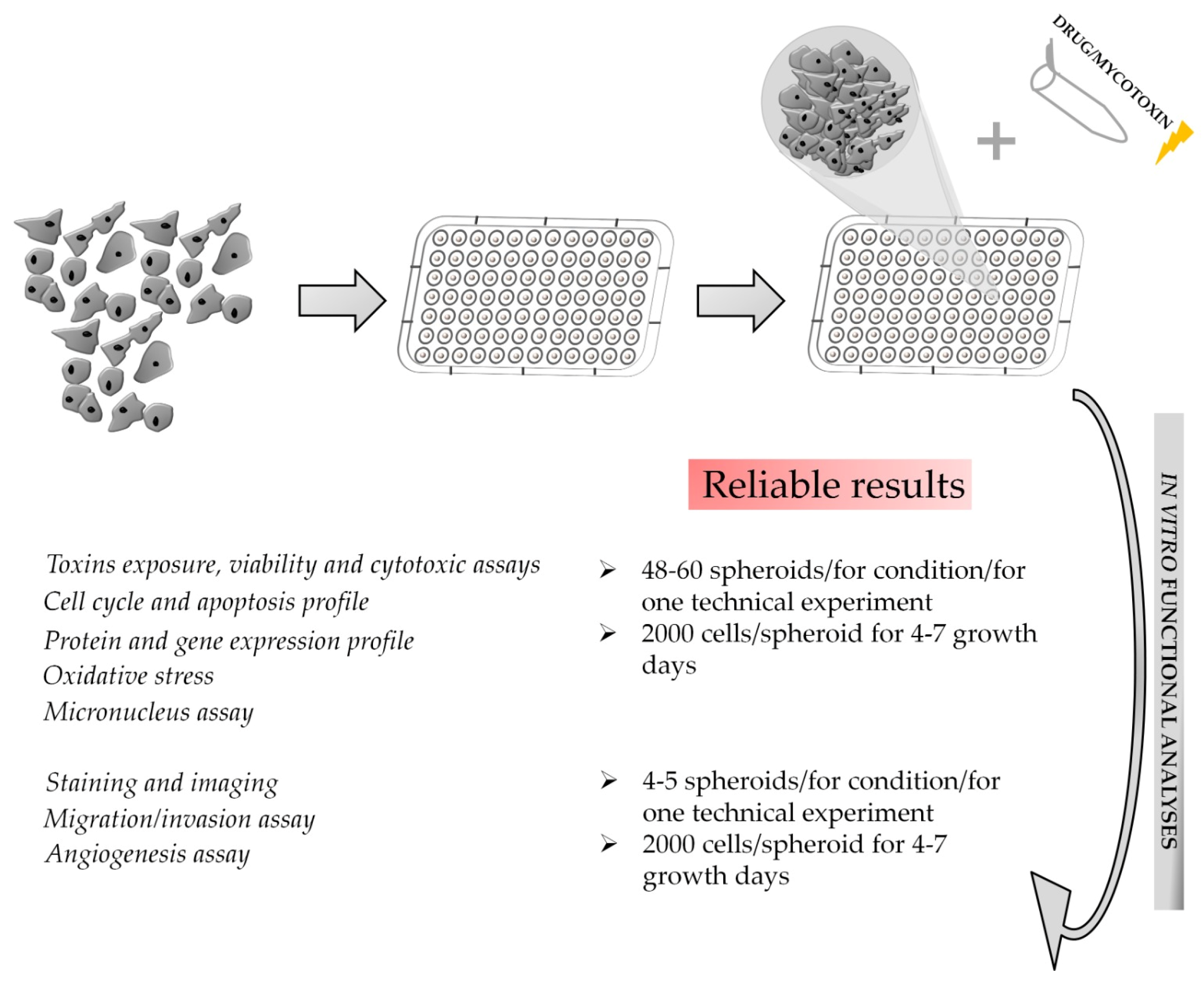
3.2. Downstream Functional Assays on 3D Spheroid Model: Advantages and Critical Issues
4. Organ-on-a-Chip (OoC) Systems
4.1. Considerations for OoC Handling
4.1.1. Material Selection
4.1.2. Selection of Cell Culture
4.2. Downstream Function Assays on OoC Model: Advantages and Critical Issues
5. Applications of Advanced In Vitro Models for Mycotoxin Assessment
| Mycotoxin | 3D Model | Endpoint | Reference |
|---|---|---|---|
| AFB1 | Hepatic spheroids (HepG2 cells) | DNA damage | [72] |
| Hepatic spheroids (HepG2 and HepaRG cells) | Cytotoxicity, liver functionality, genotoxicity | [71] | |
| Human hepatocytes cultured in a MPS | LDH release | [106] | |
| Mono-type spheroids (HepG2 cells); co-cultured spheroids (HepG2 cells + EA.hy 926 cells); triple co-cultured spheroids (HepG2 cells + EA.hy 926 cells + LX-2 cells) | Cell viability, mitochondria, oxidative stress, cell membrane | [133] | |
| Normal human bronchial epithelial (NHBE) cells cultured at the air–liquid interface (ALI); lung/liver-on-a-chip (NHBE ALI + HepaRG spheroids) | Transepithelial electrical resistance (TEER), ATP content | [136] | |
| Lung/liver-on-a-chip (Bronchial MucilAir + HepaRG and HHSteCs spheroids) | Intracellular ATP levels, LDH release | [137] | |
| Paper-based 3D HepG2 culture | Hepatotoxicity at different oxygen tensions | [142] | |
| AFB1, CPA | Triple co-cultured spheroids (HepG2 cells + EA.hy 926 cells + LX-2 cells) | Individual and combined cell viability, mitochondria, oxidative stress, metabolomic analysis | [134] |
| STE | Human neuroblastoma spheroids (SH-SY5Y and SK-N-DZ cells) | Cell viability, oxidative stress, apoptosis, DNA damage, migration | [47] |
| CIT, OTs, | Canine kidney spheroids (MDCK cells) | Individual and combined cytotoxicity | [129] |
| DON | Mouse enteroids | Intestinal barrier function | [139] |
| Porcine enteroids | Intestinal stem cells activity | [140] | |
| 3-layered 3D gut-on-a-chip Caco-2 cell culture | Intestinal barrier function | [141] | |
| FB1 | Rat hepatic spheroids | Cytotoxicity | [130] |
| 3D human esophageal epithelial cells (HEEC) | Cell viability | [143] | |
| OTA | 3D human kidney proximal tubule microphysiological system | Cytotoxicity, analysis of kidney injury biomarkers, OTA transport, detoxification, and bioactivation | [138] |
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wrzesinski, K.; Fey, S.J. From 2D to 3D—A New Dimension for Modelling the Effect of Natural Products on Human Tissue. Curr. Pharm. Des. 2015, 21, 5605–5616. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.P.; Gaspar, V.M.; Mano, J.F. Design of spherically structured 3D in vitro tumor models—Advances and prospects. Acta Biomater. 2018, 75, 11–34. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartung, T. From alternative methods to a new toxicology. Eur. J. Pharm. Biopharm. 2011, 77, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Plevkova, J.; Brozmanova, M.; Matloobi, A.; Poliacek, I.; Honetschlager, J.; Buday, T. Animal models of cough. Respir. Physiol. Neurobiol. 2021, 290, 103656. [Google Scholar] [CrossRef] [PubMed]
- Milani-Nejad, N.; Janssen, P.M. Small and large animal models in cardiac contraction research: Advantages and disadvantages. Pharmacol. Ther. 2014, 141, 235–249. [Google Scholar] [CrossRef] [Green Version]
- Loder, N. UK researchers call for limits on animal experiment ‘red tape’. Nature 2000, 405, 725. [Google Scholar] [CrossRef] [Green Version]
- National Research Council, Committee on Toxicity Testing and Assessment of Environmental Agents. Toxicity Testing in the 21st Century: A Vision and a Strategy; Academic Press: Cambridge, MA, USA, 2007; 178p. [Google Scholar]
- Firestone, M.; Kavlock, R.; Zenick, H.; Kramer, M.; the, U.S. EPA Working Group on the Future of Toxicity Testing. The U.S. Environmental Protection Agency strategic plan for evaluating the toxicity of chemicals. J. Toxicol. Environ. Health B Crit. Rev. 2010, 13, 139–162. [Google Scholar] [CrossRef]
- EPA. Available online: https://www.epa.gov/assessing-and-managing-chemicals-under-tsca/alternative-test-methods-and-strategies-reduce (accessed on 7 December 2022).
- EEC. Council Directive 86/609/EEC of 24 November 1986 on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes. Off. J. Eur. Union. 1986, L 358, pp. 1–29. Available online: http://data.europa.eu/eli/dir/1986/609/oj (accessed on 7 December 2022).
- The European Parlament; Council of the European Union. Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for Scientific purposes. Off. J. Eur. Union 2010, 276, 33–79. [Google Scholar]
- Gunness, P.; Mueller, D.; Shevchenko, V.; Heinzle, E.; Ingelman-Sundberg, M.; Noor, F. 3D organotypic cultures of human HepaRG cells: A tool for in vitro toxicity studies. Toxicol. Sci. 2013, 133, 67–78. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, S.T. High-throughput 3-D cell-based proliferation and cytotoxicity assays for drug screening and bioprocess development. J. Biotechnol. 2011, 151, 186–193. [Google Scholar] [CrossRef]
- Abbott, A. Cell culture: Biology’s new dimension. Nature 2003, 424, 870–872. [Google Scholar] [CrossRef]
- Ramaiahgari, S.C.; den Braver, M.W.; Herpers, B.; Terpstra, V.; Commandeur, J.N.; van de Water, B.; Price, L.S. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch. Toxicol. 2014, 88, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Augustyniak, J.; Bertero, A.; Coccini, T.; Baderna, D.; Buzanska, L.; Caloni, F. Organoids are promising tools for species-specific in vitro toxicological studies. J. Appl. Toxicol. 2019, 39, 1610–1622. [Google Scholar] [CrossRef]
- Iakobachvili, N.; Peters, P.J. Humans in a Dish: The Potential of Organoids in Modeling Immunity and Infectious Diseases. Front. Microbiol. 2017, 8, 2402. [Google Scholar] [CrossRef] [Green Version]
- Viravaidya, K.; Sin, A.; Shuler, M.L. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol. Prog. 2004, 20, 316–323. [Google Scholar] [CrossRef]
- Marx, U.; Andersson, T.B.; Bahinski, A.; Beilmann, M.; Beken, S.; Cassee, F.R.; Cirit, M.; Daneshian, M.; Fitzpatrick, S.; Frey, O.; et al. Biology-inspired microphysiological system approaches to solve the prediction dilemma of substance testing. ALTEX 2016, 33, 272–321. [Google Scholar] [CrossRef]
- Materne, E.M.; Maschmeyer, I.; Lorenz, A.K.; Horland, R.; Schimek, K.M.; Busek, M.; Sonntag, F.; Lauster, R.; Marx, U. The multi-organ chip—A microfluidic platform for long-term multi-tissue coculture. J. Vis. Exp. 2015, 28, e52526. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Sung, J.H. Microtechnology-Based Multi-Organ Models. Bioengineering 2017, 4, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choe, A.; Ha, S.K.; Choi, I.; Choi, N.; Sung, J.H. Microfluidic Gut-liver chip for reproducing the first pass metabolism. Biomed. Microdevices 2017, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef]
- Riffle, S.; Hegde, R.S. Modeling tumor cell adaptations to hypoxia in multicellular tumor spheroids. J. Exp. Clin. Cancer Res. 2017, 36, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouroupis, D.; Correa, D. Increased Mesenchymal Stem Cell Functionalization in Three-Dimensional Manufacturing Settings for Enhanced Therapeutic Applications. Front. Bioeng. Biotechnol. 2021, 9, 621748. [Google Scholar] [CrossRef]
- Han, S.J.; Kwon, S.; Kim, K.S. Challenges of applying multicellular tumor spheroids in preclinical phase. Cancer Cell Int. 2021, 21, 152. [Google Scholar] [CrossRef]
- Pinto, B.; Henriques, A.C.; Silva, P.M.A.; Bousbaa, H. Three-Dimensional Spheroids as In Vitro Preclinical Models for Cancer Research. Pharmaceutics 2020, 12, 1186. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, M.; Piccinini, F.; Arienti, C.; Zamagni, A.; Santi, S.; Polico, R.; Bevilacqua, A.; Tesei, A. 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016, 6, 19103. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kumacheva, E. Hydrogel microenvironments for cancer spheroid growth and drug screening. Sci. Adv. 2018, 4, eaas8998. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.H.; Kim, T.H. Recent Advances in Multicellular Tumor Spheroid Generation for Drug Screening. Biosensors 2021, 11, 445. [Google Scholar] [CrossRef]
- Ikada, Y. Challenges in tissue engineering. J. R. Soc. Interface 2006, 3, 589–601. [Google Scholar] [CrossRef]
- Bova, L.; Maggiotto, F.; Micheli, S.; Giomo, M.; Sgarbossa, P.; Gagliano, O.; Falcone, D.; Cimetta, E. A Porous Gelatin Methacrylate-Based Material for 3D Cell-Laden Constructs. Macromol. Biosci. 2023, 23, e2200357. [Google Scholar] [CrossRef] [PubMed]
- Alghuwainem, A.; Alshareeda, A.T.; Alsowayan, B. Scaffold-Free 3-D Cell Sheet Technique Bridges the Gap between 2-D Cell Culture and Animal Models. Int. J. Mol. Sci. 2019, 20, 4926. [Google Scholar] [CrossRef] [Green Version]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Catoira, M.C.; Fusaro, L.; Di Francesco, D.; Ramella, M.; Boccafoschi, F. Overview of natural hydrogels for regenerative medicine applications. J. Mater. Sci. Mater. Med. 2019, 30, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jubelin, C.; Munoz-Garcia, J.; Griscom, L.; Cochonneau, D.; Ollivier, E.; Heymann, M.F.; Vallette, F.M.; Oliver, L.; Heymann, D. Three-dimensional in vitro culture models in oncology research. Cell Biosci. 2022, 12, 155. [Google Scholar] [CrossRef]
- Kelm, J.M.; Timmins, N.E.; Brown, C.J.; Fussenegger, M.; Nielsen, L.K. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 2003, 83, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Razian, G.; Yu, Y.; Ungrin, M. Production of large numbers of size-controlled tumor spheroids using microwell plates. J. Vis. Exp. 2013, 81, e50665. [Google Scholar] [CrossRef] [Green Version]
- Habanjar, O.; Diab-Assaf, M.; Caldefie-Chezet, F.; Delort, L. 3D Cell Culture Systems: Tumor Application, Advantages, and Disadvantages. Int. J. Mol. Sci. 2021, 22, 12200. [Google Scholar] [CrossRef]
- Bartosh, T.J.; Ylostalo, J.H. Preparation of anti-inflammatory mesenchymal stem/precursor cells (MSCs) through sphere formation using hanging-drop culture technique. Curr. Protoc. Stem Cell Biol. 2014, 28, 2B.6.1–2B.6.23. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. 2017, 22, 456–472. [Google Scholar] [CrossRef] [Green Version]
- Ryu, N.E.; Lee, S.H.; Park, H. Spheroid Culture System Methods and Applications for Mesenchymal Stem Cells. Cells 2019, 8, 1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-based drug screen: Considerations and practical approach. Nat. Protoc. 2009, 4, 309–324. [Google Scholar] [CrossRef]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [Green Version]
- Zingales, V.; Torriero, N.; Zanella, L.; Fernandez-Franzon, M.; Ruiz, M.J.; Esposito, M.R.; Cimetta, E. Development of an in vitro neuroblastoma 3D model and its application for sterigmatocystin-induced cytotoxicity testing. Food Chem. Toxicol. 2021, 157, 112605. [Google Scholar] [CrossRef]
- Fuentes, P.; Torres, M.J.; Arancibia, R.; Aulestia, F.; Vergara, M.; Carrion, F.; Osses, N.; Altamirano, C. Dynamic Culture of Mesenchymal Stromal/Stem Cell Spheroids and Secretion of Paracrine Factors. Front. Bioeng. Biotechnol. 2022, 10, 916229. [Google Scholar] [CrossRef] [PubMed]
- Niibe, K.; Ohori-Morita, Y.; Zhang, M.; Mabuchi, Y.; Matsuzaki, Y.; Egusa, H. A Shaking-Culture Method for Generating Bone Marrow Derived Mesenchymal Stromal/Stem Cell-Spheroids with Enhanced Multipotency in vitro. Front. Bioeng. Biotechnol. 2020, 8, 590332. [Google Scholar] [CrossRef] [PubMed]
- Petry, F.; Salzig, D. Large-Scale Production of Size-Adjusted β-Cell Spheroids in a Fully Controlled Stirred-Tank Reactor. Processes 2022, 10, 861. [Google Scholar] [CrossRef]
- Phelan, M.A.; Gianforcaro, A.L.; Gerstenhaber, J.A.; Lelkes, P.I. An Air Bubble-Isolating Rotating Wall Vessel Bioreactor for Improved Spheroid/Organoid Formation. Tissue Eng. Part C Methods 2019, 25, 479–488. [Google Scholar] [CrossRef]
- Thoma, C.R.; Zimmermann, M.; Agarkova, I.; Kelm, J.M.; Krek, W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 2014, 69–70, 29–41. [Google Scholar] [CrossRef]
- Weiswald, L.B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Santo, V.E.; Estrada, M.F.; Rebelo, S.P.; Abreu, S.; Silva, I.; Pinto, C.; Veloso, S.C.; Serra, A.T.; Boghaert, E.; Alves, P.M.; et al. Adaptable stirred-tank culture strategies for large scale production of multicellular spheroid-based tumor cell models. J. Biotechnol. 2016, 221, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Kunz-Schughart, L.A. Multicellular tumor spheroids: Intermediates between monolayer culture and in vivo tumor. Cell Biol. Int. 1999, 23, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Bartosh, T.J.; Ylostalo, J.H.; Mohammadipoor, A.; Bazhanov, N.; Coble, K.; Claypool, K.; Lee, R.H.; Choi, H.; Prockop, D.J. Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their antiinflammatory properties. Proc. Natl. Acad. Sci. USA 2010, 107, 13724–13729. [Google Scholar] [CrossRef] [Green Version]
- Salehi, F.; Behboudi, H.; Kavoosi, G.; Ardestani, S.K. Monitoring ZEO apoptotic potential in 2D and 3D cell cultures and associated spectroscopic evidence on mode of interaction with DNA. Sci. Rep. 2017, 7, 2553. [Google Scholar] [CrossRef] [Green Version]
- Booij, T.H.; Price, L.S.; Danen, E.H.J. 3D Cell-Based Assays for Drug Screens: Challenges in Imaging, Image Analysis, and High-Content Analysis. SLAS Discov. 2019, 24, 615–627. [Google Scholar] [CrossRef] [Green Version]
- Vermes, I.; Haanen, C.; Steffens-Nakken, H.; Reutelingsperger, C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 1995, 184, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmeliet, P. Mechanisms of angiogenesis and arteriogenesis. Nat. Med. 2000, 6, 389–395. [Google Scholar] [CrossRef]
- Korff, T.; Augustin, H.G. Tensional forces in fibrillar extracellular matrices control directional capillary sprouting. J. Cell Sci. 1999, 112 Pt 19, 3249–3258. [Google Scholar] [CrossRef]
- Heiss, M.; Hellstrom, M.; Kalen, M.; May, T.; Weber, H.; Hecker, M.; Augustin, H.G.; Korff, T. Endothelial cell spheroids as a versatile tool to study angiogenesis in vitro. FASEB J. 2015, 29, 3076–3084. [Google Scholar] [CrossRef]
- Tetzlaff, F.; Fischer, A. Human Endothelial Cell Spheroid-based Sprouting Angiogenesis Assay in Collagen. Bio-Protocol 2018, 8, e2995. [Google Scholar] [CrossRef]
- Nazari, S.S. Generation of 3D Tumor Spheroids with Encapsulating Basement Membranes for Invasion Studies. Curr. Protoc. Cell Biol. 2020, 87, e105. [Google Scholar] [CrossRef]
- Carey, S.P.; Martin, K.E.; Reinhart-King, C.A. Three-dimensional collagen matrix induces a mechanosensitive invasive epithelial phenotype. Sci. Rep. 2017, 7, 42088. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Chang, J.K.; Dominguez, A.A.; Lee, H.P.; Nam, S.; Chang, J.; Varma, S.; Qi, L.S.; West, R.B.; Chaudhuri, O. YAP-independent mechanotransduction drives breast cancer progression. Nat. Commun. 2019, 10, 1848. [Google Scholar] [CrossRef] [Green Version]
- Kanopoulos, N.; Vasanthavada, N.; Baker, R.L. Design of an image edge detection filter using the Sobel operator. IEEE J. Solid State Circuits 1988, 23, 358–367. [Google Scholar] [CrossRef]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Evans, S.J.; Clift, M.J.; Singh, N.; de Oliveira Mallia, J.; Burgum, M.; Wills, J.W.; Wilkinson, T.S.; Jenkins, G.J.; Doak, S.H. Critical review of the current and future challenges associated with advanced in vitro systems towards the study of nanoparticle (secondary) genotoxicity. Mutagenesis 2017, 32, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Corvi, R.; Madia, F. In vitro genotoxicity testing-Can the performance be enhanced? Food Chem. Toxicol. 2017, 106, 600–608. [Google Scholar] [CrossRef]
- Conway, G.E.; Shah, U.K.; Llewellyn, S.; Cervena, T.; Evans, S.J.; Al Ali, A.S.; Jenkins, G.J.; Clift, M.J.D.; Doak, S.H. Adaptation of the in vitro micronucleus assay for genotoxicity testing using 3D liver models supporting longer-term exposure durations. Mutagenesis 2020, 35, 319–330. [Google Scholar] [CrossRef]
- Stampar, M.; Tomc, J.; Filipic, M.; Zegura, B. Development of in vitro 3D cell model from hepatocellular carcinoma (HepG2) cell line and its application for genotoxicity testing. Arch. Toxicol. 2019, 93, 3321–3333. [Google Scholar] [CrossRef] [PubMed]
- Reisinger, K.; Blatz, V.; Brinkmann, J.; Downs, T.R.; Fischer, A.; Henkler, F.; Hoffmann, S.; Krul, C.; Liebsch, M.; Luch, A.; et al. Validation of the 3D Skin Comet assay using full thickness skin models: Transferability and reproducibility. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018, 827, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.E.; He, X.; Muskhelishvili, L.; Malhi, P.; Mei, N.; Manjanatha, M.; Bryant, M.; Zhou, T.; Robison, T.; Guo, X. Evaluation of an in vitro three-dimensional HepaRG spheroid model for genotoxicity testing using the high-throughput CometChip platform. ALTEX 2022, 39, 583–604. [Google Scholar] [CrossRef]
- Shaw, P.; Kumar, N.; Privat-Maldonado, A.; Smits, E.; Bogaerts, A. Cold Atmospheric Plasma Increases Temozolomide Sensitivity of Three-Dimensional Glioblastoma Spheroids via Oxidative Stress-Mediated DNA Damage. Cancers 2021, 13, 1780. [Google Scholar] [CrossRef]
- McDonald, J.C.; Whitesides, G.M. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc. Chem. Res. 2002, 35, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Toepke, M.W.; Beebe, D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef]
- Wang, J.D.; Douville, N.J.; Takayama, S.; ElSayed, M. Quantitative analysis of molecular absorption into PDMS microfluidic channels. Ann. Biomed. Eng. 2012, 40, 1862–1873. [Google Scholar] [CrossRef]
- Iliescu, C.; Taylor, H.; Avram, M.; Miao, J.; Franssila, S. A practical guide for the fabrication of microfluidic devices using glass and silicon. Biomicrofluidics 2012, 6, 016505. [Google Scholar] [CrossRef] [Green Version]
- Schulze, T.; Mattern, K.; Fruh, E.; Hecht, L.; Rustenbeck, I.; Dietzel, A. A 3D microfluidic perfusion system made from glass for multiparametric analysis of stimulus-secretioncoupling in pancreatic islets. Biomed. Microdevices 2017, 19, 47. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lim, J.; Yu, J.; Ahn, J.; Lee, Y.; Jeon, N.L. Engineering tumor vasculature on an injection-molded plastic array 3D culture (IMPACT) platform. Lab Chip 2019, 19, 2071–2080. [Google Scholar] [CrossRef]
- Mottet, G.; Perez-Toralla, K.; Tulukcuoglu, E.; Bidard, F.C.; Pierga, J.Y.; Draskovic, I.; Londono-Vallejo, A.; Descroix, S.; Malaquin, L.; Louis Viovy, J. A three dimensional thermoplastic microfluidic chip for robust cell capture and high resolution imaging. Biomicrofluidics 2014, 8, 024109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, K.; Zhou, J.; Wu, H. Materials for microfluidic chip fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef]
- Gencturk, E.; Mutlu, S.; Ulgen, K.O. Advances in microfluidic devices made from thermoplastics used in cell biology and analyses. Biomicrofluidics 2017, 11, 051502. [Google Scholar] [CrossRef] [PubMed]
- Seliktar, D. Designing cell-compatible hydrogels for biomedical applications. Science 2012, 336, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, L.; Luo, C. Gel integration for microfluidic applications. Lab Chip 2016, 16, 1757–1776. [Google Scholar] [CrossRef]
- Annabi, N.; Selimovic, S.; Acevedo Cox, J.P.; Ribas, J.; Afshar Bakooshli, M.; Heintze, D.; Weiss, A.S.; Cropek, D.; Khademhosseini, A. Hydrogel-coated microfluidic channels for cardiomyocyte culture. Lab Chip 2013, 13, 3569–3577. [Google Scholar] [CrossRef] [Green Version]
- Antoine, E.E.; Vlachos, P.P.; Rylander, M.N. Review of collagen I hydrogels for bioengineered tissue microenvironments: Characterization of mechanics, structure, and transport. Tissue Eng. Part B Rev. 2014, 20, 683–696. [Google Scholar] [CrossRef] [Green Version]
- Goy, C.B.; Chaile, R.E.; Madrid, R.E. Microfluidics and hydrogel: A powerful combination. React. Funct. Polym. 2019, 145, 104314. [Google Scholar] [CrossRef]
- Leung, C.M.; De Haan, P.; Ronaldson-Bouchard, K.; Kim, G.-A.; Ko, J.; Rho, H.S.; Chen, Z.; Habibovic, P.; Jeon, N.L.; Takayama, S. A guide to the organ-on-a-chip. Nat. Rev. Methods Prim. 2022, 2, 33. [Google Scholar] [CrossRef]
- Gillet, J.P.; Varma, S.; Gottesman, M.M. The clinical relevance of cancer cell lines. J. Natl. Cancer Inst. 2013, 105, 452–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristofalo, V.J.; Lorenzini, A.; Allen, R.G.; Torres, C.; Tresini, M. Replicative senescence: A critical review. Mech. Ageing Dev. 2004, 125, 827–848. [Google Scholar] [CrossRef]
- Diederichs, S.; Tuan, R.S. Functional comparison of human-induced pluripotent stem cell-derived mesenchymal cells and bone marrow-derived mesenchymal stromal cells from the same donor. Stem Cells Dev. 2014, 23, 1594–1610. [Google Scholar] [CrossRef] [Green Version]
- Wnorowski, A.; Yang, H.; Wu, J.C. Progress, obstacles, and limitations in the use of stem cells in organ-on-a-chip models. Adv. Drug Deliv. Rev. 2019, 140, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Naumovska, E.; Aalderink, G.; Wong Valencia, C.; Kosim, K.; Nicolas, A.; Brown, S.; Vulto, P.; Erdmann, K.S.; Kurek, D. Direct On-Chip Differentiation of Intestinal Tubules from Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2020, 21, 4964. [Google Scholar] [CrossRef]
- Punt, A.; Bouwmeester, H.; Blaauboer, B.J.; Coecke, S.; Hakkert, B.; Hendriks, D.F.G.; Jennings, P.; Kramer, N.I.; Neuhoff, S.; Masereeuw, R.; et al. New approach methodologies (NAMs) for human-relevant biokinetics predictions. Meeting the paradigm shift in toxicology towards an animal-free chemical risk assessment. ALTEX 2020, 37, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Lasli, S.; Kim, H.J.; Lee, K.; Suurmond, C.E.; Goudie, M.; Bandaru, P.; Sun, W.; Zhang, S.; Zhang, N.; Ahadian, S.; et al. A Human Liver-on-a-Chip Platform for Modeling Nonalcoholic Fatty Liver Disease. Adv. Biosyst. 2019, 3, e1900104. [Google Scholar] [CrossRef]
- Jang, K.J.; Otieno, M.A.; Ronxhi, J.; Lim, H.K.; Ewart, L.; Kodella, K.R.; Petropolis, D.B.; Kulkarni, G.; Rubins, J.E.; Conegliano, D.; et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 2019, 11, eaax5516. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewski, T.; Cornforth, T.; Snow, S.A.; Ouro-Gnao, L.; Rowe, C.; Large, E.M.; Hughes, D.J. Three-dimensional perfused human in vitro model of non-alcoholic fatty liver disease. World J. Gastroenterol. 2017, 23, 204–215. [Google Scholar] [CrossRef]
- Tao, T.P.; Brandmair, K.; Gerlach, S.; Przibilla, J.; Genies, C.; Jacques-Jamin, C.; Schepky, A.; Marx, U.; Hewitt, N.J.; Maschmeyer, I.; et al. Demonstration of the first-pass metabolism in the skin of the hair dye, 4-amino-2-hydroxytoluene, using the Chip2 skin-liver microphysiological model. J. Appl. Toxicol. 2021, 41, 1553–1567. [Google Scholar] [CrossRef]
- Yang, F.; Cohen, R.N.; Brey, E.M. Optimization of Co-Culture Conditions for a Human Vascularized Adipose Tissue Model. Bioengineering 2020, 7, 114. [Google Scholar] [CrossRef]
- Chang, S.Y.; Weber, E.J.; Sidorenko, V.S.; Chapron, A.; Yeung, C.K.; Gao, C.; Mao, Q.; Shen, D.; Wang, J.; Rosenquist, T.A.; et al. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight 2017, 2, e95978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhao, Z.; Abdul Rahim, N.A.; van Noort, D.; Yu, H. Towards a human-on-chip: Culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip 2009, 9, 3185–3192. [Google Scholar] [CrossRef]
- Junaid, A.; Mashaghi, A.; Hankemeier, T.; Vulto, P. An end-user perspective on Organ-on-a-Chip: Assays and usability aspects. Curr. Opin. Biomed. Eng. 2017, 1, 15–22. [Google Scholar] [CrossRef]
- Chang, S.Y.; Voellinger, J.L.; Van Ness, K.P.; Chapron, B.; Shaffer, R.M.; Neumann, T.; White, C.C.; Kavanagh, T.J.; Kelly, E.J.; Eaton, D.L. Characterization of rat or human hepatocytes cultured in microphysiological systems (MPS) to identify hepatotoxicity. Toxicol. In Vitro 2017, 40, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Wevers, N.R.; van Vught, R.; Wilschut, K.J.; Nicolas, A.; Chiang, C.; Lanz, H.L.; Trietsch, S.J.; Joore, J.; Vulto, P. High-throughput compound evaluation on 3D networks of neurons and glia in a microfluidic platform. Sci. Rep. 2016, 6, 38856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Cai, C.; Yu, Z.; Pang, Y.; Zhou, Y.; Qian, L.; Wei, W.; Huang, Y. A microfluidic live cell assay to study anthrax toxin induced cell lethality assisted by conditioned medium. Sci. Rep. 2015, 5, 8651. [Google Scholar] [CrossRef] [Green Version]
- Sobrino, A.; Phan, D.T.; Datta, R.; Wang, X.; Hachey, S.J.; Romero-Lopez, M.; Gratton, E.; Lee, A.P.; George, S.C.; Hughes, C.C. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 2016, 6, 31589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, O.Y.F.; Villenave, R.; Cronce, M.J.; Leineweber, W.D.; Benz, M.A.; Ingber, D.E. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip 2017, 17, 2264–2271. [Google Scholar] [CrossRef]
- van der Helm, M.W.; Odijk, M.; Frimat, J.P.; van der Meer, A.D.; Eijkel, J.C.T.; van den Berg, A.; Segerink, L.I. Direct quantification of transendothelial electrical resistance in organs-on-chips. Biosens. Bioelectron. 2016, 85, 924–929. [Google Scholar] [CrossRef] [Green Version]
- Elbakary, B.; Badhan, R.K.S. A dynamic perfusion based blood-brain barrier model for cytotoxicity testing and drug permeation. Sci. Rep. 2020, 10, 3788. [Google Scholar] [CrossRef] [Green Version]
- Yalcin, Y.D.; Luttge, R. Electrical monitoring approaches in 3-dimensional cell culture systems: Toward label-free, high spatiotemporal resolution, and high-content data collection in vitro. Organs-on-a-Chip 2021, 3, 100006. [Google Scholar] [CrossRef]
- Ramadan, Q.; Ting, F.C. In vitro micro-physiological immune-competent model of the human skin. Lab Chip 2016, 16, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Zeller, P.; Legendre, A.; Jacques, S.; Fleury, M.J.; Gilard, F.; Tcherkez, G.; Leclerc, E. Hepatocytes cocultured with Sertoli cells in bioreactor favors Sertoli barrier tightness in rat. J. Appl. Toxicol. 2017, 37, 287–295. [Google Scholar] [CrossRef]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jager, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Kim, D.S.; Ha, S.K.; Choi, I.; Lee, J.M.; Sung, J.H. A pumpless multi-organ-on-a-chip (MOC) combined with a pharmacokinetic-pharmacodynamic (PK-PD) model. Biotechnol. Bioeng. 2017, 114, 432–443. [Google Scholar] [CrossRef]
- Wang, X.; Yi, L.; Mukhitov, N.; Schrell, A.M.; Dhumpa, R.; Roper, M.G. Microfluidics-to-mass spectrometry: A review of coupling methods and applications. J. Chromatogr. A 2015, 1382, 98–116. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.-C.; Zhang, W.-Z.; Chen, W.-R.; Jair, Y.-C.; Wu, Y.-H.; Liu, Y.-H.; Chen, P.-Z.; Chen, L.-Y.; Chen, P.-S. Engineering an integrated system with a high pressure polymeric microfluidic chip coupled to liquid chromatography-mass spectrometry (LC-MS) for the analysis of abused drugs. Sens. Actuators B Chem. 2022, 350, 130888. [Google Scholar] [CrossRef]
- Varuni, K.; Nivetha, V.; Gandhimathi, R. A Role of Microfluidic Chip Technology in Analytical Method Development: An Overview. NeuroQuantology 2022, 20, 2125. [Google Scholar]
- Vit, F.F.; Nunes, R.; Wu, Y.T.; Prado Soares, M.C.; Godoi, N.; Fujiwara, E.; Carvalho, H.F.; Gaziola de la Torre, L. A modular, reversible sealing, and reusable microfluidic device for drug screening. Anal. Chim. Acta 2021, 1185, 339068. [Google Scholar] [CrossRef]
- Hartung, T. Food for thought…on alternative methods for chemical safety testing. ALTEX 2010, 27, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Biomin. The Global Mycotoxin Threat. 2021. Available online: https://www.biomin.net/science-hub/world-mycotoxin-survey-impact-2021/ (accessed on 14 December 2022).
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [Green Version]
- Baek, N.; Seo, O.W.; Kim, M.; Hulme, J.; An, S.S. Monitoring the effects of doxorubicin on 3D-spheroid tumor cells in real-time. OncoTargets Ther. 2016, 9, 7207–7218. [Google Scholar] [CrossRef] [Green Version]
- De Simone, U.; Roccio, M.; Gribaldo, L.; Spinillo, A.; Caloni, F.; Coccini, T. Human 3D Cultures as Models for Evaluating Magnetic Nanoparticle CNS Cytotoxicity after Short- and Repeated Long-Term Exposure. Int. J. Mol. Sci. 2018, 19, 1993. [Google Scholar] [CrossRef] [Green Version]
- Goodman, T.T.; Ng, C.P.; Pun, S.H. 3-D tissue culture systems for the evaluation and optimization of nanoparticle-based drug carriers. Bioconjug. Chem. 2008, 19, 1951–1959. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Lilly, G.D.; Doty, R.C.; Podsiadlo, P.; Kotov, N.A. In vitro toxicity testing of nanoparticles in 3D cell culture. Small 2009, 5, 1213–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csenki, Z.; Garai, E.; Faisal, Z.; Csepregi, R.; Garai, K.; Sipos, D.K.; Szabo, I.; Koszegi, T.; Czeh, A.; Czompoly, T.; et al. The individual and combined effects of ochratoxin A with citrinin and their metabolites (ochratoxin B, ochratoxin C, and dihydrocitrinone) on 2D/3D cell cultures, and zebrafish embryo models. Food Chem. Toxicol. 2021, 158, 112674. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Ejaz, S.; Chekarova, I.; Sukura, A.; Ashraf, M.; Lim, C.W. Cytotoxicity of fumonisin B(1) in spheroid and monolayer cultures of rat hepatocytes. Drug Chem. Toxicol. 2008, 31, 339–352. [Google Scholar] [CrossRef]
- Bell, C.C.; Hendriks, D.F.; Moro, S.M.; Ellis, E.; Walsh, J.; Renblom, A.; Fredriksson Puigvert, L.; Dankers, A.C.; Jacobs, F.; Snoeys, J.; et al. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 2016, 6, 25187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, U.K.; Mallia, J.O.; Singh, N.; Chapman, K.E.; Doak, S.H.; Jenkins, G.J.S. Reprint of: A three-dimensional in vitro HepG2 cells liver spheroid model for genotoxicity studies. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018, 834, 35–41. [Google Scholar] [CrossRef]
- Ma, X.; Sun, J.; Ye, Y.; Ji, J.; Sun, X. Application of triple co-cultured cell spheroid model for exploring hepatotoxicity and metabolic pathway of AFB1. Sci. Total. Environ. 2022, 807, 150840. [Google Scholar] [CrossRef]
- Ma, X.; Ye, Y.; Sun, J.; Ji, J.; Wang, J.S.; Sun, X. Coexposure of Cyclopiazonic Acid with Aflatoxin B1 Involved in Disrupting Amino Acid Metabolism and Redox Homeostasis Causing Synergistic Toxic Effects in Hepatocyte Spheroids. J. Agric. Food Chem. 2022, 70, 5166–5176. [Google Scholar] [CrossRef]
- Neal, G.E.; Eaton, D.L.; Judah, D.J.; Verma, A. Metabolism and toxicity of aflatoxins M1 and B1 in human-derived in vitro systems. Toxicol. Appl. Pharmacol. 1998, 151, 152–158. [Google Scholar] [CrossRef]
- Bovard, D.; Sandoz, A.; Luettich, K.; Frentzel, S.; Iskandar, A.; Marescotti, D.; Trivedi, K.; Guedj, E.; Dutertre, Q.; Peitsch, M.C.; et al. A lung/liver-on-a-chip platform for acute and chronic toxicity studies. Lab Chip 2018, 18, 3814–3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schimek, K.; Frentzel, S.; Luettich, K.; Bovard, D.; Rutschle, I.; Boden, L.; Rambo, F.; Erfurth, H.; Dehne, E.M.; Winter, A.; et al. Human multi-organ chip co-culture of bronchial lung culture and liver spheroids for substance exposure studies. Sci. Rep. 2020, 10, 7865. [Google Scholar] [CrossRef]
- Imaoka, T.; Yang, J.; Wang, L.; McDonald, M.G.; Afsharinejad, Z.; Bammler, T.K.; Van Ness, K.; Yeung, C.K.; Rettie, A.E.; Himmelfarb, J.; et al. Microphysiological system modeling of ochratoxin A-associated nephrotoxicity. Toxicology 2020, 444, 152582. [Google Scholar] [CrossRef] [PubMed]
- Hanyu, H.; Yokoi, Y.; Nakamura, K.; Ayabe, T.; Tanaka, K.; Uno, K.; Miyajima, K.; Saito, Y.; Iwatsuki, K.; Shimizu, M.; et al. Mycotoxin Deoxynivalenol Has Different Impacts on Intestinal Barrier and Stem Cells by Its Route of Exposure. Toxins 2020, 12, 610. [Google Scholar] [CrossRef]
- Li, X.G.; Zhu, M.; Chen, M.X.; Fan, H.B.; Fu, H.L.; Zhou, J.Y.; Zhai, Z.Y.; Gao, C.Q.; Yan, H.C.; Wang, X.Q. Acute exposure to deoxynivalenol inhibits porcine enteroid activity via suppression of the Wnt/beta-catenin pathway. Toxicol. Lett. 2019, 305, 19–31. [Google Scholar] [CrossRef]
- Poschl, F.; Hoher, T.; Pirklbauer, S.; Wolinski, H.; Lienhart, L.; Ressler, M.; Riederer, M. Dose and route dependent effects of the mycotoxin deoxynivalenol in a 3D gut-on-a-chip model with flow. Toxicol. In Vitro 2023, 88, 105563. [Google Scholar] [CrossRef]
- DiProspero, T.J.; Dalrymple, E.; Lockett, M.R. Physiologically relevant oxygen tensions differentially regulate hepatotoxic responses in HepG2 cells. Toxicol. In Vitro 2021, 74, 105156. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Jia, B.; Liu, N.; Yu, D.; Zhang, S.; Wu, A. Fumonisin B1 triggers carcinogenesis via HDAC/PI3K/Akt signalling pathway in human esophageal epithelial cells. Sci. Total. Environ. 2021, 787, 147405. [Google Scholar] [CrossRef]
- Hardwick, R.N.; Betts, C.J.; Whritenour, J.; Sura, R.; Thamsen, M.; Kaufman, E.H.; Fabre, K. Drug-induced skin toxicity: Gaps in preclinical testing cascade as opportunities for complex in vitro models and assays. Lab Chip 2020, 20, 199–214. [Google Scholar] [CrossRef]
- Gordon, S.; Daneshian, M.; Bouwstra, J.; Caloni, F.; Constant, S.; Davies, D.E.; Dandekar, G.; Guzman, C.A.; Fabian, E.; Haltner, E.; et al. Non-animal models of epithelial barriers (skin, intestine and lung) in research, industrial applications and regulatory toxicology. ALTEX 2015, 32, 327–378. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zingales, V.; Esposito, M.R.; Torriero, N.; Taroncher, M.; Cimetta, E.; Ruiz, M.-J. The Growing Importance of Three-Dimensional Models and Microphysiological Systems in the Assessment of Mycotoxin Toxicity. Toxins 2023, 15, 422. https://doi.org/10.3390/toxins15070422
Zingales V, Esposito MR, Torriero N, Taroncher M, Cimetta E, Ruiz M-J. The Growing Importance of Three-Dimensional Models and Microphysiological Systems in the Assessment of Mycotoxin Toxicity. Toxins. 2023; 15(7):422. https://doi.org/10.3390/toxins15070422
Chicago/Turabian StyleZingales, Veronica, Maria Rosaria Esposito, Noemi Torriero, Mercedes Taroncher, Elisa Cimetta, and María-José Ruiz. 2023. "The Growing Importance of Three-Dimensional Models and Microphysiological Systems in the Assessment of Mycotoxin Toxicity" Toxins 15, no. 7: 422. https://doi.org/10.3390/toxins15070422
APA StyleZingales, V., Esposito, M. R., Torriero, N., Taroncher, M., Cimetta, E., & Ruiz, M. -J. (2023). The Growing Importance of Three-Dimensional Models and Microphysiological Systems in the Assessment of Mycotoxin Toxicity. Toxins, 15(7), 422. https://doi.org/10.3390/toxins15070422







