Recombination in Bacterial Genomes: Evolutionary Trends
Abstract
1. Introduction
2. Functional Impact of Recombination
2.1. Ecological Adaptation
2.2. Symbiotic Relationships
2.3. Pathogenesis
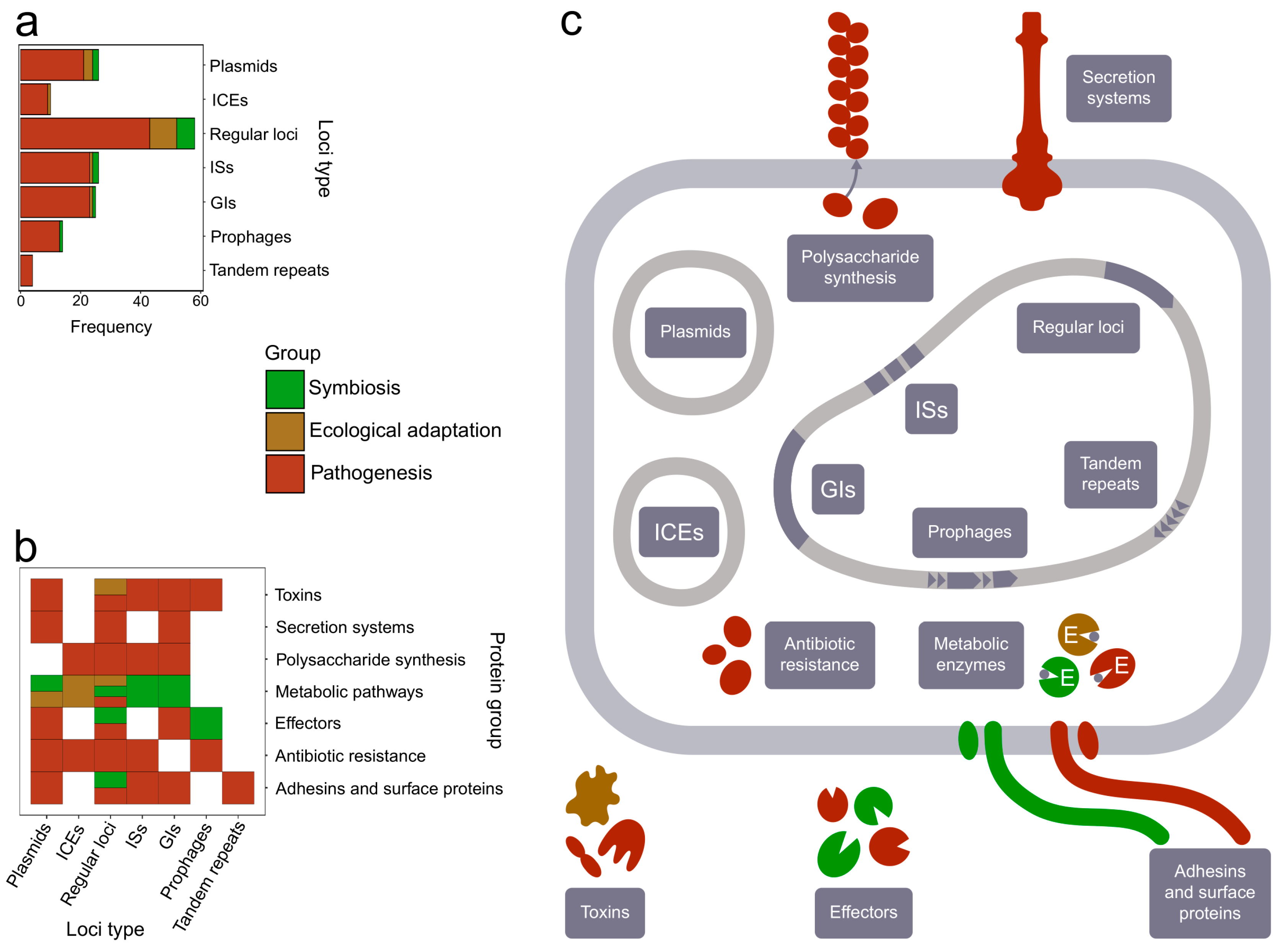
3. The Distribution of Recombination Events among the Bacterial Genome
3.1. Plasmids
3.2. Insertion Sequences
3.3. Long Genomic Regions
3.4. Repeats
3.5. Genomic Islands and Integrative Elements
3.6. Prophages
4. Functional Characteristics of Genes Subjected to Recombination
4.1. Surface Proteins and Adhesion Factors
4.2. Secretion Systems
4.3. Infection Effectors
4.4. Toxins
4.5. Antibiotic Resistance Genes
4.6. Polysaccharide Synthesis
4.7. Metabolic Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HGT | Horizontal gene transfer |
| LGT | Lateral gene transfer |
| HR | Homologous recombination |
| ICEs | Integrative and conjugative elements |
| MGEs | Mobile genetic elements |
| SSB | Single-stranded DNA-binding protein |
| SCCmec | Staphylococcal cassette chromosome element |
| AICEs | Actinomycete integrative and conjugative elements |
| SI | Symbiotic island |
| GTAs | Gene transfer agents |
| GIs | Genomic islands |
References
- Ivars-Martínez, E.; D’Auria, G.; Rodríguez-Valera, F.; Sánchez-Porro, C.; Ventosa, A.; Joint, I.; Mühling, M. Biogeography of the Ubiquitous Marine Bacterium Alteromonas macleodii Determined by Multilocus Sequence Analysis. Mol. Ecol. 2008, 17, 4092–4106. [Google Scholar] [CrossRef] [PubMed]
- Chochua, S.; Rivers, J.; Mathis, S.; Li, Z.; Velusamy, S.; McGee, L.; Van Beneden, C.; Li, Y.; Metcalf, B.J.; Beall, B. Emergent Invasive Group A Streptococcus dysgalactiae subsp. equisimilis, United States, 2015–2018. Emerg. Infect. Dis. 2019, 25, 1543–1547. [Google Scholar] [CrossRef]
- Emamalipour, M.; Seidi, K.; Zununi Vahed, S.; Jahanban-Esfahlan, A.; Jaymand, M.; Majdi, H.; Amoozgar, Z.; Chitkushev, L.T.; Javaheri, T.; Jahanban-Esfahlan, R.; et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front. Cell Dev. Biol. 2020, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.R.; Cornejo, O.E. The Population and Evolutionary Dynamics of Homologous Gene Recombination in Bacteria. PLoS Genet. 2009, 5, e1000601. [Google Scholar] [CrossRef]
- Shikov, A.E.; Malovichko, Y.V.; Nizhnikov, A.A.; Antonets, K.S. Current Methods for Recombination Detection in Bacteria. Int. J. Mol. Sci. 2022, 23, 6257. [Google Scholar] [CrossRef] [PubMed]
- Fondi, M.; Bacci, G.; Brilli, M.; Papaleo, M.C.; Mengoni, A.; Vaneechoutte, M.; Dijkshoorn, L.; Fani, R. Exploring the Evolutionary Dynamics of Plasmids: The Acinetobacter Pan-Plasmidome. BMC Evol. Biol. 2010, 10, 59. [Google Scholar] [CrossRef]
- Blakely, G.W. Mechanisms of Horizontal Gene Transfer and DNA Recombination; Academic Press: Boston, MA, USA, 2015; Volume 1–3, ISBN 9780123971692. [Google Scholar]
- Wang, J.; Li, Y.; Pinto-Tomás, A.A.; Cheng, K.; Huang, Y. Habitat Adaptation Drives Speciation of a Streptomyces Species with Distinct Habitats and Disparate Geographic Origins. mBio 2022, 13, e0278121. [Google Scholar] [CrossRef]
- Nudel, K.; Zhao, X.; Basu, S.; Dong, X.; Hoffmann, M.; Feldgarden, M.; Allard, M.; Klompas, M.; Bry, L. Genomics of Corynebacterium striatum, an Emerging Multidrug-Resistant Pathogen of Immunocompromised Patients. Clin. Microbiol. Infect. 2018, 24, 1016.e7–1016.e13. [Google Scholar] [CrossRef]
- Hao, L.; Holden, M.T.G.; Wang, X.; Andrew, L.; Wellnitz, S.; Hu, F.; Whaley, M.; Sammons, S.; Knipe, K.; Frace, M.; et al. Distinct Evolutionary Patterns of Neisseria meningitidis Serogroup B Disease Outbreaks at Two Universities in the USA. Microb. Genom. 2018, 4, e000155. [Google Scholar] [CrossRef]
- Guo, Q.; Mustapha, M.M.; Chen, M.; Qu, D.; Zhang, X.; Chen, M.; Doi, Y.; Wang, M.; Harrison, L.H. Evolution of Sequence Type 4821 Clonal Complex Meningococcal Strains in China from Prequinolone to Quinolone Era, 1972–2013. Emerg. Infect. Dis. 2018, 24, 683–690. [Google Scholar] [CrossRef]
- Tong, W.; Li, X.; Wang, E.; Cao, Y.; Chen, W.; Tao, S.; Wei, G. Genomic Insight into the Origins and Evolution of Symbiosis Genes in Phaseolus vulgaris Microsymbionts. BMC Genom. 2020, 21, 186. [Google Scholar] [CrossRef]
- Bosch, R.; GarcıÍa-Valdés, E.; Moore, E.R.B. Complete Nucleotide Sequence and Evolutionary Significance of a Chromosomally Encoded Naphthalene-Degradation Lower Pathway from Pseudomonas stutzeri AN10. Gene 2000, 245, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Didelot, X.; Maiden, M.C.J. Impact of Recombination on Bacterial Evolution. Trends Microbiol. 2010, 18, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral Gene Transfer and the Nature of Bacterial Innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Chun, J.; Grim, C.J.; Hasan, N.A.; Lee, J.H.; Choi, S.Y.; Haley, B.J.; Taviani, E.; Jeon, Y.-S.; Kim, D.-W.; Lee, J.-H.; et al. Comparative Genomics Reveals Mechanism for Short-Term and Long-Term Clonal Transitions in Pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2009, 106, 15442–15447. [Google Scholar] [CrossRef] [PubMed]
- Julin, D.A. Recombination: Mechanisms, Pathways, and Applications; Wells, R., Bond, J., Klinman, J., Masters, B., Bell, E., Eds.; Molecular Life Sciences; Springer: New York, NY, USA, 2017; pp. 1–28. [Google Scholar] [CrossRef]
- Rocha, E.P.C.; Cornet, E.; Michel, B. Comparative and Evolutionary Analysis of the Bacterial Homologous Recombination Systems. PLoS Genet. 2005, 1, e15. [Google Scholar] [CrossRef] [PubMed]
- Gil, R.; Sabater-Muñoz, B.; Perez-Brocal, V.; Silva, F.J.; Latorre, A. Plasmids in the Aphid Endosymbiont Buchnera aphidicola with the Smallest Genomes. A Puzzling Evolutionary Story. Gene 2006, 370, 17–25. [Google Scholar] [CrossRef]
- Williams, L.E.; Wernegreen, J.J. Genome Evolution in an Ancient Bacteria-Ant Symbiosis: Parallel Gene Loss among Blochmannia Spanning the Origin of the Ant Tribe Camponotini. PeerJ 2015, 3, e881. [Google Scholar] [CrossRef]
- Zhou, K.; Aertsen, A.; Michiels, C.W. The Role of Variable DNA Tandem Repeats in Bacterial Adaptation. FEMS Microbiol. Rev. 2014, 38, 119–141. [Google Scholar] [CrossRef]
- Iranzo, J.; Wolf, Y.I.; Koonin, E.V.; Sela, I. Gene Gain and Loss Push Prokaryotes beyond the Homologous Recombination Barrier and Accelerate Genome Sequence Divergence. Nat. Commun. 2019, 10, 5376. [Google Scholar] [CrossRef] [PubMed]
- Ely, B. Recombination and Gene Loss Occur Simultaneously during Bacterial Horizontal Gene Transfer. PLoS ONE 2020, 15, e0227987. [Google Scholar] [CrossRef] [PubMed]
- Gratia, J.-P. Genetic Recombinational Events in Prokaryotes and Their Viruses: Insight into the Study of Evolution and Biodiversity. Antonie Leeuwenhoek 2017, 110, 1493–1514. [Google Scholar] [CrossRef]
- Spagnoletti, M.; Ceccarelli, D.; Rieux, A.; Fondi, M.; Taviani, E.; Fani, R.; Colombo, M.M.; Colwell, R.R.; Balloux, F. Acquisition and Evolution of SXT-R391 Integrative Conjugative Elements in the Seventh-Pandemic Vibrio cholerae Lineage. mBio 2014, 5, e01356-14. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.R.; Sekulovic, O.; Dupuy, B.; Soutourina, O.; Monot, M.; Fortier, L.-C. High Prevalence and Genetic Diversity of Large PhiCD211 (PhiCDIF1296T)-Like Prophages in Clostridioides difficile. Appl. Environ. Microbiol. 2018, 84, e02164-17. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Wang, J.; Bertels, F.; Giordano, P.R.; Chilvers, M.I.; Huntley, R.B.; Vargas, J.M.; Sundin, G.W.; Jacobs, J.L.; Yang, C.-H. Recombination of Virulence Genes in Divergent Acidovorax avenae Strains That Infect a Common Host. Mol. Plant-Microbe Interact. 2017, 30, 813–828. [Google Scholar] [CrossRef]
- Vos, M.; Didelot, X. A Comparison of Homologous Recombination Rates in Bacteria and Archaea. ISME J. 2009, 3, 199–208. [Google Scholar] [CrossRef]
- González-Torres, P.; Rodríguez-Mateos, F.; Antón, J.; Gabaldón, T. Impact of Homologous Recombination on the Evolution of Prokaryotic Core Genomes. mBio 2019, 10, e02494-18. [Google Scholar] [CrossRef]
- Helgason, E.; Caugant, D.A.; Lecadet, M.M.; Chen, Y.; Mahillon, J.; Lövgren, A.; Hegna, I.; Kvaløy, K.; Kolstø, A.B. Genetic Diversity of Bacillus cereus/B. thuringiensis Isolates from Natural Sources. Curr. Microbiol. 1998, 37, 80–87. [Google Scholar] [CrossRef]
- Liu, Q.; Xin, Y.H.; Zhou, Y.G.; Chen, W.X. Multilocus Sequence Analysis of Homologous Recombination and Diversity in Arthrobacter sensu lato Named Species and Glacier-Inhabiting Strains. Syst. Appl. Microbiol. 2018, 41, 23–29. [Google Scholar] [CrossRef]
- Hoetzinger, M.; Hahn, M.W. Genomic Divergence and Cohesion in a Species of Pelagic Freshwater Bacteria. BMC Genom. 2017, 18, 794. [Google Scholar] [CrossRef] [PubMed]
- Nesbø, C.L.; Swithers, K.S.; Dahle, H.; Haverkamp, T.H.A.; Birkeland, N.K.; Sokolova, T.; Kublanov, I.; Zhaxybayeva, O. Evidence for Extensive Gene Flow and Thermotoga Subpopulations in Subsurface and Marine Environments. ISME J. 2015, 9, 1532–1542. [Google Scholar] [CrossRef] [PubMed]
- Mihasan, M.; Brandsch, R. PAO1 of Arthrobacter nicotinovorans and the Spread of Catabolic Traits by Horizontal Gene Transfer in Gram-Positive Soil Bacteria. J. Mol. Evol. 2013, 77, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Sogge, H.; Rohrlack, T.; Rounge, T.B.; Sønstebø, J.H.; Tooming-Klunderud, A.; Kristensen, T.; Jakobsena, K.S. Gene Flow, Recombination, and Selection in Cyanobacteria: Population Structure of Geographically Related Planktothrix Freshwater Strains. Appl. Environ. Microbiol. 2013, 79, 508–515. [Google Scholar] [CrossRef]
- Rounge, T.B.; Rohrlack, T.; Kristensen, T.; Jakobsen, K.S. Recombination and Selectional Forces in Cyanopeptolin NRPS Operons from Highly Similar, but Geographically Remote Planktothrix Strains. BMC Microbiol. 2008, 8, 141. [Google Scholar] [CrossRef]
- Tidjani, A.R.; Lorenzi, J.N.; Toussaint, M.; Van Dijk, E.; Naquin, D.; Lespinet, O.; Bontemps, C.; Leblond, P. Massive Gene Flux Drives Genome Diversity between Sympatric Streptomyces Conspecifics. mBio 2019, 10, e01533-19. [Google Scholar] [CrossRef]
- Mouton, L.; Thierry, M.; Henri, H.; Baudin, R.; Gnankine, O.; Reynaud, B.; Zchori-Fein, E.; Becker, N.; Fleury, F.; Delatte, H. Evidence of Diversity and Recombination in Arsenophonus Symbionts of the Bemisia tabaci Species Complex. BMC Microbiol. 2012, 12, S10. [Google Scholar] [CrossRef]
- Naamala, J.; Jaiswal, S.K.; Dakora, F.D. Microsymbiont Diversity and Phylogeny of Native Bradyrhizobia Associated with Soybean (Glycine max L. Merr.) Nodulation in South African Soils. Syst. Appl. Microbiol. 2016, 39, 336–344. [Google Scholar] [CrossRef]
- Parker, M.A. Legumes Select Symbiosis Island Sequence Variants in Bradyrhizobium. Mol. Ecol. 2012, 21, 1769–1778. [Google Scholar] [CrossRef]
- Guo, H.J.; Wang, E.T.; Zhang, X.X.; Li, Q.Q.; Zhang, Y.M.; Tian, C.F.; Chen, W.X. Replicon-Dependent Differentiation of Symbiosis-Related Genes in Sinorhizobium Strains Nodulating Glycine Max. Appl. Environ. Microbiol. 2014, 80, 1245–1255. [Google Scholar] [CrossRef]
- Altamia, M.A.; Wood, N.; Fung, J.M.; Dedrick, S.; Linton, E.W.; Concepcion, G.P.; Haygood, M.G.; Distel, D.L. Genetic Differentiation among Isolates of Teredinibacter turnerae, a Widely Occurring Intracellular Endosymbiont of Shipworms. Mol. Ecol. 2014, 23, 1418–1432. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiong, X.; Cao, W.; Zhang, C.; Werren, J.H.; Wang, X. Phylogenomic Analysis of Wolbachia Strains Reveals Patterns of Genome Evolution and Recombination. Genome Biol. Evol. 2020, 12, 2508–2520. [Google Scholar] [CrossRef] [PubMed]
- Siozios, S.; Ioannidis, P.; Klasson, L.; Andersson, S.G.E.; Braig, H.R.; Bourtzis, K. The Diversity and Evolution of Wolbachia Ankyrin Repeat Domain Genes. PLoS ONE 2013, 8, e55390. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, M.; Atyame, C.; Beji, M.; Justy, F.; Cohen-Gonsaud, M.; Sicard, M.; Weill, M. Culex pipiens Crossing Type Diversity Is Governed by an Amplified and Polymorphic Operon of Wolbachia. Nat. Commun. 2018, 9, 319. [Google Scholar] [CrossRef]
- Sánchez-Busó, L.; Comas, I.; Jorques, G.; González-Candelas, F. Recombination Drives Genome Evolution in Outbreak-Related Legionella pneumophila Isolates. Nat. Genet. 2014, 46, 1205–1211. [Google Scholar] [CrossRef]
- Hao, W.; Allen, V.G.; Jamieson, F.B.; Low, D.E.; Alexander, D.C. Phylogenetic Incongruence in E. coli O104: Understanding the Evolutionary Relationships of Emerging Pathogens in the Face of Homologous Recombination. PLoS ONE 2012, 7, e33971. [Google Scholar] [CrossRef]
- Orsi, R.H.; Borowsky, M.L.; Lauer, P.; Young, S.K.; Nusbaum, C.; Galagan, J.E.; Birren, B.W.; Ivy, R.A.; Sun, Q.; Graves, L.M.; et al. Short-term Genome Evolution of Listeria monocytogenes in a Non-Controlled Environment. BMC Genom. 2008, 9, 539. [Google Scholar] [CrossRef]
- Lamelas, A.; Harris, S.R.; Röltgen, K.; Dangy, J.P.; Hauser, J.; Kingsley, R.A.; Connor, T.R.; Sie, A.; Hodgson, A.; Dougan, G.; et al. Emergence of a New Epidemic Neisseria meningitidis Serogroup a Clone in the African Meningitis Belt: High-Resolution Picture of Genomic Changes That Mediate Immune Evasion. mBio 2014, 5, e01974-14. [Google Scholar] [CrossRef]
- Caimi, K.; Repetto, S.A.; Varni, V.; Ruybal, P. Leptospira Species Molecular Epidemiology in the Genomic Era. Infect. Genet. Evol. 2017, 54, 478–485. [Google Scholar] [CrossRef]
- Kim, G.; Ha, N.Y.; Min, C.K.; Kim, H.I.; Yen, N.T.H.; Lee, K.H.; Oh, I.; Kang, J.S.; Choi, M.S.; Kim, I.S.; et al. Diversification of Orientia tsutsugamushi Genotypes by Intragenic Recombination and Their Potential Expansion in Endemic Areas. PLoS Neglected Trop. Dis. 2017, 11, e5408. [Google Scholar] [CrossRef]
- Donati, C.; Hiller, N.L.; Tettelin, H.; Muzzi, A.; Croucher, N.J.; Angiuoli, S.V.; Oggioni, M.; Dunning Hotopp, J.C.; Hu, F.Z.; Riley, D.R.; et al. Structure and Dynamics of the Pan-Genome of Streptococcus pneumoniae and Closely Related Species. Genome Biol. 2010, 11, R107. [Google Scholar] [CrossRef] [PubMed]
- Bisharat, N.; Cohen, D.I.; Maiden, M.C.; Crook, D.W.; Peto, T.; Harding, R.M. The Evolution of Genetic Structure in the Marine Pathogen, Vibrio vulnificus. Infect. Genet. Evol. 2007, 7, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Ch’Ng, S.L.; Octavia, S.; Xia, Q.; Duong, A.; Tanaka, M.M.; Fukushima, H.; Lan, R. Population Structure and Evolution of Pathogenicity of Yersinia pseudotuberculosis. Appl. Environ. Microbiol. 2011, 77, 768–775. [Google Scholar] [CrossRef]
- Waine, D.J.; Honeybourne, D.; Smith, E.G.; Whitehouse, J.L.; Dowson, C.G. Cross-Sectional and Longitudinal Multilocus Sequence Typing of Pseudomonas aeruginosa in Cystic Fibrosis Sputum Samples. J. Clin. Microbiol. 2009, 47, 3444–3448. [Google Scholar] [CrossRef]
- Feng, L.; Reeves, P.R.; Lan, R.; Ren, Y.; Gao, C.; Zhou, Z.; Ren, Y.; Cheng, J.; Wang, W.; Wang, J.; et al. A Recalibrated Molecular Clock and Independent Origins for the Cholera Pandemic Clones. PLoS ONE 2008, 3, e4053. [Google Scholar] [CrossRef]
- Biggs, P.J.; Fearnhead, P.; Hotter, G.; Mohan, V.; Collins-Emerson, J.; Kwan, E.; Besser, T.E.; Cookson, A.; Carter, P.E.; French, N.P. Whole-Genome Comparison of Two Campylobacter jejuni Isolates of the Same Sequence Type Reveals Multiple Loci of Different Ancestral Lineage. PLoS ONE 2011, 6, e27121. [Google Scholar] [CrossRef]
- Štaudová, B.; Strouhal, M.; Zobaníková, M.; Čejková, D.; Fulton, L.L.; Chen, L.; Giacani, L.; Centurion-Lara, A.; Bruisten, S.M.; Sodergren, E.; et al. Whole Genome Sequence of the Treponema pallidum subsp. endemicum Strain Bosnia A: The Genome Is Related to Yaws Treponemes but Contains Few Loci Similar to Syphilis Treponemes. PLoS Neglected Trop. Dis. 2014, 8, e3261. [Google Scholar] [CrossRef] [PubMed]
- Aujoulat, F.; Romano-Bertrand, S.; Masnou, A.; Marchandin, H.; Jumas-Bilak, E. Niches, Population Structure and Genome Reduction in Ochrobactrum intermedium: Clues to Technology-Driven Emergence of Pathogens. PLoS ONE 2014, 9, e83376. [Google Scholar] [CrossRef]
- McMillan, D.J.; Kaul, S.Y.; Bramhachari, P.V.; Smeesters, P.R.; Vu, T.; Karmarkar, M.G.; Shaila, M.S.; Sriprakash, K.S. Recombination Drives Genetic Diversification of Streptococcus dysgalactiae subspecies equisimilis in a Region of Streptococcal Endemicity. PLoS ONE 2011, 6, e21346. [Google Scholar] [CrossRef]
- Desai, P.T.; Porwollik, S.; Long, F.; Cheng, P.; Wollam, A.; Clifton, S.W.; Weinstock, G.M.; McClelland, M. EvolutionaryGenomics of Salmonella enterica Subspecies. mBio 2013, 4, e00579-12. [Google Scholar] [CrossRef]
- Didelot, X.; Achtman, M.; Parkhill, J.; Thomson, N.R.; Falush, D. A Bimodal Pattern of Relatedness between the Salmonella Paratyphi A and Typhi Genomes: Convergence or Divergence by Homologous Recombination? Genome Res. 2007, 17, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Holt, K.E.; Thomson, N.R.; Wain, J.; Langridge, G.C.; Hasan, R.; Bhutta, Z.A.; Quail, M.A.; Norbertczak, H.; Walker, D.; Simmonds, M.; et al. Pseudogene Accumulation in the Evolutionary Histories of Salmonella enterica Serovars Paratyphi A and Typhi. BMC Genom. 2009, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.J.; Didelot, X.; Rothschild, J.; De Vries, H.J.C.; Morré, S.A.; Read, T.D.; Dean, D. Population Genomics of Chlamydia trachomatis: Insights on Drift, Selection, Recombination, and Population Structure. Mol. Biol. Evol. 2012, 29, 3933–3946. [Google Scholar] [CrossRef] [PubMed]
- Sawires, Y.S.; Songer, J.G. Clostridium perfringens: Insight into Virulence Evolution and Population Structure. Anaerobe 2006, 12, 23–43. [Google Scholar] [CrossRef]
- Wu, J.; Yu, T.; Bao, Q.; Zhao, F. Evidence of Extensive Homologous Recombination in the Core Genome of Rickettsia. Comp. Funct. Genom. 2009, 2009, 510270. [Google Scholar] [CrossRef]
- Hernández-López, A.; Chabrol, O.; Royer-Carenzi, M.; Merhej, V.; Pontarotti, P.; Raoult, D. To Tree or Not to Tree? Genome-Wide Quantification of Recombination and Reticulate Evolution during the Diversification of Strict Intracellular Bacteria. Genome Biol. Evol. 2013, 5, 2305–2317. [Google Scholar] [CrossRef]
- Didelot, X.; Nell, S.; Yang, I.; Woltemate, S.; Van Der Merwe, S.; Suerbaum, S. Genomic Evolution and Transmission of Helicobacter pylori in Two South African Families. Proc. Natl. Acad. Sci. USA 2013, 110, 13880–13885. [Google Scholar] [CrossRef]
- Krebes, J.; Didelot, X.; Kennemann, L.; Suerbaum, S. Bidirectional Genomic Exchange between Helicobacter pylori Strains from a Family in Coventry, United Kingdom. Int. J. Med. Microbiol. 2014, 304, 1135–1146. [Google Scholar] [CrossRef]
- Lara-Ramírez, E.E.; Segura-Cabrera, A.; Guo, X.; Yu, G.; García-Pérez, C.A.; Rodríguez-Pérez, M.A. New Implications on Genomic Adaptation Derived from the Helicobacter pylori Genome Comparison. PLoS ONE 2011, 6, e17300. [Google Scholar] [CrossRef]
- Paziewska, A.; Harris, P.D.; Zwolińska, L.; Bajer, A.; Siński, E. Recombination Within and Between Species of the Alpha Proteobacterium Bartonella Infecting Rodents. Microb. Ecol. 2011, 61, 134–145. [Google Scholar] [CrossRef]
- Bai, Y.; Hayman, D.T.S.; McKee, C.D.; Kosoy, M.Y. Classification of Bartonella Strains Associated with Straw-Colored Fruit Bats (Eidolon helvum) across Africa Using a Multi-Locus Sequence Typing Platform. PLoS Neglected Trop. Dis. 2015, 9, e3478. [Google Scholar] [CrossRef] [PubMed]
- Berglund, E.C.; Ellegaard, K.; Granberg, F.; Xie, Z.; Maruyama, S.; Kosoy, M.Y.; Birtles, R.J.; Andersson, S.G.E. Rapid Diversification by Recombination in Bartonella grahamii from Wild Rodents in Asia Contrasts with Low Levels of Genomic Divergence in Northern Europe and America. Mol. Ecol. 2010, 19, 2241–2255. [Google Scholar] [CrossRef] [PubMed]
- Knupp, C.; Wiens, G.D.; Faisal, M.; Call, D.R.; Cain, K.D.; Nicolas, P.; Van Vliet, D.; Yamashita, C.; Ferguson, J.A.; Meuninck, D.; et al. Large-Scale Analysis of Flavobacterium psychrophilum Multilocus Sequence Typing Genotypes Recovered from North American Salmonids Indicates That Both Newly Identified and Recurrent Clonal Complexes Are Associated with Disease. Appl. Environ. Microbiol. 2019, 85, e02305-18. [Google Scholar] [CrossRef] [PubMed]
- Athey, T.B.T.; Auger, J.-P.; Teatero, S.; Dumesnil, A.; Takamatsu, D.; Wasserscheid, J.; Dewar, K.; Gottschalk, M.; Fittipaldi, N. Complex Population Structure and Virulence Differences among Serotype 2 Streptococcus suis Strains Belonging to Sequence Type 28. PLoS ONE 2015, 10, e137760. [Google Scholar] [CrossRef]
- Cangi, N.; Gordon, J.L.; Bournez, L.; Pinarello, V.; Aprelon, R.; Huber, K.; Lefrançois, T.; Neves, L.; Meyer, D.F.; Vachiéry, N. Recombination Is a Major Driving Force of Genetic Diversity in the Anaplasmataceae Ehrlichia ruminantium. Front. Cell. Infect. Microbiol. 2016, 6, 111. [Google Scholar] [CrossRef]
- Adakal, H.; Meyer, D.F.; Carasco-Lacombe, C.; Pinarello, V.; Allègre, F.; Huber, K.; Stachurski, F.; Morand, S.; Martinez, D.; Lefrançois, T.; et al. MLST Scheme of Ehrlichia ruminantium: Genomic Stasis and Recombination in Strains from Burkina-Faso. Infect. Genet. Evol. 2009, 9, 1320–1328. [Google Scholar] [CrossRef]
- Gordon, J.L.; Lefeuvre, P.; Escalon, A.; Barbe, V.; Cruveiller, S.; Gagnevin, L.; Pruvost, O. Comparative Genomics of 43 Strains of Xanthomonas citri pv. Citri Reveals the Evolutionary Events Giving Rise to Pathotypes with Different Host Ranges. BMC Genom. 2015, 16, 1098. [Google Scholar] [CrossRef]
- Timilsina, S.; Jibrin, M.O.; Potnis, N.; Minsavage, G.V.; Kebede, M.; Schwartz, A.; Bart, R.; Staskawicz, B.; Boyer, C.; Vallad, G.E.; et al. Multilocus Sequence Analysis of Xanthomonads Causing Bacterial Spot of Tomato and Pepper Plants Reveals Strains Generated by Recombination among Species and Recent Global Spread of Xanthomonas gardneri. Appl. Environ. Microbiol. 2015, 81, 1520–1529. [Google Scholar] [CrossRef]
- Newberry, E.A.; Bhandari, R.; Minsavage, G.V.; Timilsina, S.; Jibrin, M.O.; Kemble, J.; Sikora, E.J.; Jones, J.B.; Potnis, N. Independent Evolution with the Gene Flux Originating from Multiple Xanthomonas Species Explains Genomic Heterogeneity in Xanthomonas perforans. Appl. Environ. Microbiol. 2019, 85, e00885-19. [Google Scholar] [CrossRef]
- Timilsina, S.; Pereira-Martin, J.A.; Minsavage, G.V.; Iruegas-Bocardo, F.; Abrahamian, P.; Potnis, N.; Kolaczkowski, B.; Vallad, G.E.; Goss, E.M.; Jones, J.B. Multiple Recombination Events Drive the Current Genetic Structure of Xanthomonas perforans in Florida. Front. Microbiol. 2019, 10, 448. [Google Scholar] [CrossRef]
- Potnis, N.; Kandel, P.P.; Merfa, M.V.; Retchless, A.C.; Parker, J.K.; Stenger, D.C.; Almeida, R.P.P.; Bergsma-Vlami, M.; Westenberg, M.; Cobine, P.A.; et al. Patterns of Inter- and Intrasubspecific Homologous Recombination Inform Eco-Evolutionary Dynamics of Xylella fastidiosa. ISME J. 2019, 13, 2319–2333. [Google Scholar] [CrossRef]
- Jacques, M.A.; Denancé, N.; Legendre, B.; Morel, E.; Briand, M.; Mississipi, S.; Durand, K.; Olivier, V.; Portier, P.; Poliakoff, F.; et al. New Coffee Plant-Infecting Xylella fastidiosa Variants Derived via Homologous Recombination. Appl. Environ. Microbiol. 2016, 82, 1556–1568. [Google Scholar] [CrossRef]
- Höfte, H.; De Greve, H.; Seurinck, J.; Jansens, S.; Mahillon, J.; Ampe, C.; Vandekerckhove, J.; Vanderbruggen, H.; Van Montagu, M.; Zabeau, M.; et al. Structural and Functional Analysis of a Cloned Delta Endotoxin of Bacillus thuringiensis Berliner 1715. Eur. J. Biochem. 1986, 161, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Zhang, F.; Chen, G.; Joseph, L.; Barqawi, A.; Evans, J.; Song, F.; Li, G.; Zhang, J.; Crickmore, N. A Natural Hybrid of a Bacillus thuringiensis Cry2A Toxin Implicates Domain I in Specificity Determination. J. Invertebr. Pathol. 2017, 150, 35–40. [Google Scholar] [CrossRef] [PubMed]
- De Maagd, R.A.; Bravo, A.; Crickmore, N. How Bacillus thuringiensis Has Evolved Specific Toxins to Colonize the Insect World. Trends Genet. 2001, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Toro, N.; Martínez-Abarca, F.; Fernández-López, M. The Early Events Underlying Genome Evolution in a Localized Sinorhizobium meliloti Population. BMC Genom. 2016, 17, 556. [Google Scholar] [CrossRef]
- Eto, K.Y.; Firth, N.; Davis, A.M.; Kwong, S.M.; Krysiak, M.; Lee, Y.T.; O’Brien, F.G.; Grubb, W.B.; Coombs, G.W.; Bond, C.S.; et al. Evolution of a 72-Kilobase Cointegrant, Conjugative Multiresistance Plasmid in Community-Associated Methicillin-Resistant Staphylococcus aureus Isolates from the Early 1990s. Antimicrob. Agents Chemother. 2019, 63, e01560-19. [Google Scholar] [CrossRef]
- Cazares, A.; Moore, M.P.; Hall, J.P.J.; Wright, L.L.; Grimes, M.; Emond-Rhéault, J.-G.; Pongchaikul, P.; Santanirand, P.; Levesque, R.C.; Fothergill, J.L.; et al. A Megaplasmid Family Driving Dissemination of Multidrug Resistance in Pseudomonas. Nat. Commun. 2020, 11, 1370. [Google Scholar] [CrossRef]
- Schink, A.K.; Kadlec, K.; Kaspar, H.; Mankertz, J.; Schwarz, S. Analysis of Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolates Collected in the GERM-Vet Monitoring Programme. J. Antimicrob. Chemother. 2013, 68, 1741–1749. [Google Scholar] [CrossRef]
- Yabuki, M.; Nakao, M.; Fukunaga, M. Genetic Diversity and the Absence of Regional Differences of Borrelia garinii as Demonstrated by ospA and ospB Gene Sequence Analysis. Microbiol. Immunol. 1999, 43, 1097–1102. [Google Scholar] [CrossRef][Green Version]
- Taylor, J.C.; Martin, H.C.; Lise, S.; Broxholme, J.; Cazier, J.-B.; Rimmer, A.; Kanapin, A.; Lunter, G.; Fiddy, S.; Allan, C.; et al. Factors Influencing Success of Clinical Genome Sequencing across a Broad Spectrum of Disorders. Nat. Genet. 2015, 47, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, B.; Casjens, S.; Rosa, P. Evidence of Past Recombination Events among the Genes Encoding the Erp Antigens of Borrelia burgdorferi. Microbiology 1998, 144, 1869–1879. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.R.; Di, L.; Akther, S.; Mongodin, E.F.; Luft, B.J.; Schutzer, S.E.; Fraser, C.M.; Qiu, W.G. Primordial Origin and Diversification of Plasmids in Lyme Disease Agent Bacteria. BMC Genom. 2018, 19, 218. [Google Scholar] [CrossRef]
- Murawska, E.; Fiedoruk, K.; Swiecicka, I. Modular Genetic Architecture of the Toxigenic Plasmid pIS56-63 Harboring cry1Ab21 in Bacillus thuringiensis subsp. thuringiensis Strain IS5056. Pol. J. Microbiol. 2014, 63, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, C.; Zhu, Y.; Deng, Y.; Guo, S.; Peng, D.; Ruan, L.; Sun, M. The Resolution and Regeneration of a Cointegrate Plasmid Reveals a Model for Plasmid Evolution Mediated by Conjugation and OriT Site-Specific Recombination. Environ. Microbiol. 2013, 15, 3305–3318. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, K.; Bravo, A.; Soberón, M.; Cai, J.; Shu, C.; Zhang, J. Coexistence of Cry9 with the Vip3A Gene in an Identical Plasmid of Bacillus thuringiensis Indicates Their Synergistic Insecticidal Toxicity. J. Agric. Food Chem. 2020, 68, 14081–14090. [Google Scholar] [CrossRef]
- Wardal, E.; Kuch, A.; Gawryszewska, I.; Żabicka, D.; Hryniewicz, W.; Sadowy, E. Diversity of Plasmids and Tn1546-Type Transposons among VanA Enterococcus faecium in Poland. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 313–328. [Google Scholar] [CrossRef]
- van Hal, S.J.; Beukers, A.G.; Timms, V.J.; Ellem, J.A.; Taylor, P.; Maley, M.W.; Newton, P.J.; Ferguson, J.K.; Lee, A.; Chen, S.C.-A.; et al. Relentless Spread and Adaptation of Non-Typeable VanA Vancomycin-resistant Enterococcus faecium: A Genome-Wide Investigation. J. Antimicrob. Chemother. 2018, 73, 1487–1491. [Google Scholar] [CrossRef]
- Hudson, C.M.; Bent, Z.W.; Meagher, R.J.; Williams, K.P. Resistance Determinants and Mobile Genetic Elements of an NDM-1-Encoding Klebsiella pneumoniae Strain. PLoS ONE 2014, 9, e99209. [Google Scholar] [CrossRef]
- Venturini, C.; Hassan, K.A.; Chowdhury, P.R.; Paulsen, I.T.; Walker, M.J.; Djordjevic, S.P. Sequences of Two Related Multiple Antibiotic Resistance Virulence Plasmids Sharing a Unique IS26-Related Molecular Signature Isolated from Different Escherichia coli Pathotypes from Different Hosts. PLoS ONE 2013, 8, e78862. [Google Scholar] [CrossRef]
- Hong, S.F.; Chiu, C.H.; Chu, C.; Feng, Y.; Ou, J.T. Complete Nucleotide Sequence of a Virulence Plasmid of Salmonella enterica Serovar Dublin and Its Phylogenetic Relationship to the Virulence Plasmids of Serovars Choleraesuis, Enteritidis and Typhimurium. FEMS Microbiol. Lett. 2008, 282, 39–43. [Google Scholar] [CrossRef]
- Chu, C.; Feng, Y.; Chien, A.C.; Hu, S.; Chu, C.H.; Chiu, C.H. Evolution of Genes on the Salmonella Virulence Plasmid Phylogeny Revealed from Sequencing of the Virulence Plasmids of S. enterica Serotype Dublin and Comparative Analysis. Genomics 2008, 92, 339–343. [Google Scholar] [CrossRef]
- Dong, H.; Chen, T.; Dewhirst, F.E.; Fleischmann, R.D.; Fraser, C.M.; Duncan, M.J. Genomic Loci of the Porphyromonas gingivalis Insertion Element IS 1126. Infect. Immun. 1999, 67, 3416–3423. [Google Scholar] [CrossRef]
- Bouchami, O.; De Lencastre, H.; Miragaia, M. Impact of Insertion Sequences and Recombination on the Population Structure of Staphylococcus haemolyticus. PLoS ONE 2016, 11, e0156653. [Google Scholar] [CrossRef] [PubMed]
- Bayjanov, J.R.; Baan, J.; Rogers, M.R.C.; Troelstra, A.; Willems, R.J.L.; van Schaik, W. Enterococcus faecium Genome Dynamics during Long-Term Asymptomatic Patient Gut Colonization. Microb. Genom. 2019, 5, e000277. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Gorrie, C.; Edwards, D.J.; Wertheim, H.F.L.; Hsu, L.Y.; Van Kinh, N.; Zadoks, R.; Baker, S.; Holt, K.E. Extensive Capsule Locus Variation and Large-Scale Genomic Recombination within the Klebsiella pneumoniae Clonal Group 258. Genome Biol. Evol. 2015, 7, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Mathema, B.; Pitout, J.D.D.; DeLeo, F.R.; Kreiswirth, B.N. Epidemic Klebsiella pneumoniae ST258 Is a Hybrid Strain. mBio 2014, 5, e01355-14. [Google Scholar] [CrossRef]
- Hill, K.K.; Xie, G.; Foley, B.T.; Smith, T.J.; Munk, A.C.; Bruce, D.; Smith, L.A.; Brettin, T.S.; Detter, J.C. Recombination and Insertion Events Involving the Botulinum Neurotoxin Complex Genes in Clostridium botulinum Types A, B, E and F and Clostridium butyricum Type E Strains. BMC Biol. 2009, 7, 66. [Google Scholar] [CrossRef]
- Sharma, V.K.; Akavaram, S.; Schaut, R.G.; Bayles, D.O. Comparative Genomics Reveals Structural and Functional Features Specific to the Genome of a Foodborne Escherichia coli O157:H7. BMC Genom. 2019, 20, 196. [Google Scholar] [CrossRef]
- Losada, L.; Ronning, C.M.; Deshazer, D.; Woods, D.; Fedorova, N.; Kim, H.S.; Shabalina, S.A.; Pearson, T.R.; Brinkac, L.; Tan, P.; et al. Continuing Evolution of Burkholderia mallei through Genome Reduction and Large-Scale Rearrangements. Genome Biol. Evol. 2010, 2, 102–116. [Google Scholar] [CrossRef]
- Chen, Y.; Stine, O.C.; Badger, J.H.; Gil, A.I.; Nair, G.B.; Nishibuchi, M.; Fouts, D.E. Comparative Genomic Analysis of Vibrio parahaemolyticus: Serotype Conversion and Virulence. BMC Genom. 2011, 12, 294. [Google Scholar] [CrossRef] [PubMed]
- Comandatore, F.; Sassera, D.; Bayliss, S.C.; Scaltriti, E.; Gaiarsa, S.; Cao, X.; Gales, A.C.; Saito, R.; Pongolini, S.; Brisse, S.; et al. Gene Composition as a Potential Barrier to Large Recombinations in the Bacterial Pathogen Klebsiella pneumoniae. Genome Biol. Evol. 2019, 11, 3240–3251. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Valero, L.; Rusniok, C.; Jarraud, S.; Vacherie, B.; Rouy, Z.; Barbe, V.; Medigue, C.; Etienne, J.; Buchrieser, C. Extensive Recombination Events and Horizontal Gene Transfer Shaped the Legionella pneumophila Genomes. BMC Genom. 2011, 12, 536. [Google Scholar] [CrossRef]
- Campisi, E.; Rinaudo, C.D.; Donati, C.; Barucco, M.; Torricelli, G.; Edwards, M.S.; Baker, C.J.; Margarit, I.; Rosini, R. Serotype IV Streptococcus agalactiae ST-452 Has Arisen from Large Genomic Recombination Events between CC23 and the Hypervirulent CC17 Lineages. Sci. Rep. 2016, 6, 29799. [Google Scholar] [CrossRef] [PubMed]
- Gawlik, D.; Ruppelt-Lorz, A.; Müller, E.; Reißig, A.; Hotzel, H.; Braun, S.D.; Söderquist, B.; Ziegler-Cordts, A.; Stein, C.; Pletz, M.W.; et al. Molecular Investigations on a Chimeric Strain of Staphylococcus aureus Sequence Type 80. PLoS ONE 2020, 15, e0232071. [Google Scholar] [CrossRef]
- Spoor, L.E.; Richardson, E.; Richards, A.C.; Wilson, G.J.; Mendonca, C.; Gupta, R.K.; McAdam, P.R.; Nutbeam-Tuffs, S.; Black, N.S.; O’gara, J.P.; et al. Recombination-Mediated Remodelling of Host–Pathogen Interactions during Staphylococcus aureus Niche Adaptation. Microb. Genom. 2015, 1, e36. [Google Scholar] [CrossRef]
- Lista, F.; Faggioni, G.; Valjevac, S.; Ciammaruconi, A.; Vaissaire, J.; Le Doujet, C.; Gorgé, O.; De Santis, R.; Carattoli, A.; Ciervo, A.; et al. Genotyping of Bacillus anthracis Strains Based on Automated Capillary 25-Loci Multiple Locus Variable-Number Tandem Repeats Analysis. BMC Microbiol. 2006, 6, 33. [Google Scholar] [CrossRef]
- Spuesens, E.B.M.; Oduber, M.; Hoogenboezem, T.; Stuijter, M.; Hartwig, N.G.; van Rossum, A.M.C.; Vink, C. Sequence Variations in RepMP2/3 and RepMP4 Elements Reveal Intragenomic Homologous DNA Recombination Events in Mycoplasma pneumoniae. Microbiology 2009, 155, 2182–2196. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Croucher, N.J.; Hanage, W.P.; Harris, S.R.; McGee, L.; van der Linden, M.; de Lencastre, H.; Sá-Leão, R.; Song, J.-H.; Ko, K.S.; Beall, B.; et al. Variable Recombination Dynamics during the Emergence, Transmission and “disarming” of a Multidrug-Resistant Pneumococcal Clone. BMC Biol. 2014, 12, 49. [Google Scholar] [CrossRef]
- Rolo, J.; Worning, P.; Nielsen, J.B.; Bowden, R.; Bouchami, O.; Damborg, P.; Guardabassi, L.; Perreten, V.; Tomasz, A.; Westh, H.; et al. Evolutionary Origin of the Staphylococcal Cassette Chromosome Mec (SCC Mec). Antimicrob. Agents Chemother. 2017, 61, e02302-16. [Google Scholar] [CrossRef] [PubMed]
- Araki, H.; Innan, H.; Kreitman, M.; Bergelson, J. Molecular Evolution of Pathogenicity-Island Genes in Pseudomonas viridiflava. Genetics 2007, 177, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Shen, X.; Yan, J.; Han, H.; Zheng, B.; Liu, D.; Cheng, H.; Zhao, Y.; Rao, X.; Wang, C.; et al. GI-Type T4SS-Mediated Horizontal Transfer of the 89K Pathogenicity Island in Epidemic Streptococcus suis Serotype 2. Mol. Microbiol. 2011, 79, 1670–1683. [Google Scholar] [CrossRef]
- Elliott, B.; Dingle, K.E.; Didelot, X.; Crook, D.W.; Riley, T.V. The Complexity and Diversity of the Pathogenicity Locus in Clostridium difficile Clade 5. Genome Biol. Evol. 2014, 6, 3159–3170. [Google Scholar] [CrossRef] [PubMed]
- Planet, P.J.; Kachlany, S.C.; Fine, D.H.; DeSalle, R.; Figurski, D.H. The Widespread Colonization Island of Actinobacillus actinomycetemcomitans. Nat. Genet. 2003, 34, 193–198. [Google Scholar] [CrossRef]
- Schubert, S.; Nörenberg, D.; Clermont, O.; Magistro, G.; Wieser, A.; Romann, E.; Hoffmann, C.; Weinert, K.; Denamur, E. Prevalence and Phylogenetic History of the TcpC Virulence Determinant in Escherichia coli. Int. J. Med. Microbiol. 2010, 300, 429–434. [Google Scholar] [CrossRef]
- Thrane, S.W.; Taylor, V.L.; Freschi, L.; Kukavica-Ibrulj, I.; Boyle, B.; Laroche, J.; Pirnay, J.P.; Lévesque, R.C.; Lam, J.S.; Jelsbaka, L. The Widespread Multidrug-Resistant Serotype O12 Pseudomonas aeruginosa Clone Emerged through Concomitant Horizontal Transfer of Serotype Antigen and Antibiotic Resistance Gene Clusters. mBio 2015, 6, e01396-15. [Google Scholar] [CrossRef]
- Liao, J.; Orsi, R.H.; Carroll, L.M.; Kovac, J.; Ou, H.; Zhang, H.; Wiedmann, M. Serotype-Specific Evolutionary Patterns of Antimicrobial-Resistant Salmonella enterica. BMC Evol. Biol. 2019, 19, 132. [Google Scholar] [CrossRef]
- Morales, M.; García, P.; De La Campa, A.G.; LinÍares, J.; Ardanuy, C.; GarciÍa, E. Evidence of Localized Prophage-Host Recombination in the lytA Gene, Encoding the Major Pneumococcal Autolysin. J. Bacteriol. 2010, 192, 2624–2632. [Google Scholar] [CrossRef]
- Tamarit, D.; Neuvonen, M.M.; Engel, P.; Guy, L.; Andersson, S.G.E. Origin and Evolution of the Bartonella Gene Transfer Agent. Mol. Biol. Evol. 2018, 35, 451–464. [Google Scholar] [CrossRef]
- Verne, S.; Johnson, M.; Bouchon, D.; Grandjean, F. Evidence for Recombination between Feminizing Wolbachia in the Isopod Genus Armadillidium. Gene 2007, 397, 58–66. [Google Scholar] [CrossRef]
- Miller, J.C.; Stevenson, B. Immunological and Genetic Characterization of Borrelia burgdorferi BapA and EppA Proteins. Microbiology 2003, 149, 1113–1125. [Google Scholar] [CrossRef][Green Version]
- Akins, D.R.; Caimano, M.J.; Yang, X.; Cerna, F.; Norgard, M.V.; Radolf, J.D. Molecular and Evolutionary Analysis of Borrelia burgdorferi 297 Circular Plasmid-Encoded Lipoproteins with OspE- and OspF-like Leader Peptides. Infect. Immun. 1999, 67, 1526–1532. [Google Scholar] [CrossRef]
- Joseph, S.J.; Didelot, X.; Gandhi, K.; Dean, D.; Read, T.D. Interplay of Recombination and Selection in the Genomes of Chlamydia trachomatis. Biol. Direct 2011, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Gomes, J.P.; Bruno, W.J.; Nunes, A.; Santos, N.; Florindo, C.; Borrego, M.J.; Dean, D. Evolution of Chlamydia trachomatis Diversity Occurs by Widespread Interstrain Recombination Involving Hotspots. Genome Res. 2007, 17, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Smelov, V.; Vrbanac, A.; van Ess, E.F.; Noz, M.P.; Wan, R.; Eklund, C.; Morgan, T.; Shrier, L.A.; Sanders, B.; Dillner, J.; et al. Chlamydia trachomatis Strain Types Have Diversified Regionally and Globally with Evidence for Recombination across Geographic Divides. Front. Microbiol. 2017, 8, 2195. [Google Scholar] [CrossRef]
- Roulis, E.; Bachmann, N.; Humphrys, M.; Myers, G.; Huston, W.; Polkinghorne, A.; Timms, P. Phylogenetic Analysis of Human Chlamydia pneumoniae Strains Reveals a Distinct Australian Indigenous Clade That Predates European Exploration of the Continent. BMC Genom. 2015, 16, 1094. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Elizalde, S.; Cortés-Márquez, A.C.; Zuñiga, G.; Cerritos, R.; Valencia-Mayoral, P.; Sánchez, A.C.; Olivares-Clavijo, H.; Velázquez-Guadarrama, N. Inference from the Analysis of Genetic Structure of Helicobacter pylori Strains Isolates from Two Paediatric Patients with Recurrent Infection. BMC Microbiol. 2019, 19, 184. [Google Scholar] [CrossRef]
- Tsai, Y.H.L.; Orsi, R.H.; Nightingale, K.K.; Wiedmann, M. Listeria monocytogenes Internalins Are Highly Diverse and Evolved by Recombination and Positive Selection. Infect. Genet. Evol. 2006, 6, 378–389. [Google Scholar] [CrossRef]
- Hill, D.J.; Whittles, C.; Virji, M. A Novel Group of Moraxella catarrhalis UspA Proteins Mediates Cellular Adhesion via CEACAMs and Vitronectin. PLoS ONE 2012, 7, e45452. [Google Scholar] [CrossRef]
- Muzzi, A.; Moschioni, M.; Covacci, A.; Rappuoli, R.; Donati, C. Pilus Operon Evolution in Streptococcus pneumoniae Is Driven by Positive Selection and Recombination. PLoS ONE 2008, 3, e3660. [Google Scholar] [CrossRef] [PubMed]
- Pandya, G.A.; McEllistrem, M.C.; Venepally, P.; Holmes, M.H.; Jarrahi, B.; Sanka, R.; Liu, J.; Karamycheva, S.A.; Bai, Y.; Fleischmann, R.D.; et al. Monitoring the Long-Term Molecular Epidemiology of the Pneumococcus and Detection of Potential “vaccine Escape” Strains. PLoS ONE 2011, 6, e15950. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rahbar, M.R.; Zarei, M.; Jahangiri, A.; Khalili, S.; Nezafat, N.; Negahdaripour, M.; Fattahian, Y.; Ghasemi, Y. Trimeric Autotransporter Adhesins in Acinetobacter baumannii, Coincidental Evolution at Work. Infect. Genet. Evol. 2019, 71, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Achtman, M. Clonal Spread of Serogroup A Meningococci: A Paradigm for the Analysis of Microevolution in Bacteria. Mol. Microbiol. 1994, 11, 15–22. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Sánchez-Busó, L.; Harris, S.R.; Marttinen, P.; Rusniok, C.; Buchrieser, C.; Harrison, T.G.; Parkhill, J. Dynamics and Impact of Homologous Recombination on the Evolution of Legionella pneumophila. PLoS Genet. 2017, 13, e1006855. [Google Scholar] [CrossRef]
- Tsuru, T.; Kobayashi, I. Multiple Genome Comparison within a Bacterial Species Reveals a Unit of Evolution Spanning Two Adjacent Genes in a Tandem Paralog Cluster. Mol. Biol. Evol. 2008, 25, 2457–2473. [Google Scholar] [CrossRef]
- Brochet, M.; Couvé, E.; Zouine, M.; Vallaeys, T.; Rusniok, C.; Lamy, M.C.; Buchrieser, C.; Trieu-Cuot, P.; Kunst, F.; Poyart, C.; et al. Genomic Diversity and Evolution within the Species Streptococcus agalactiae. Microbes Infect. 2006, 8, 1227–1243. [Google Scholar] [CrossRef]
- Whatmore, A.M.; Kapur, V.; Sullivan, D.J.; Musser, J.M.; Kehoe, M.A. Non-congruent Relationships between Variation in Emm Gene Sequences and the Population Genetic Structure of Group A Streptococci. Mol. Microbiol. 1994, 14, 619–631. [Google Scholar] [CrossRef]
- McBride, A.J.A.; Cerqueira, G.M.; Suchard, M.A.; Moreira, A.N.; Zuerner, R.L.; Reis, M.G.; Haake, D.A.; Ko, A.I.; Dellagostin, O.A. Genetic Diversity of the Leptospiral Immunoglobulin-like (Lig) Genes in Pathogenic Leptospira spp. Infect. Genet. Evol. 2009, 9, 196–205. [Google Scholar] [CrossRef]
- Sabat, A.J.; Wladyka, B.; Kosowska-Shick, K.; Grundmann, H.; Van Dijl, J.M.; Kowal, J.; Appelbaum, P.C.; Dubin, A.; Hryniewicz, W. Polymorphism, Genetic Exchange and Intragenic Recombination of the Aureolysin Gene among Staphylococcus aureus Strains. BMC Microbiol. 2008, 8, 129. [Google Scholar] [CrossRef]
- Watanabe, S.; Ito, T.; Sasaki, T.; Li, S.; Uchiyama, I.; Kishii, K.; Kikuchi, K.; Skov, R.L.; Hiramatsu, K. Genetic Diversity of Staphylocoagulase Genes (coa): Insight into the Evolution of Variable Chromosomal Virulence Factors in Staphylococcus aureus. PLoS ONE 2009, 4, e5714. [Google Scholar] [CrossRef][Green Version]
- Muzzi, A.; Mora, M.; Pizza, M.; Rappuoli, R.; Donati, C. Conservation of Meningococcal Antigens in the Genus Neisseria. mBio 2013, 4, e00163-13. [Google Scholar] [CrossRef]
- Gray, R.R.; Mulligan, C.J.; Molini, B.J.; Sun, E.S.; Giacani, L.; Godornes, C.; Kitchen, A.; Lukehart, S.A.; Centurion-Lara, A. Molecular Evolution of the tprC, D, I, K, G, and J Genes in the Pathogenic Genus Treponema. Mol. Biol. Evol. 2006, 23, 2220–2233. [Google Scholar] [CrossRef]
- Guttman, D.S.; Gropp, S.J.; Morgan, R.L.; Wang, P.W. Diversifying Selection Drives the Evolution of the Type III Secretion System Pilus of Pseudomonas syringae. Mol. Biol. Evol. 2006, 23, 2342–2354. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.; Teixeira, P.G.; D’Avó, A.F.; Júnior, C.S.; Veríssimo, A. Intragenic Recombination Has a Critical Role on the Evolution of Legionella pneumophila Virulence-Related Effector SidJ. PLoS ONE 2014, 9, e109840. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.; Tiago, I.; da Costa, M.S.; Veríssimo, A. Molecular Evolution of Legionella pneumophila DotA Gene, the Contribution of Natural Environmental Strains. Environ. Microbiol. 2010, 12, 2711–2729. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Davies, R.L. Evidence for a Common Gene Pool and Frequent Recombinational Exchange of the tbpBA Operon in Mannheimia haemolytica, Mannheimia glucosida and Bibersteinia trehalosi. Microbiology 2011, 157, 123–135. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Harrison, O.B.; Maiden, M.C.J.; Rokbi, B. Distribution of Transferrin Binding Protein B Gene (tbpB) Variants among Neisseria Species. BMC Microbiol. 2008, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Giffard, P.M.; Allen, D.M.; Milward, C.P.; Simpson, C.L.; Jacques, N.A. Sequence of the GtfK Gene of Streptococcus salivarius ATCC 25975 and Evolution of the gtf Genes of Oral Streptococci. J. Gen. Microbiol. 1993, 139, 1511–1522. [Google Scholar] [CrossRef]
- Ng, V.; Lin, W.J. Comparison of Assembled Clostridium botulinum A1 Genomes Revealed Their Evolutionary Relationship. Genomics 2014, 103, 94–106. [Google Scholar] [CrossRef][Green Version]
- Prisilla, A.; Prathiviraj, R.; Chellapandi, P. Molecular Evolutionary Constraints That Determine the Avirulence State of Clostridium botulinum C2 Toxin. J. Mol. Evol. 2017, 84, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, M.J.; Tremblay, B.J.M.; Zeng, J.; Wei, X.; Hodgins, H.; Worley, J.; Bry, L.; Dong, M.; Doxey, A.C. Phylogenomics of 8,839 Clostridioides difficile Genomes Reveals Recombination-Driven Evolution and Diversification of Toxin A and B. PLoS Pathog. 2020, 16, e1009181. [Google Scholar] [CrossRef]
- Davies, R.L.; Campbell, S.; Whittam, T.S. Mosaic Structure and Molecular Evolution of the Leukotoxin Operon (lktCABD) in Mannheimia (Pasteurella) haemolytica, Mannheimia glucosida, and Pasteurella trehalosi. J. Bacteriol. 2002, 184, 266–277. [Google Scholar] [CrossRef]
- Turner, C.E.; Holden, M.T.G.; Blane, B.; Horner, C.; Peacock, S.J.; Sriskandan, S. The Emergence of Successful Streptococcus pyogenes Lineages through Convergent Pathways of Capsule Loss and Recombination Directing High Toxin Expression. mBio 2019, 10, e02521-19. [Google Scholar] [CrossRef] [PubMed]
- Jobling, M.G.; Holmes, R.K. Type II Heat-Labile Enterotoxins from 50 Diverse Escherichia coli Isolates Belong Almost Exclusively to the LT-IIc Family and May Be Prophage Encoded. PLoS ONE 2012, 7, e29898. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.S.; Valk, P.L.; Shaffer, C.L.; Bordenstein, S.R.; Cover, T.L. J-Western Forms of Helicobacter pylori cagA Constitute a Distinct Phylogenetic Group with a Widespread Geographic Distribution. J. Bacteriol. 2012, 194, 1593–1604. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Poirel, L.; Nordmann, P. New Delhi Metallo-β-Lactamase-Producing Acinetobacter baumannii: A Novel Paradigm for Spreading Antibiotic Resistance Genes. Future Microbiol. 2014, 9, 33–41. [Google Scholar] [CrossRef]
- Liu, L.; Cui, Y.; Zheng, B.; Jiang, S.; Yu, W.; Shen, P.; Ji, J.; Li, L.; Qin, N.; Xiao, Y. Analysis of Tigecycline Resistance Development in Clinical Acinetobacter baumannii Isolates through a Combined Genomic and Transcriptomic Approach. Sci. Rep. 2016, 6, 26930. [Google Scholar] [CrossRef]
- Diaz Caballero, J.; Clark, S.T.; Wang, P.W.; Donaldson, S.L.; Coburn, B.; Tullis, D.E.; Yau, Y.C.W.; Waters, V.J.; Hwang, D.M.; Guttman, D.S. A Genome-Wide Association Analysis Reveals a Potential Role for Recombination in the Evolution of Antimicrobial Resistance in Burkholderia multivorans. PLoS Pathog. 2018, 14, e1007453. [Google Scholar] [CrossRef]
- Tchesnokova, V.; Radey, M.; Chattopadhyay, S.; Larson, L.; Weaver, J.L.; Kisiela, D.; Sokurenko, E.V. Pandemic Fluoroquinolone Resistant Escherichia coli Clone ST1193 Emerged via Simultaneous Homologous Recombinations in 11 Gene Loci. Proc. Natl. Acad. Sci. USA 2019, 116, 14740–14748. [Google Scholar] [CrossRef]
- De Been, M.; Van Schaik, W.; Cheng, L.; Corander, J.; Willems, R.J. Recent Recombination Events in the Core Genome Are Associated with Adaptive Evolution in Enterococcus faecium. Genome Biol. Evol. 2013, 5, 1524–1535. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Kido, N.; Kato, Y.; Koide, N.; Yoshida, T.; Yokochi, T. Evolutionary Relationship among rfb Gene Clusters Synthesizing Mannose Homopolymer as O-Specific Polysaccharides in Escherichia coli and Klebsiella. Gene 1997, 198, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Yahara, K.; Kawai, M.; Furuta, Y.; Takahashi, N.; Handa, N.; Tsuru, T.; Oshima, K.; Yoshida, M.; Azuma, T.; Hattori, M.; et al. Genome-Wide Survey of Mutual Homologous Recombination in a Highly Sexual Bacterial Species. Genome Biol. Evol. 2012, 4, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Holt, K.; Kenyon, J.J.; Hamidian, M.; Schultz, M.B.; Pickard, D.J.; Dougan, G.; Hall, R. Five Decades of Genome Evolution in the Globally Distributed, Extensively Antibiotic-Resistant Acinetobacter baumannii Global Clone 1. Microb. Genom. 2016, 2, e000052. [Google Scholar] [CrossRef]
- Rishishwar, L.; Katz, L.S.; Sharma, N.V.; Rowe, L.; Frace, M.; Thomas, J.D.; Harcourt, B.H.; Mayer, L.W.; Jordana, I.K. Genomic Basis of a Polyagglutinating Isolate of Neisseria meningitidis. J. Bacteriol. 2012, 194, 5649–5656. [Google Scholar] [CrossRef]
- Berti, F.; Campisi, E.; Toniolo, C.; Morelli, L.; Crotti, S.; Rosini, R.; Romano, M.R.; Pinto, V.; Brogioni, B.; Torricelli, G.; et al. Structure of the Type IX Group B Streptococcus Capsular Polysaccharide and Its Evolutionary Relationship with Types V and VII. J. Biol. Chem. 2014, 289, 23437–23448. [Google Scholar] [CrossRef]
- Neemuchwala, A.; Teatero, S.; Athey, T.B.T.; McGeer, A.; Fittipaldi, N. Capsular Switching and Other Large-Scale Recombination Events in Invasive Sequence Type 1 Group B Streptococcus. Emerg. Infect. Dis. 2016, 22, 1941–1944. [Google Scholar] [CrossRef]
- Croucher, N.J.; Kagedan, L.; Thompson, C.M.; Parkhill, J.; Bentley, S.D.; Finkelstein, J.A.; Lipsitch, M.; Hanage, W.P. Selective and Genetic Constraints on Pneumococcal Serotype Switching. PLoS Genet. 2015, 11, e1005095. [Google Scholar] [CrossRef]
- Alqasim, A.; Scheutz, F.; Zong, Z.; McNally, A. Comparative Genome Analysis Identifies Few Traits Unique to the Escherichia coli ST131 H30Rx Clade and Extensive Mosaicism at the Capsule Locus. BMC Genom. 2014, 15, 830. [Google Scholar] [CrossRef]
- Starcevic, A.; Diminic, J.; Zucko, J.; Elbekali, M.; Schlosser, T.; Lisfi, M.; Vukelic, A.; Long, P.F.; Hranueli, D.; Cullum, J. A Novel Docking Domain Interface Model Predicting Recombination between Homoeologous Modular Biosynthetic Gene Clusters. J. Ind. Microbiol. Biotechnol. 2011, 38, 1295–1304. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shikov, A.E.; Savina, I.A.; Nizhnikov, A.A.; Antonets, K.S. Recombination in Bacterial Genomes: Evolutionary Trends. Toxins 2023, 15, 568. https://doi.org/10.3390/toxins15090568
Shikov AE, Savina IA, Nizhnikov AA, Antonets KS. Recombination in Bacterial Genomes: Evolutionary Trends. Toxins. 2023; 15(9):568. https://doi.org/10.3390/toxins15090568
Chicago/Turabian StyleShikov, Anton E., Iuliia A. Savina, Anton A. Nizhnikov, and Kirill S. Antonets. 2023. "Recombination in Bacterial Genomes: Evolutionary Trends" Toxins 15, no. 9: 568. https://doi.org/10.3390/toxins15090568
APA StyleShikov, A. E., Savina, I. A., Nizhnikov, A. A., & Antonets, K. S. (2023). Recombination in Bacterial Genomes: Evolutionary Trends. Toxins, 15(9), 568. https://doi.org/10.3390/toxins15090568





