Revitalizing the Gut Microbiome in Chronic Kidney Disease: A Comprehensive Exploration of the Therapeutic Potential of Physical Activity
Abstract
1. A Healthy Gut Microbiome
1.1. Characteristics of a Healthy Gut Microbiome
1.2. An Active Lifestyle, a Healthy Gut?
| Physical activity (PA) [40] | Any skeletal movement that results in energy expenditure, including all bodily movements produced during recreational time, transport, or occupational activities |
| Exercise [40] | A subset of PA that is planned, structured, and repetitive and has the objective to improve or maintain physical fitness |
| Exercise intensity [41] | PA can be classified based on the amount of energy expenditure or metabolic equivalent of the task (MET), into low intensity PA (<3 METs), moderate intensity PA (3–6 METs) or high intensity PA (>6 METs) |
| Aerobic exercise [42,43] | Involve continuous rhythmic movements that elevate breathing frequency and heart rate, also referred to as endurance exercise |
| Strength exercises [44] | Aim to enhance muscle strength and mass, typically through resistance training where muscles work against a force |
| Balance exercises [43] | Help improve stability and coordination by challenging the body’s ability to maintain equilibrium |
| Flexibility exercises [43] | Focus on improving range of motion in muscles and joints |
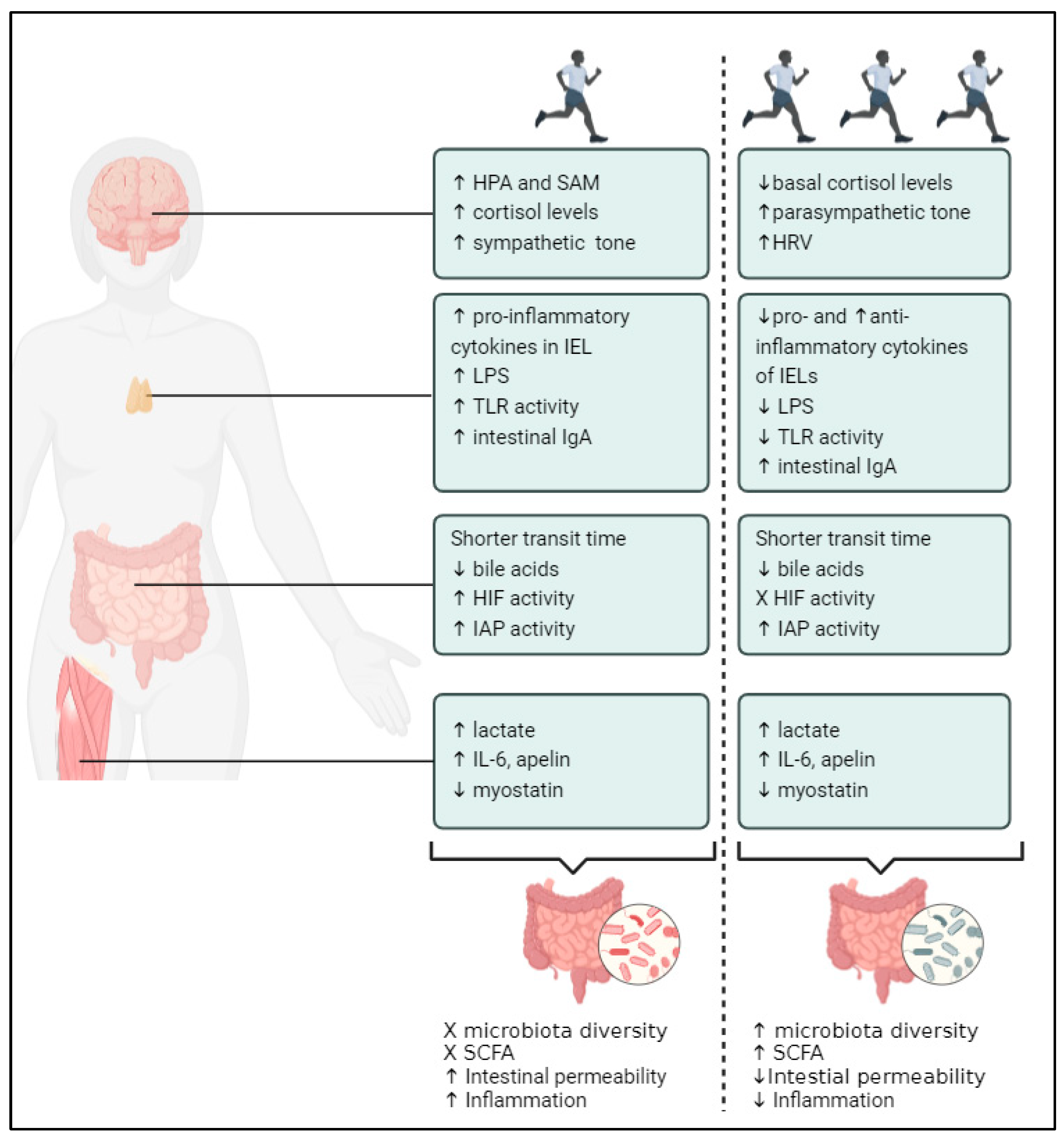
1.3. Mechanisms via Which PA Modifies the Gut Microbiome
1.3.1. PA and Gastrointestinal Fitness
1.3.2. PA and Fitness of the Immune System
1.3.3. PA and Fitness of the Nervous System
1.3.4. PA and Muscle Fitness
2. CKD
2.1. Gut Dysbiosis in CKD: The Gut–Kidney Axis
2.2. PA and the Gut in CKD: A Model of Physical Inactivity
2.3. Mechanisms via Which PA Could Restore Gut Dysbiosis in CKD
2.3.1. CKD, PA and Gastrointestinal Fitness
2.3.2. CKD, PA and Fitness of the Immune System
2.3.3. CKD, PA and Fitness of the Nervous System
2.3.4. CKD, PA, and Muscle Fitness
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, C.; Zhu, H.; Yao, Y.; Zeng, R. Gut Dysbiosis and Kidney Diseases. Front. Med. 2022, 9, 829349. [Google Scholar] [CrossRef] [PubMed]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef]
- Gilbert, J.; Blaser, M.; Capraso, G.; Jansson, J.K. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Krukowski, H.; Valkenburg, S.; Madella, A.-M.; Garssen, J.; van Bergenhenegouwen, J.; Overbeek, S.A.; Huys, G.R.B.; Raes, J.; Glorieux, G. Gut microbiome studies in CKD: Opportunities, pitfalls and therapeutic potential. Nat. Rev. Nephrol. 2022, 19, 87–101. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Correction to: Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 1–22. [Google Scholar] [CrossRef]
- Dorelli, B.; Gallè, F.; De Vito, C.; Duranti, G.; Iachini, M.; Zaccarin, M.; Standoli, J.P.; Ceci, R.; Romano, F.; Liguori, G.; et al. Can physical activity influence human gut microbiota composition independently of diet? A systematic review. Nutrients 2021, 13, 1890. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Cerdá, B.; Pérez, M.; Pérez-Santiago, J.D.; Tornero-Aguilera, J.F.; González-Soltero, R.; Larrosa, M. Gut microbiota modification: Another piece in the puzzle of the benefits of physical exercise in health? Front. Physiol. 2016, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 1–28. [Google Scholar] [CrossRef]
- Young, V.B.; Schmidt, T.M. Overview of the gastrointestinal microbiota. Adv. Exp. Med. Biol. 2008, 635, 29–40. [Google Scholar]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the healthy gut microbiota composition? a changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Nogal, A.; Valdes, A.M.; Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1897212. [Google Scholar] [CrossRef] [PubMed]
- Vinelli, V.; Biscotti, P.; Martini, D.; Del Bo’, C.; Marino, M.; Meroño, T.; Nikoloudaki, O.; Calabrese, F.M.; Turroni, S.; Taverniti, V.; et al. Effects of Dietary Fibers on Short-Chain Fatty Acids and Gut Microbiota Composition in Healthy Adults: A Systematic Review. Nutrients 2022, 14, 2559. [Google Scholar] [CrossRef] [PubMed]
- Aya, V.; Jimenez, P.; Muñoz, E.; Ramírez, J.D. Effects of exercise and physical activity on gut microbiota composition and function in older adults: A systematic review. BMC Geriatr. 2023, 23, 364. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Tsolis, R.M.; Bäumler, A.J. The microbiome and gut homeostasis. Science 2022, 377, 44. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current understanding of dysbiosis in disease in human and animal models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Estaki, M.; Pither, J.; Baumeister, P.; Little, J.P.; Gill, S.K.; Ghosh, S.; Ahmadi-Vand, Z.; Marsden, K.R.; Gibson, D.L. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome 2016, 4, 42. [Google Scholar] [CrossRef]
- Wegierska, A.E.; Charitos, I.A.; Topi, S.; Potenza, M.A.; Montagnani, M.; Santacroce, L. The Connection Between Physical Exercise and Gut Microbiota: Implications for Competitive Sports Athletes. Sports Med. 2022, 52, 2355–2369. [Google Scholar] [CrossRef]
- Walker, A.W.W.; Hoyles, L. Human microbiome myths and misconceptions. Nat. Microbiol. 2023, 8, 1392–1396. [Google Scholar] [CrossRef]
- Mailing, L.J.; Allen, J.M.; Buford, T.W.; Fields, C.J.; Woods, J.A. Exercise and the Gut Microbiome: A Review of the Evidence, Potential Mechanisms, and Implications for Human Health. Exerc. Sport Sci. Rev. 2019, 47, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.C.; Wisniewski, P.J.; Noji, M.; Lightfoot, S.A. The effect of diet and exercise on intestinal integrity and microbial diversity in mice. PLoS ONE 2016, 11, e0150502. [Google Scholar] [CrossRef] [PubMed]
- Batacan, A.; Fenning, V.J.; Dalbo, A.T.; Duncan, M.J.; Moore, R.J. A gut reaction: The combined influence of exercise and diet on gastrointestinal microbiota in rats. Int. J. Lab. Hematol. 2016, 38, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Inoue, R.; Tsukahara, T.; Ushida, K.; Chiji, H.; Matsubara, N.; Hara, H. Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci. Biotechnol. Biochem. 2008, 72, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.C.; LePard, K.J.; Kwak, J.W.; Stancukas, M.C.; Laskowski, S.; Dougherty, J.; Moulton, L.; Glawe, A.; Wang, Y.; Leone, V.; et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS ONE 2014, 9, e92193. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef]
- Monda, V.; Villano, I.; Messina, A.; Valenzano, A.; Esposito, T.; Moscatelli, F.; Viggiano, A.; Cibelli, G.; Chieffi, S.; Monda, M.; et al. Exercise modifies the gut microbiota with positive health effects. Oxidative Med. Cell Longev. 2017, 2017, 3831972. [Google Scholar] [CrossRef] [PubMed]
- Keirns, B.H.; Koemel, N.A.; Sciarrillo, C.M.; Anderson, K.L.; Emerson, S.R. Exercise and intestinal permeability: Another form of exercise-induced hormesis? Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G512–G518. [Google Scholar] [CrossRef]
- Ribeiro, F.M.; Petriz, B.; Marques, G.; Kamilla, L.H.; Franco, O.L. Is There an Exercise-Intensity Threshold Capable of Avoiding the Leaky Gut? Front. Nutr. 2021, 8, 62789. [Google Scholar] [CrossRef]
- Lira, F.S.; Rosa, J.C.; Pimentel, G.D.; A Souza, H.; Caperuto, E.C.; CarnevaliJr, L.C.; Seelaender, M.; Damaso, A.R.; Oyama, L.M.; de Mello, M.T.; et al. Endotoxin levels correlate positively with a sedentary lifestyle and negatively with highly trained subjects. Lipids Health Dis. 2010, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Cullen, J.M.A.; Shahzad, S.; Dhillon, J. A systematic review on the effects of exercise on gut microbial diversity, taxonomic composition, and microbial metabolites: Identifying research gaps and future directions. Front. Physiol. 2023, 14, 1292673. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.M.; Mailing, L.J.; Niemiro, G.M.; Moore, R.; Cook, M.D.; White, B.A.; Holscher, H.D.; Woods, J.A. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med. Sci. Sports Exerc. 2018, 50, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Kern, T.; Blond, M.B.; Hansen, T.H.; Rosenkilde, M.; Quist, J.S.; Gram, A.S.; Ekstrom, C.T.; Hansen, T.; Stallknecht, B. Structured exercise alters the gut microbiota in humans with overweight and obesity—A randomized controlled trial. Int. J. Obes. 2020, 44, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Ni, Y.; Cheung, C.K.Y.; Lam, K.S.L.; Wang, Y.; Xia, Z.; Ye, D.; Guo, J.; Tse, M.A.; et al. Gut Microbiome Fermentation Determines the Efficacy of Exercise for Diabetes Prevention. Cell Metab. 2020, 31, 77–91.e5. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.S.C.; Seguro, C.S.; Naves, M.M. Gut microbiota and physical exercise in obesity and diabetes- A systematic review. Nutr. Metab. Cardiovasc. Dis. 2022, 32, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Cataldi, S.; Bonavolontà, V.; Poli, L.; Clemente, F.M.; De Candia, M.; Carvutto, R.; Silva, A.F.; Badicu, G.; Greco, G.; Fischetti, F. The Relationship between Physical Activity, Physical Exercise, and Human Gut Microbiota in Healthy and Unhealthy Subjects: A Systematic Review. Biology 2022, 11, 479. [Google Scholar] [CrossRef]
- WHO. WHO Guidelines on Physical Activity and Sedentary Behaviour; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Bonomini-Gnutzmann, R.; Plaza-Díaz, J.; Jorquera-Aguilera, C.; Rodríguez-Rodríguez, A.; Rodríguez-Rodríguez, F. Effect of Intensity and Duration of Exercise on Gut Microbiota in Humans: A Systematic Review. Int. J. Environ. Res. Public Health 2022, 19, 9518. [Google Scholar] [CrossRef]
- Caspersen, C.J.; Powell, K.E.; Christenson, G.M. Physical Activity, Exercise, and Physical Fitness: Definitions and Distinctions for Health-Related Research. Public Health Rep. 1985, 100, 127–131. [Google Scholar]
- Ainsworth, B.E.; Haskell, W.L.; Leon, A.S.; Cheng, G.S. Compendium of Physical Activities. Med. Sci. Sports Exerc. 1983, 25, 71–80. [Google Scholar] [CrossRef]
- Patel, H.; Alkhawam, H.; Madanieh, R.; Shah, N. Aerobic vs anaerobic exercise training effects on the cardiovascular system. World J. Cardiol. 2017, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.E.; Rejeski, W.J.; Blair, S.N.; Fu, G.X. Physical activity and public health in older adults: Recommendation from the American College of Sports Medicine and the American Heart Association. Circulation 2007, 116, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.C.; Ellefsen, S.; Baar, K. Adaptations to endurance and strength training. Cold Spring Harb. Perspect. Med. 2017, 8, a029769. [Google Scholar] [CrossRef] [PubMed]
- Procházková, N.; Falony, G.; Dragsted, L.O.; Licht, T.R.; Raes, J.; Roager, H.M. Advancing human gut microbiota research by considering gut transit time. Gut 2023, 72, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.M.; Pedersen, H.E.; Clemmensen, K.K.; Ekblond, T.S.; Ried-Larsen, M.; Færch, K.; Brock, C.; Quist, J.S. Associations Between Physical Activity and Gastrointestinal Transit Times in People with Normal Weight, Overweight, and Obesity. J. Nutr. 2023, 154, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, P.C.; Marcobal, A.; Ursell, L.K.; Larauche, M.; Duboc, H.; Earle, K.A.; Sonnenburg, E.D.; Ferreyra, J.A.; Higginbottom, S.K.; Million, M.; et al. Complex Interactions Among Diet, Gastrointestinal Transit, and Gut Microbiota in Humanized Mice. Gastroenterology 2014, 144, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Asnicar, F.; Leeming, E.R.; Dimidi, E.; Mazidi, M.; Franks, P.W.; Al Khatib, H.; Valdes, A.M.; Davies, R.; Bakker, E.; Francis, L.; et al. Blue poo: Impact of gut transit time on the gut microbiome using a novel marker. Gut 2021, 70, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Hansen, L.B.S.; Bahl, M.I.; Frandsen, H.L.; Carvalho, V.; Gøbel, R.J.; Dalgaard, M.D.; Plichta, D.R.; Sparholt, M.H.; Vestergaard, H.; et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 2016, 1, 16093. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.J.; Heaton, K.W. Increasing butyrate concentration in the distal colon by accelerating intestinal transit. Gut 1997, 41, 245–251. [Google Scholar] [CrossRef]
- Rauch, B.; Davos, C.H.; Doherty, P.; Saure, D.; Metzendorf, M.-I.; Salzwedel, A.; Völler, H.; Jensen, K.; Schmid, J.-P. The prognostic effect of cardiac rehabilitation in the era of acute revascularisation and statin therapy: A systematic review and meta-analysis of randomized and non-randomized studies—The Cardiac Rehabilitation Outcome Study (CROS). Eur. J. Prev. Cardiol. 2016, 23, 1914–1939. [Google Scholar] [CrossRef]
- Esgalhado, M.; Borges, N.A.; Mafra, D. Could physical exercise help modulate the gut microbiota in chronic kidney disease? Future Microbiol. 2016, 11, 699–707. [Google Scholar] [CrossRef] [PubMed]
- De Schryver, A.M.; Keulemans, Y.C.; Peters, H.P.; Akkermans, L.M.; Smout, A.J.; De Vries, W.R.; Van Berge-Henegouwen, G.P. Effects of regular physical activity on defecation pattern in middle-aged patients complaining of chronic constipation. Scand. J. Gastroenterol. 2005, 40, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Koffler, K.H.; Menkes, A.; Redmond, R.A.; Whitehead, W.E.; Pratley, R.E.; Hurley, B.F. Strength training accelerates gastrointestinal transit in middle-aged and older men. Med. Sci. Sports Exerc. 1992, 24, 7823–7830. [Google Scholar] [CrossRef]
- Oettlé, G.J. Effect of moderate exercise on bowel habit. Gut 1991, 32, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Strid, H.; Simrén, M.; Störsrud, S.; Stotzer, P.-O.; Sadik, R. Effect of heavy exercise on gastrointestinal transit in endurance athletes. Scand. J. Gastroenterol. 2011, 46, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Sun, Y.; Zhang, X. Combined Physical Exercise and Diet: Regulation of Gut Microbiota to Prevent and Treat of Metabolic Disease: A Review. Nutrients 2022, 14, 4774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-L.; Li, Z.-J.; Gou, H.-Z.; Song, X.-J.; Zhang, L. The gut microbiota–bile acid axis: A potential therapeutic target for liver fibrosis. Front. Cell Infect. Microbiol. 2022, 12, 945368. [Google Scholar] [CrossRef]
- Collins, S.L.; Stine, J.G.; Bisanz, J.E.; Okafor, C.D.; Patterson, A.D. Bile acids and the gut microbiota: Metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 2023, 21, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Umar, S.; Rust, B.; Lazarova, D.; Bordonaro, M. Secondary bile acids and short chain fatty acids in the colon: A focus on colonic microbiome, cell proliferation, inflammation, and cancer. Int. J. Mol. Sci. 2019, 20, 1214. [Google Scholar] [CrossRef]
- Kriaa, A.; Bourgin, M.; Potiron, A.; Mkaouar, H.; Jablaoui, A.; Gérard, P.; Maguin, E.; Rhimi, M. Microbial impact on cholesterol and bile acid metabolism: Current status and future prospects. J. Lipid Res. 2019, 60, 323–332. [Google Scholar] [CrossRef]
- Wertheim, B.C.; Martínez, M.E.; Ashbeck, E.L.; Roe, D.J.; Jacobs, E.T.; Alberts, D.S.; Thompson, P.A. Physical activity as a determinant of fecal bile acid levels. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1591–1598. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, W.H.F.; Nye, E.R.; Macfarlane, D.J.; Robertson, M.C.; Williamson, S.A. Fecal bile acid concentration in distance runners. Int. J. Sports Med. 1991, 12, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Danese, E.; Salvagno, G.L.; Tarperi, C.; Negrini, D.; Montagnana, M.; Festa, L.; Sanchis-Gomar, F.; Schena, F. Middle-distance running acutely influences the concentration and composition of serum bile acids: Potential implications for cancer risk? Oncotarget 2017, 8, 52775–52782. [Google Scholar] [CrossRef] [PubMed]
- Osuna-Prieto, F.J.; Jurado-Fasoli, L.; Plaza-Florido, A.; Yang, W.; Kohler, I.; Di, X.; Rubio-López, J.; Sanchez-Delgado, G.; Rensen, P.C.N.; Ruiz, J.R.; et al. A bout of endurance and resistance exercise transiently decreases plasma levels of bile acids in young, sedentary adults. Scand. J. Med. Sci. Sports 2023, 33, 1607–1620. [Google Scholar] [CrossRef] [PubMed]
- Morville, T.; Sahl, R.E.; Trammell, S.A.J.; Svenningsen, J.S.; Gillum, M.P.; Helge, J.W.; Clemmensen, C. Divergent effects of resistance and endurance exercise on plasma bile acids, FGF19, and FGF21 in humans. J. Clin. Investig. 2018, 3, e122737. [Google Scholar] [CrossRef]
- Maurer, A.; Ward, J.L.; Dean, K.; Billinger, S.A.; Lin, H.; Mercer, K.E.; Adams, S.H.; Thyfault, J.P. Divergence in aerobic capacity impacts bile acid metabolism in young women. J. Appl. Physiol. 2020, 129, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Cao, W.; Xiang, D.; Hu, Y.-P.; Luo, B.; Chen, P. Exercise induces tissue hypoxia and HIF-1α redistribution in the small intestine. J. Sport Health Sci. 2020, 9, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Kelly, C.J.; Colgan, S.P. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am. J. Physiol. Cell Physiol. 2015, 309, C350–C360. [Google Scholar] [CrossRef]
- Pral, L.P.; Fachi, J.L.; Corrêa, R.O.; Colonna, M.; Vinolo, M.A. Hypoxia and HIF-1 as key regulators of gut microbiota and host interactions. Physiol. Behav. 2016, 176, 100–106. [Google Scholar] [CrossRef]
- Singhal, R.; Shah, Y.M. Oxygen battle in the gut: Hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine. J. Biol. Chem. 2020, 295, 10493–10505. [Google Scholar] [CrossRef]
- Bressa, C.; Bailén-Andrino, M.; Pérez-Santiago, J.; González-Soltero, R.; Pérez, M.; Montalvo-Lominchar, M.G.; Maté-Muñoz, J.L.; Domínguez, R.; Moreno, D.; Larrosa, M. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS ONE 2017, 12, e0171352. [Google Scholar] [CrossRef] [PubMed]
- Saranya, G.R.; Viswanathan, P. Gut microbiota dysbiosis in AKI to CKD transition. Biomed. Pharmacother. 2023, 161, 114447. [Google Scholar] [CrossRef]
- Tierney, S.; Elwers, H.; Sange, C.; Mamas, M.; Rutter, M.K.; Gibson, M.; Neyses, L.; Deaton, C. What influences physical activity in people with heart failure? A qualitative study. Int. J. Nurs. Stud. 2011, 48, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.B.; Carroll-Portillo, A.; Coffman, C.; Ritz, N.L.; Lin, H.C. Intestinal Alkaline Phosphatase Exerts Anti-Inflammatory Effects Against Lipopolysaccharide by Inducing Autophagy. Sci. Rep. 2020, 10, 3107. [Google Scholar] [CrossRef]
- Malo, M.S.; Alam, S.N.; Mostafa, G.; Zeller, S.J.; Johnson, P.V.; Mohammad, N.; Chen, K.T.; Moss, A.K.; Ramasamy, S.; Faruqui, A.; et al. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut 2010, 59, 1476–1484. [Google Scholar] [CrossRef]
- Sun, J.N.; Hou, B.; Ai, M.; Yu, X.Y.; Cai, W.W.; Qiu, L.Y. The effect of different types of exercise on the intestinal mechanical barrier and related regulatory factors in type 2 diabetic mice. Sheng Li Xue Bao 2022, 74, 237–245. [Google Scholar] [PubMed]
- Wojcik-Grzybek, D.; Hubalewska-Mazgaj, M.; Surmiak, M.; Sliwowski, Z.; Dobrut, A.; Mlodzinska, A.; Wojcik, A.; Kwiecien, S.; Magierowski, M.; Mazur-Bialy, A.; et al. The Combination of Intestinal Alkaline Phosphatase Treatment with Moderate Physical Activity Alleviates the Severity of Experimental Colitis in Obese Mice via Modulation of Gut Microbiota, Attenuation of Proinflammatory Cytokines, Oxidative Stress Biomarkers and DNA Oxidative Damage in Colonic Mucosa. Int. J. Mol. Sci. 2022, 23, 2964. [Google Scholar] [CrossRef]
- Ruiz-Iglesias, P.; Estruel-Amades, S.; Camps-Bossacoma, M.; Massot-Cladera, M.; Castell, M.; Pérez-Cano, F.J. Alterations in the mucosal immune system by a chronic exhausting exercise in Wistar rats. Sci. Rep. 2020, 10, 17950. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Ramos, R.; Martnez-Carrillo, B.E.; Aranda-González, I.I.; Guadarrama, A.L. Diet, exercise and gut mucosal immunity. Proc. Nutr. Soc. 2010, 69, 644–650. [Google Scholar] [CrossRef]
- Stahl, M.G.; Belkind-Gerson, J. Development of the Gastrointestinal Tract, 6th ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2020. [Google Scholar] [CrossRef]
- Hernández-Urbán, A.J.; Drago-Serrano, M.E.; Cruz-Baquero, A.; García-Hernández, A.L.; Arciniega-Martínez, I.M.; Pacheco-Yépez, J.; Guzmán-Mejía, F.; Godínez-Victoria, M. Exercise improves intestinal IgA production by T-dependent cell pathway in adults but not in aged mice. Front. Endocrinol. 2023, 14, 1190547. [Google Scholar] [CrossRef]
- Hoffman-Goetz, L.; Pervaiz, N.; Guan, J. Voluntary exercise training in mice increases the expression of antioxidant enzymes and decreases the expression of TNF-α in intestinal lymphocytes. Brain Behav. Immun. 2009, 23, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Cheroutre, H.; Lambolez, F.; Mucida, D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 2011, 11, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Yang, H. Effects of intraepithelial lymphocyte-derived cytokines on intestinal mucosal barrier function. J. Interf. Cytokine Res. 2013, 33, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Qiu, Y.; Yang, H. Intestinal intraepithelial lymphocytes: Maintainers of intestinal immune tolerance and regulators of intestinal immunity. J. Leukoc. Biol. 2021, 109, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Packer, N.; Hoffman-Goetz, L. Exercise training reduces inflammatory mediators in the intestinal tract of healthy older adult mice. Can. J. Aging 2012, 31, 161–171. [Google Scholar] [CrossRef]
- Hoffman-Goetz, L.; Pervaiz, N.; Packer, N.; Guan, J. Freewheel training decreases pro- and increases anti-inflammatory cytokine expression in mouse intestinal lymphocytes. Brain Behav. Immun. 2010, 24, 1105–1115. [Google Scholar] [CrossRef]
- Yang, Y.; Palm, N.W. Immunoglobulin A and the microbiome. Curr. Opin. Microbiol. 2020, 56, 89–96. [Google Scholar] [CrossRef]
- Pabst, O.; Slack, E. IgA and the intestinal microbiota: The importance of being specific. Mucosal Immunol. 2020, 13, 12–21. [Google Scholar] [CrossRef]
- Nakajima, A.; Vogelzang, A.; Maruya, M.; Miyajima, M.; Murata, M.; Son, A.; Kuwahara, T.; Tsuruyama, T.; Yamada, S.; Matsuura, M.; et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J. Exp. Med. 2018, 215, 2019–2034. [Google Scholar] [CrossRef]
- Viloria, M.; Lara-Padilla, E.; Campos-Rodríguez, R.; Jarillo-Luna, A.; Reyna-Garfias, H.; López-Sánchez, P.; Rivera-Aguilar, V.; Salas-Casas, A.; de la Rosa, F.J.B.; García-Latorre, E. Effect of moderate exercise on iga levels and lymphocyte count in mouse intestine. Immunol. Investig. 2011, 40, 640–656. [Google Scholar] [CrossRef]
- Drago-Serrano, M.-E.; Godínez-Victoria, M.; Lara-Padilla, E.; Resendiz-Albor, A.A.; Reyna-Garfias, H.; Arciniega-Martínez, I.M.; Kormanovski-Kovsova, A.; Campos-Rodriguez, R. Moderate exercise enhances expression of siga in mouse ileum. Int. J. Sports Med. 2012, 33, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Allgrove, J.E.; Gomes, E.; Hough, J.; Gleeson, M. Effects of exercise intensity on salivary antimicrobial proteins and markers of stress in active men. J. Sports Sci. 2008, 26, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of Commensal Microflora by Toll-Like Receptors Is Required for Intestinal Homeostasis. J. Torrey Bot. Soc. 2003, 130, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Burgueno, J.F.; Abreu, M.T. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Yiu, J.H.C.; Dorweiler, B.; Woo, C.W. Interaction between gut microbiota and toll-like receptor: From immunity to metabolism. J. Mol. Med. 2017, 95, 13–20. [Google Scholar] [CrossRef]
- Uchida, M.; Oyanagi, E.; Kawanishi, N.; Iemitsu, M.; Miyachi, M.; Kremenik, M.J.; Onodera, S.; Yano, H. Exhaustive exercise increases the TNF-α production in response to flagellin via the upregulation of toll-like receptor 5 in the large intestine in mice. Immunol. Lett. 2014, 158, 151–158. [Google Scholar] [CrossRef]
- Manilla, V.; Di Tommaso, N.; Santopaolo, F.; Gasbarrini, A.; Ponziani, F.R. Endotoxemia and Gastrointestinal Cancers: Insight into the Mechanisms Underlying a Dangerous Relationship. Microorganisms 2023, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Guo, Y.; Gui, Y.; Xu, D. Physical exercise, gut, gut microbiota, and atherosclerotic cardiovascular diseases. Lipids Health Dis. 2018, 17, 1–7. [Google Scholar] [CrossRef]
- Carvalho, F.A.; Koren, O.; Goodrich, J.K.; Johansson, M.E.; Nalbantoglu, I.; Aitken, J.D.; Su, Y.; Chassaing, B.; Walters, W.A.; González, A.; et al. Transient Inability to Manage Proteobacteria Promotes Chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 2012, 12, 139–152. [Google Scholar] [CrossRef]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science 2013, 328, 228–231. [Google Scholar] [CrossRef]
- Dheer, R.; Santaolalla, R.; Davies, J.M.; Lang, J.K.; Phillips, M.C.; Pastorini, C.; Vazquez-Pertejo, M.T.; Abreu, M.T. Function and Colonic Microbiota and Promotes a Risk for Transmissible Colitis. Infect. Immun. 2016, 84, 798–810. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, X.; Wu, Y.; Tian, Q. Aerobic exercise improves intestinal mucosal barrier dysfunction through TLR4/MyD88/NF-κB signaling pathway in diabetic rats. Biochem. Biophys. Res. Commun. 2022, 634, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.; McFarlin, B.; Flynn, M. Exercise and toll-like receptors. Exerc. Immunol. Rev. 2006, 12, 34–53. [Google Scholar] [PubMed]
- Lancaster, G.I.; Khan, Q.; Drysdale, P.; Wallace, F.; Jeukendrup, A.E.; Drayson, M.T.; Gleeson, M. The physiological regulation of toll-like receptor expression and function in humans. J. Physiol. 2005, 563, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Flynn, M.G.; McFarlin, B.K.; Phillips, M.D.; Stewart, L.K.; Timmerman, K.L. Toll-like receptor 4 and CD14 mRNA expression are lower in resistive exercise-trained elderly women. J. Appl. Physiol. 2003, 95, 1833–1842. [Google Scholar] [CrossRef] [PubMed]
- McFarlin, B.K.; Flynn, M.G.; Campbell, W.W.; Craig, B.A.; Robinson, J.P.; Stewart, L.K. Physical activity status, but not age, influences inflammatory biomarkers and toll-like receptor 4. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2006, 61, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.K.; Flynn, M.G.; Campbell, W.W.; Craig, B.A.; Robinson, J.P.; McFarlin, B.K.; Timmerman, K.L.; Coen, P.M.; Felker, J.; Talbert, E. Influence of exercise training and age on CD14+ cell-surface expression of toll-like receptor 2 and 4. Brain Behav. Immun. 2005, 19, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Morais, L.H.; Schreiber, H.L., IV; Mazmanian, S.K. The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef]
- Zhu, X.; Han, Y.; Du, J.; Liu, R.; Jin, K.; Yi, W. Microbiota-gut-brain axis and the central nervous system. Oncotarget 2017, 8, 53829–53838. [Google Scholar] [CrossRef]
- Dalton, A.; Mermier, C.; Zuhl, M. Exercise influence on the microbiome-gut-brain axis. Gut Microbes 2019, 10, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. Q. Publ. Hell. Soc. Gastroenterol. 2015, 28, 203–209. [Google Scholar]
- Clark, A.; Mach, N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: A systematic review for athletes. J. Int. Soc. Sports Nutr. 2016, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Bonaz, B.; Bazin, T.; Pellissier, S. The vagus nerve at the interface of the microbiota-gut-brain axis. Front. Neurosci. 2018, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, O.; Cronin, O.; Clarke, S.F.; Murphy, E.F.; Molloy, M.G.; Shanahan, F.; Cotter, P.D. Exercise and the microbiota. Gut Microbes 2015, 6, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Carnevali, L.; Sgoifo, A. Vagal modulation of resting heart rate in rats: The role of stress, psychosocial factors, and physical exercise. Front. Physiol. 2014, 5, 118. [Google Scholar] [CrossRef] [PubMed]
- Kai, S.; Nagino, K.; Ito, T.; Oi, R.; Nishimura, K.; Morita, S.; Yaoi, R. Effectiveness of Moderate Intensity Interval Training as an Index of Autonomic Nervous Activity. Rehabil. Res. Prac. 2016, 2016, 6209671. [Google Scholar] [CrossRef] [PubMed]
- Grässler, B.; Thielmann, B.; Böckelmann, I.; Hökelmann, A. Effects of Different Training Interventions on Heart Rate Variability and Cardiovascular Health and Risk Factors in Young and Middle-Aged Adults: A Systematic Review. Front. Physiol. 2021, 12, 657274. [Google Scholar] [CrossRef] [PubMed]
- Tsubokawa, M.; Nishimura, M.; Mikami, T.; Ishida, M.; Hisada, T.; Tamada, Y. Association of Gut Microbial Genera with Heart Rate Variability in the General Japanese Population: The Iwaki Cross-Sectional Research Study. Metabolites 2022, 12, 730. [Google Scholar] [CrossRef]
- Mörkl, S.; Oberascher, A.; Tatschl, J.M.; Lackner, S.; Bastiaanssen, T.F.S.; Butler, M.I.; Moser, M.; Frühwirth, M.; Mangge, H.; Cryan, J.F.; et al. Cardiac vagal activity is associated with gut-microbiome patterns in women—An exploratory pilot study. Dialogues Clin. Neurosci. 2022, 24, 1–9. [Google Scholar] [CrossRef]
- Yang, F.; Ma, Y.; Liang, S.; Shi, Y.; Wang, C. Effect of Exercise Modality on Heart Rate Variability in Adults: A Systematic Review and Network Meta-Analysis. Rev. Cardiovasc. Med. 2024, 25, 9. [Google Scholar] [CrossRef]
- Athanasiou, N.; Bogdanis, G.C.; Mastorakos, G. Endocrine responses of the stress system to different types of exercise. Rev. Endocr. Metab. Disord. 2023, 24, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, S.; Hommel, B.; Kibele, A.; Colzato, L.S. Neuromodulation of Aerobic Exercise—A Review. Front. Psychol. 2016, 6, 1890. [Google Scholar] [CrossRef] [PubMed]
- Paudel, D.; Uehara, O.; Giri, S.; Yoshida, K.; Morikawa, T.; Kitagawa, T.; Matsuoka, H.; Miura, H.; Toyofuku, A.; Kuramitsu, Y.; et al. Effect of psychological stress on the oral-gut microbiota and the potential oral-gut-brain axis. Jpn. Dent. Sci. Rev. 2022, 58, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Keskitalo, A.; Aatsinki, A.-K.; Kortesluoma, S.; Pelto, J.; Korhonen, L.; Lahti, L.; Lukkarinen, M.; Munukka, E.; Karlsson, H.; Karlsson, L. Gut microbiota diversity but not composition is related to saliva cortisol stress response at the age of 2.5 months. Stress 2021, 24, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, S.A.; Koufaki, P.; Mercer, T.H.; Rush, R.; O’connor, E.; Tuffnell, R.; Lindup, H.; Haggis, L.; Dew, T.; Abdulnassir, L.; et al. Aerobic or resistance training and pulse wave velocity in kidney transplant recipients: A 12-week pilot randomized controlled trial (the exercise in renal transplant [ExeRT] Trial). Am. J. Kidney Dis. 2015, 66, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Marullo, A.L.; O’Halloran, K.D. Microbes, metabolites and muscle: Is the gut–muscle axis a plausible therapeutic target in Duchenne muscular dystrophy? Exp. Physiol. 2023, 108, 1132–1143. [Google Scholar] [CrossRef] [PubMed]
- Leal, L.G.; Lopes, M.A.; Batista, M.L., Jr. Physical exercise-induced myokines and muscle-adipose tissue crosstalk: A review of current knowledge and the implications for health and metabolic diseases. Front. Physiol. 2018, 9, 1307. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.H.; Moon, K.M.; Min, K.-W. Exercise-induced myokines can explain the importance of physical activity in the elderly: An overview. Healthcare 2020, 8, 378. [Google Scholar] [CrossRef]
- Nara, H.; Watanabe, R. Anti-inflammatory effect of muscle-derived interleukin-6 and its involvement in lipid metabolism. Int. J. Mol. Sci. 2021, 22, 9889. [Google Scholar] [CrossRef]
- Pedersen, B.K.; Steensberg, A.; Schjerling, P. Muscle-derived interleukin-6: Possible biological effects. J. Physiol. 2001, 536, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Ellingsgaard, H.; Hojman, P.; Pedersen, B.K. Exercise and health—Emerging roles of IL-6. Curr. Opin. Physiol. 2019, 10, 49–54. [Google Scholar] [CrossRef]
- Docherty, S.; Harley, R.; McAuley, J.J.; Crowe, L.A.N.; Pedret, C.; Kirwan, P.D.; Siebert, S.; Millar, N.L. The effect of exercise on cytokines: Implications for musculoskeletal health: A narrative review. BMC Sports Sci. Med. Rehabil. 2022, 14, 5. [Google Scholar] [CrossRef] [PubMed]
- Ellingsgaard, H.; Hauselmann, I.; Schuler, B.; Habib, A.M.; Baggio, L.L.; Meier, D.T.; Eppler, E.; Bouzakri, K.; Wueest, S.; Muller, Y.D.; et al. Interleukin-6 enhances insulin secretion by increasing glucagon-like peptide-1 secretion from L cells and alpha cells. Nat. Med. 2011, 17, 1481–1489. [Google Scholar] [CrossRef]
- Chae, S.A.; Du, M.; Son, J.S.; Zhu, M.-J. Exercise improves homeostasis of the intestinal epithelium by activation of apelin receptor-AMP-activated protein kinase signalling. J. Physiol. 2023, 601, 2371–2389. [Google Scholar] [CrossRef]
- Bonnieu, A.; Carnac, G.; Vernus, B. Myostatin in the Pathophysiology of Skeletal Muscle. J. Cachexia Sarcopenia Muscle 2018, 9, 1213–1234. [Google Scholar] [CrossRef]
- Wang, X.H.; Mitch, W.E. Mechanisms of muscle wasting in chronic kidney disease. Nat. Rev. Nephrol. 2014, 10, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.B.; Han, S.; Yin, X.J.; Liu, H.; Wang, J.; Xuan, M. Myostatin gene deletion alters gut microbiota stimulating fast-twitch glycolytic muscle growth Short. BioRxiv 2022, 2022, 501334. [Google Scholar]
- Pei, Y.; Chen, C.; Mu, Y.; Yang, Y.; Feng, Z.; Li, B.; Li, H.; Li, K. Integrated Microbiome and Metabolome Analysis Reveals a Positive Change in the Intestinal Environment of Myostatin Edited Large White Pigs. Front. Microbiol. 2021, 12, 628685. [Google Scholar] [CrossRef]
- Khalafi, M.; Aria, B.; E Symonds, M.; Rosenkranz, S.K. The effects of resistance training on myostatin and follistatin in adults: A systematic review and meta-analysis. Physiol. Behav. 2023, 269, 114272. [Google Scholar] [CrossRef]
- Clark, A.; Mach, N. The Crosstalk between the Gut Microbiota and Mitochondria during Exercise. Front. Physiol. 2017, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Zhang, B.; Lin, X.; Fu, X.; An, Y.; Zou, Y.; Wang, J.-X.; Wang, Z.; Yu, T. Lactate metabolism in human health and disease. Signal Transduct. Target. Ther. 2022, 7, 305. [Google Scholar] [CrossRef] [PubMed]
- Nalbandian, M.; Takeda, M. Lactate as a signaling molecule that regulates exercise-induced adaptations. Biology 2016, 5, 38. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.P.; Rubio, L.A.; Duncan, S.H.; Donachie, G.E.; Holtrop, G.; Lo, G.; Farquharson, F.M.; Wagner, J.; Parkhill, J.; Louis, P.; et al. Pivotal Roles for pH, Lactate, and Lactate-Utilizing Bacteria in the Stability of a Human Colonic Microbial Ecosystem. MSystems 2020, 5, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Suryani, D.; Alfaqih, M.S.; Gunadi, J.W.; Sylviana, N.; Goenawan, H.; Megantara, I.; Lesmana, R. Type, Intensity, and Duration of Exercise as Regulator of Gut Microbiome Profile. Curr. Sports Med. Rep. 2022, 21, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Scheiman, J.; Luber, J.M.; Chavkin, T.A.; MacDonald, T.; Tung, A.; Pham, L.-D.L.-D.; Wibowo, M.C.; Wurth, R.C.; Punthambaker, S.; Tierney, B.T.; et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 2019, 25, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Valverde, D.; Bonilla, D.A.; Gómez-Miranda, L.M.; Calleja-Núñez, J.J.; Arias, N.; Martínez-Guardado, I. Examining the Interaction between Exercise, Gut Microbiota, and Neurodegeneration: Future Research Directions. Biomedicines 2023, 11, 2267. [Google Scholar] [CrossRef] [PubMed]
- Rysz, J.; Franczyk, B.; Lawisnki, J.; Barreiros, L.; Segundo, M. Uremic toxin levels are associated with gut microbiome changes in kidney transplant recipients. Transpl. Int. 2021, 34, 231. [Google Scholar]
- Lau, W.L.; Savoj, J.; Nakata, M.B.; Vaziri, N.D. Altered microbiome in chronic kidney disease: Systemic effects of gut-derived uremic toxins. Clin. Sci. 2018, 132, 509–522. [Google Scholar] [CrossRef]
- Dziewiecka, H.; Buttar, H.S.; Kasperska, A.; Ostapiuk–Karolczuk, J.; Domagalska, M.; Cichoń, J.; Skarpańska-Stejnborn, A. Physical activity induced alterations of gut microbiota in humans: A systematic review. BMC Sports Sci. Med. Rehabil. 2022, 14, 122. [Google Scholar] [CrossRef]
- Lim, Y.J.; Sidor, N.A.; Tonial, N.C.; Che, A.; Urquhart, B.L. Uremic Toxins in the Progression of Chronic Kidney Disease and Cardiovascular Disease: Mechanisms and Therapeutic Targets. Toxins 2021, 13, 142. [Google Scholar] [CrossRef]
- Bikbov, B.; Purcell, C.A.; Levey, A.S.; Smith, M.; Abdoli, A.; Abebe, M. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020, 395, 709–733. [Google Scholar] [CrossRef]
- Kovesdy, C.P. Epidemiology of chronic kidney disease: An update 2022. Kidney Int. Suppl. 2022, 12, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Lohia, S.; Vlahou, A.; Zoidakis, J. Microbiome in Chronic Kidney Disease: An Omics Perspective. Toxins 2022, 14, 176. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, M.; Wang, J.; Li, R.; Zhang, Y. Alterations to the gut microbiota and their correlation with inflammatory factors in chronic kidney disease. Front. Cell Infect. Microbiol. 2019, 9, 206. [Google Scholar] [CrossRef] [PubMed]
- Meijers, B.; Evenepoel, P.; Anders, H.-J. Intestinal microbiome and fitness in kidney disease. Nat. Rev. Nephrol. 2019, 15, 531–545. [Google Scholar] [CrossRef]
- Di Vincenzo, F.; Del Gaudio, A.; Petito, V.; Lopetuso, L.R.; Scaldaferri, F. Gut microbiota, intestinal permeability, and systemic inflammation: A narrative review. Intern. Emerg. Med. 2023, 19, 275–293. [Google Scholar] [CrossRef]
- Rodríguez-Romero, J.d.J.; Durán-Castañeda, A.C.; Cárdenas-Castro, A.P.; Sánchez-Burgos, J.A.; Zamora-Gasga, V.M.; Sáyago-Ayerdi, S.G. What we know about protein gut metabolites: Implications and insights for human health and diseases. Food Chem. X 2022, 13, 100195. [Google Scholar] [CrossRef]
- Bammens, B.; Evenepoel, P.; Verbeke, K.; Vanrenterghem, Y. Impairment of small intestinal protein assimilation in patients with end-stage renal disease: Extending the malnutrition-inflammation-atherosclerosis concept. Am. J. Clin. Nutr. 2004, 80, 1536–1543. [Google Scholar] [CrossRef]
- Graboski, A.; Redinbo, M. Gut-Derived Protein-Bound Uremic Toxins. Toxins 2020, 12, 590. [Google Scholar] [CrossRef] [PubMed]
- Gryp, T.; De Paepe, K.; Vanholder, R.; Kerckhof, F.-M.; Van Biesen, W.; Van de Wiele, T.; Verbeke, F.; Speeckaert, M.; Joossens, M.; Couttenye, M.M.; et al. Gut microbiota generation of protein -bound uremic toxins and related metabolites is not altered at different stages of chronic kidney disease. Kidney Int. 2020, 97, 1230–1242. [Google Scholar] [CrossRef]
- Vanholder, R.; De Smet, R.; Glorieux, G.; Argilés, A.; Baurmeister, U.; Brunet, P.; Clark, W.; Cohen, G.; De Deyn, P.P.; Deppisch, R.; et al. Review on uremic toxins: Classification, concentration, and interindividual variability. Kidney Int. 2003, 63, 1934–1943. [Google Scholar] [CrossRef] [PubMed]
- Meijers, B.; Glorieux, G.; Poesen, R.; Bakker, S.J. Non extracorporeal Methods for Decreasing Uremic Solute Concentration: A Future Way To Go? Semin. Nephrol. 2014, 34, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Mishima, E.; Fukuda, S.; Mukawa, C.; Yuri, A.; Kanemitsu, Y.; Matsumoto, Y.; Akiyama, Y.; Fukuda, N.N.; Tsukamoto, H.; Asaji, K.; et al. Evaluation of the impact of gut microbiota on uremic solute accumulation by a CE-TOFMS–based metabolomics approach. Kidney Int. 2017, 92, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Song, I.H. The clinical impact of gut microbiota in chronic kidney disease. Korean J. Intern. Med. 2020, 35, 1305–1316. [Google Scholar] [CrossRef] [PubMed]
- Evenepoel, P.; Meijers, B.K.; Bammens, B.R.; Verbeke, K. Uremic toxins originating from colonic microbial metabolism. Kidney Int. 2009, 76, S12–S19. [Google Scholar] [CrossRef] [PubMed]
- Beker, B.M.; Colombo, I.; Gonzalez-Torres, H.; Musso, C.G. Decreasing microbiota-derived uremic toxins to improve CKD outcomes. Clin. Kidney J. 2022, 15, 2214–2219. [Google Scholar] [CrossRef] [PubMed]
- Mafra, D.; Kalantar-Zadeh, K.; Moore, L.W. New Tricks for Old Friends: Treating Gut Microbiota of Patients With CKD. J. Ren. Nutr. 2021, 31, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Rukavina Mikusic, N.L.; Kouyoumdzian, N.M.; Choi, M.R. Gut microbiota and chronic kidney disease: Evidences and mechanisms that mediate a new communication in the gastrointestinal-renal axis. Pflug. Arch. Eur. J. Physiol. 2020, 472, 303–320. [Google Scholar] [CrossRef]
- Hu, X.; Ouyang, S.; Xie, Y.; Gong, Z.; Du, J. Characterizing the gut microbiota in patients with chronic kidney disease. Postgrad. Med. 2020, 132, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Huang, J.; Liu, T.; Hu, Y.; Shi, K.; Zhou, Y.; Zhang, N. Changes in gut microbial community upon chronic kidney disease. PLoS ONE 2023, 18, e0283389. [Google Scholar] [CrossRef] [PubMed]
- Caggiano, G.; Cosola, C.; Di Leo, V.; Gesualdo, M.; Gesualdo, L. Microbiome modulation to correct uremic toxins and to preserve kidney functions. Curr. Opin. Nephrol. Hypertens. 2020, 29, 49–56. [Google Scholar] [CrossRef] [PubMed]
- WHO. Innovative Care for Chronic Conditions: Building Blocks for Action; World Health Organization: Geneva, Switzerland, 2002; Volume 24, p. 159017. [Google Scholar]
- Zhang, F.; Yin, X.; Huang, L.; Zhang, H. The “adult inactivity triad” in patients with chronic kidney disease: A review. Front. Med. 2023, 10, 1160450. [Google Scholar] [CrossRef]
- Wilkinson, T.J.; Clarke, A.L.; Nixon, D.G.D.; Hull, K.L.; Song, Y.; O Burton, J.; Yates, T.; Smith, A.C. Prevalence and correlates of physical activity across kidney disease stages: An observational multicentre study. Nephrol. Dial. Transplant. 2021, 36, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Avesani, C.M.; Trolonge, S.; Deleaval, P.; Baria, F.; Mafra, D.; Faxen-Irving, G.; Chauveau, P.; Teta, D.; Kamimura, M.A.; Cuppari, L.; et al. Physical activity and energy expenditure in haemodialysis patients: An international survey. Nephrol. Dial. Transplant. 2012, 27, 2430–2434. [Google Scholar] [CrossRef]
- Zelle, D.M.; Klaassen, G.; van Adrichem, E.; Bakker, S.J.; Corpeleijn, E.; Navis, G. Physical inactivity: A risk factor and target for intervention in renal care. Nat. Rev. Nephrol. 2017, 13, 152–168. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, W.; Ling, L.; Zhang, Z.; Cui, Z.; Ge, J.; Wang, Y.; Meng, Q.; Wang, Y.; Liu, K.; et al. A Sedentary Lifestyle Changes the Composition and Predicted Functions of the Gut Bacterial and Fungal Microbiota of Subjects from the Same Company. Curr. Microbiol. 2023, 80, 368. [Google Scholar] [CrossRef] [PubMed]
- Jollet, M.; Nay, K.; Chopard, A.; Bareille, M.-P.; Beck, A.; Ollendorff, V.; Vernus, B.; Bonnieu, A.; Mariadassou, M.; Rué, O.; et al. Does Physical Inactivity Induce Significant Changes in Human Gut Microbiota? New Answers Using the Dry Immersion Hypoactivity Model. Nutrients 2021, 13, 3865. [Google Scholar] [CrossRef]
- de Brito, J.S.; Vargas, D.; da Silva, G.S.; Marinho, S.; Borges, N.A.; Cardozo, L.F.M.F.; Fonseca, L.; Ribeiro, M.; Chermut, T.R.; Moura, M.; et al. Uremic toxins levels from the gut microbiota seem not to be altered by physical exercise in hemodialysis patients. Int. Urol. Nephrol. 2022, 54, 687–693. [Google Scholar] [CrossRef]
- Headley, S.A.; Chapman, D.J.; Germain, M.J.; Evans, E.E.; Hutchinson, J.; Madsen, K.L.; Ikizler, T.A.; Miele, E.M.; Kirton, K.; O’Neill, E.; et al. The effects of 16-weeks of prebiotic supplementation and aerobic exercise training on inflammatory markers, oxidative stress, uremic toxins, and the microbiota in pre-dialysis kidney patients: A randomized controlled trial-protocol paper. BMC Nephrol. 2020, 21, 517. [Google Scholar] [CrossRef]
- Ikee, R.; Yano, K.; Tsuru, T. Constipation in chronic kidney disease: It is time to reconsider. Ren. Replace. Ther. 2019, 5, 221–233. [Google Scholar] [CrossRef]
- Cha, R.R.; Park, S.-Y.; Camilleri, M. The Constipation Research Group of Korean Society of Neurogastroenterology and Motility Constipation in Patients with Chronic Kidney Disease. J. Neurogastroenterol. Motil. 2023, 29, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Wing, M.R.; Patel, S.S.; Ramezani, A.; Raj, D.S. Gut microbiome in chronic kidney disease. Exp. Physiol. 2016, 101, 471–477. [Google Scholar] [CrossRef]
- Gao, R.; Tao, Y.; Zhou, C.; Li, J.; Wang, X.; Chen, L.; Li, F.; Guo, L. Exercise therapy in patients with constipation: A systematic review and meta-analysis of randomized controlled trials. Scand. J. Gastroenterol. 2019, 54, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Moorthi, R.N.; Avin, K.G. Clinical relevance of sarcopenia in chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2017, 26, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Meijers, B.; Farré, R.; Dejongh, S.; Vicario, M.; Evenepoel, P. Intestinal barrier function in chronic kidney disease. Toxins 2018, 10, 298. [Google Scholar] [CrossRef]
- Amini Khiabani, S.; Asgharzadeh, M.; Samadi Kafil, H. Chronic kidney disease and gut microbiota. Heliyon 2023, 9, e18991. [Google Scholar] [CrossRef]
- Vaziri, N.D. CKD impairs barrier function and alters microbial flora of the intestine: A major link to inflammation and uremic toxicity. Curr. Opin. Nephrol. Hypertens. 2012, 21, 587–592. [Google Scholar] [CrossRef]
- Ilhan, Z.E.; Marcus, A.K.; Kang, D.-W.; Rittmann, B.E.; Krajmalnik-Brown, R. pH-Mediated Microbial and Metabolic Interactions in Fecal Enrichment Cultures. mSphere 2017, 2, 10-1128. [Google Scholar] [CrossRef]
- Gai, Z.; Chu, L.; Hiller, C.; Arsenijevic, D.; Penno, C.A.; Montani, J.P.; Odermatt, A.; Kullak-Ublick, G.A. Effect of chronic renal failure on the hepatic, intestinal, and renal expression of bile acid transporters. Am. J. Physiol. Ren. Physiol. 2014, 306, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Shankaranarayanan, D.; Raj, D.S. Gut Microbiome and Kidney Disease Reconciling Optimism and Skepticism. Clin. J. Am. Soc. Nephrol. 2022, 17, 1694–1696. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Zhang, J.; Ji, S.; Qin, C.; Wu, Y.; Zou, Y.; Yang, J.; Zhao, Y.; Yang, Q.; Liu, F. Lower bile acids as an independent risk factor for renal outcomes in patients with type 2 diabetes mellitus and biopsy-proven diabetic kidney disease. Front. Endocrinol. 2022, 13, 1026995. [Google Scholar] [CrossRef] [PubMed]
- Frazier, R.; Cai, X.; Lee, J.; Bundy, J.D.; Jovanovich, A.; Chen, J.; Deo, R.; Lash, J.P.; Anderson, A.H.; Go, A.S.; et al. Deoxycholic Acid and Risks of Cardiovascular Events, ESKD, and Mortality in CKD: The CRIC Study. Kidney Med. 2022, 4, 100387. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Zhang, K.; Zhang, Y.; Jin, X.; Jiang, H. Mechanism underlying an elevated serum bile acid level in chronic renal failure patients. Int. Urol. Nephrol. 2015, 47, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.-L.; Cao, G.; Chen, D.-Q.; Vaziri, N.D.; Chen, L.; Zhang, J.; Wang, M.; Guo, Y.; Zhao, Y.-Y. Microbiome–metabolomics reveals gut microbiota associated with glycine-conjugated metabolites and polyamine metabolism in chronic kidney disease. Cell Mol. Life Sci. 2019, 76, 4961–4978. [Google Scholar] [CrossRef] [PubMed]
- Wu, I.-W.; Gao, S.-S.; Chou, H.-C.; Yang, H.-Y.; Chang, L.-C.; Kuo, Y.-L.; Dinh, M.C.V.; Chung, W.-H.; Yang, C.-W.; Lai, H.-C.; et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics 2020, 10, 5398–5411. [Google Scholar] [CrossRef]
- Tanaka, S.; Tanaka, T.; Nangaku, M. Hypoxia and hypoxia-inducible factors in chronic kidney disease. Ren. Replace. Ther. 2016, 2, 25. [Google Scholar] [CrossRef]
- Wang, B.; Li, Z.-L.; Zhang, Y.-L.; Wen, Y.; Gao, Y.-M.; Liu, B.-C. Hypoxia and chronic kidney disease. EBioMedicine 2022, 77, 103942. [Google Scholar] [CrossRef]
- Li, W.; Zhao, Y.; Fu, P. Hypoxia induced factor in chronic kidney disease: Friend or foe? Front. Med. 2017, 4, 317823. [Google Scholar] [CrossRef]
- Dvornikova, K.A.; Platonova, O.N.; Bystrova, E.Y. Hypoxia and Intestinal Inflammation: Common Molecular Mechanisms and Signaling Pathways. Int. J. Mol. Sci. 2023, 24, 2425. [Google Scholar] [CrossRef] [PubMed]
- Lau, W.L.; Kalantar-Zadeh, K.; Vaziri, N.D. The Gut as a Source of Inflammation in Chronic Kidney Disease. Nephron 2015, 130, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, L.; Cardozo, L.F.M.F.; Lindholm, B.; Stenvinkel, P.; Mafra, D. Intestinal alkaline phosphatase modulation by food components: Predictive, preventive, and personalized strategies for novel treatment options in chronic kidney disease. EPMA J. 2020, 11, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Fawley, J.; Gourlay, D.M. Intestinal alkaline phosphatase: A summary of its role in clinical disease. J. Surg. Res. 2016, 202, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Bover, J.; Ureña, P.; Aguilar, A.; Mazzaferro, S.; Benito, S.; López-Báez, V.; Ramos, A.; Dasilva, I.; Cozzolino, M. Alkaline Phosphatases in the Complex Chronic Kidney Disease-Mineral and Bone Disorders. Calcif. Tissue Int. 2018, 103, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Haarhaus, M.; Brandenburg, V.; Kalantar-Zadeh, K.; Stenvinkel, P.; Magnusson, P. Alkaline phosphatase: A novel treatment target for cardiovascular disease in CKD. Nat. Rev. Nephrol. 2017, 13, 429–442. [Google Scholar] [CrossRef]
- Trevaskis, N.L. Intestinal lymphatic dysfunction: A new pathway mediating gut–kidney crosstalk in kidney disease. Kidney Int. 2021, 100, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, N.D.; Yuan, J.; Norris, K. Role of urea in intestinal barrier dysfunction and dysruption of epithelial tight junction in CKD. Am. J. Nephrol. 2014, 37, 1–6. [Google Scholar] [CrossRef]
- Vaziri, N.D.; Yuan, J.; Nazertehrani, S.; Ni, Z.; Liu, S. Chronic kidney disease causes disruption of gastric and small intestinal epithelial tight junction. Am. J. Nephrol. 2013, 38, 99–103. [Google Scholar] [CrossRef]
- Vaziri, N.D.; Goshtasbi, N.; Yuan, J.; Jellbauer, S.; Moradi, H.; Raffatellu, M.; Kalantar-Zadeh, K. Uremic plasma impairs barrier function and depletes the tight junction protein constituents of intestinal epithelium. Bone 2008, 23, 438–443. [Google Scholar] [CrossRef]
- Georgopoulou, G.A.; Papasotiriou, M.; Bosgana, P.; de Lastic, A.L.; Koufou, E.E.; Papachristou, E.; Goumenos, D.S. Altered Expression of Intestinal Tight Junctions in Patients with Chronic Kidney Disease: A Pathogenetic Mechanism of Intestinal Hyperpermeability. Biomedicines 2024, 12, 368. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, C.W.; Harrison, L.E.; Eldehni, M.T.; Jefferies, H.J.; Szeto, C.C. Circulating endotoxemia: A novel factor in systemic inflammation and cardiovascular disease in chronic kidney disease. Clin. J. Am. Soc. Nephrol. 2011, 6, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Szeto, C.-C.; Kwan, B.C.-H.; Chow, K.-M.; Lai, K.-B.; Chung, K.-Y.; Leung, C.-B.; Li, P.K.-T. Endotoxemia is related to systemic inflammation and atherosclerosis in peritoneal dialysis patients. Clin. J. Am. Soc. Nephrol. 2008, 3, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Adda-Rezig, H.; Carron, C.; de Barros, J.-P.P.; Choubley, H.; Charron, E.; Rerole, A.-L.; Laheurte, C.; Louvat, P.; Gaiffe, E.; Simula-Faivre, D.; et al. New Insights on End-Stage Renal Disease and Healthy Individual Gut Bacterial Translocation: Different Carbon Composition of Lipopolysaccharides and Different Impact on Monocyte Inflammatory Response. Front. Immunol. 2021, 12, 658404. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Gautier, T.; Reocreux, G.; Pallot, G.; Maquart, G.; Bahr, P.-A.; Tavernier, A.; Grober, J.; Masson, D.; Bouhemad, B.; et al. Increased Phospholipid Transfer Protein Activity Is Associated With Markers of Enhanced Lipopolysaccharide Clearance in Human During Cardiopulmonary Bypass. Front. Cardiovasc. Med. 2021, 8, 756269. [Google Scholar] [CrossRef] [PubMed]
- Ando, M.; Shibuya, A.; Tsuchiya, K.; Akiba, T.; Nitta, K. Reduced expression of Toll-like receptor 4 contributes to impaired cytokine response of monocytes in uremic patients. Kidney Int. 2006, 70, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Koc, M.; Toprak, A.; Arikan, H.; Odabasi, Z.; Elbir, Y.; Tulunay, A.; Asicioglu, E.; Eksioglu-Demiralp, E.; Glorieux, G.; Vanholder, R.; et al. Toll-like receptor expression in monocytes in patients with chronic kidney disease and haemodialysis: Relation with inflammation. Nephrol. Dial. Transplant. 2011, 26, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Anders, H.-J.; Andersen, K.; Stecher, B. The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int. 2013, 83, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- Baião, V.M.; Cunha, V.A.; Duarte, M.P.; Andrade, F.P.; Ferreira, A.P.; Nóbrega, O.T.; Viana, J.L.; Ribeiro, H.S. Effects of Exercise on Inflammatory Markers in Individuals with Chronic Kidney Disease: A Systematic Review and Meta-Analysis. Metabolites 2023, 13, 795. [Google Scholar] [CrossRef]
- Gesualdo, L.; Di Leo, V.; Coppo, R. The mucosal immune system and IgA nephropathy. Semin. Immunopathol. 2021, 43, 657–668. [Google Scholar] [CrossRef]
- Shah, N.B.; Nigwekar, S.U.; Kalim, S.; Lelouvier, B.; Servant, F.; Dalal, M.; Krinsky, S.; Fasano, A.; Tolkoff-Rubin, N.; Allegretti, A.S. The Gut and Blood Microbiome in IgA Nephropathy and Healthy Controls. Kidney360 2021, 2, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Hilderman, M.; Bruchfeld, A. The cholinergic anti-inflammatory pathway in chronic kidney disease—Review and vagus nerve stimulation clinical pilot study. Nephrol. Dial. Transplant. 2020, 35, 1840–1852. [Google Scholar] [CrossRef] [PubMed]
- Onodugo, O.; Arodiwe, E.; Okoye, J.; Ezeala, B.; Onodugo, N.; Ulasi, I.; Ijoma, C. Prevalence of autonomic dysfunction among pre-dialysis chronic kidney disease patients in a tertiary hospital, South East Nigeria. Afr. Health Sci. 2018, 18, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Seibert, E.; Zohles, K.; Ulrich, C.; Kluttig, A.; Nuding, S.; Kors, J.A.; Swenne, C.A.; Werdan, K.; Fiedler, R.; Girndt, M. Association between autonomic nervous dysfunction and cellular inflammation in end-stage renal disease. BMC Cardiovasc. Disord. 2016, 16, 210. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.; Bagordo, D.; Perrotta, A.M.; Giante, T. Autonomic dysfunction in kidney diseases. Clin. Bioenerg. Pathophysiol. Clin. Transl. 2020, 6, 119–154. [Google Scholar]
- Brotman, D.J.; Bash, L.D.; Qayyum, R.; Crews, D.; Whitsel, E.A.; Astor, B.C.; Coresh, J. Heart rate variability predicts esrd and ckd-related hospitalization. J. Am. Soc. Nephrol. 2010, 21, 1560–1570. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Sprick, J.D.; DaCosta, D.R.; Mammino, K.; Nocera, J.R.; Park, J. Exercise modulates sympathetic and vascular function in chronic kidney disease. J. Clin. Investig. 2023, 8, 164221. [Google Scholar] [CrossRef] [PubMed]
- Barcellos, F.C.; Santos, I.S.; Umpierre, D.; Bohlke, M.; Hallal, P.C. Effects of exercise in the whole spectrum of chronic kidney disease: A systematic review. Clin. Kidney J. 2015, 8, 753–765. [Google Scholar] [CrossRef]
- Heiwe, S.; Jacobson, S.H. Exercise training in adults with ckd: A systematic review and meta-analysis. Am. J. Kidney Dis. 2014, 64, 383–393. [Google Scholar] [CrossRef]
- Clodi, M.; Riedl, M.; Schmaldienst, S.; Vychytil, A.; Kotzmann, H.; Kaider, A.; Bieglmayer, C.; Mayer, G.; Waldhausl, W.; Luger, A. Adrenal function in patients with chronic renal failure. Am. J. Kidney Dis. 1998, 32, 52–55. [Google Scholar] [CrossRef]
- Sagmeister, M.S.; Harper, L.; Hardy, R.S. Cortisol excess in chronic kidney disease—A review of changes and impact on mortality. Front. Endocrinol. 2023, 13, 1075809. [Google Scholar] [CrossRef] [PubMed]
- Chatzipetrou, V.; Bégin, M.-J.; Hars, M.; Trombetti, A. Sarcopenia in Chronic Kidney Disease: A Scoping Review of Prevalence, Risk Factors, Association with Outcomes, and Treatment. Calcif. Tissue Int. 2021, 110, 1–31. [Google Scholar] [CrossRef] [PubMed]
- De Amorim, G.J.; Calado, C.K.M.; Souza de Oliveira, B.C.; Araujo, R.P.O.; Filgueira, T.O. Sarcopenia in Non-Dialysis Chronic Kidney Disease Patients: Prevalence and Associated Factors. Front. Med. 2022, 9, 854410. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.D.; Zhang, H.Z.; Zhang, Y.; Yang, S.P.; Lin, M.; Zhang, Y.M. Relationship between chronic kidney disease and sarcopenia. Sci. Rep. 2021, 11, 20523. [Google Scholar] [CrossRef] [PubMed]
- Arazi, H.; Mohabbat, M.; Saidie, P.; Falahati, A.; Suzuki, K. Effects of Different Types of Exercise on Kidney Diseases. Sports 2022, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cheung, W.-H.; Li, J.; Chow, S.K.-H.; Yu, J.; Wong, S.H.; Ip, M.; Sung, J.J.Y.; Wong, R.M.Y. Understanding the gut microbiota and sarcopenia: A systematic review. J. Cachex. Sarcopenia Muscle 2021, 12, 1393–1407. [Google Scholar] [CrossRef] [PubMed]
- De Spiegeleer, A.; Elewaut, D.; Noortgate, N.V.D.; Janssens, Y.; Debunne, N.; Van Langenhove, S.; Govindarajan, S.; De Spiegeleer, B.; Wynendaele, E. Quorum sensing molecules as a novel microbial factor impacting muscle cells. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165646. [Google Scholar] [CrossRef]
- Wang, K.; Liu, Q.; Tang, M.; Qi, G.; Qiu, C.; Huang, Y.; Yu, W.; Wang, W.; Sun, H.; Ni, X.; et al. Chronic kidney disease-induced muscle atrophy: Molecular mechanisms and promising therapies. Biochem. Pharmacol. 2023, 208, 115407. [Google Scholar] [CrossRef]
- Kosmadakis, G.; Bevington, A.; Smith, A.; Clapp, E.; Viana, J.; Bishop, N.; Feehally, J. Physical exercise in patients with severe kidney disease. Nephron Clin. Prac. 2010, 115, c7–c16. [Google Scholar] [CrossRef]
- Clapp, E.L.; Bevington, A. Exercise-induced biochemical modifications in muscle in chronic kidney disease: Occult acidosis as a potential factor limiting the anabolic effect of exercise. J. Ren. Nutr. 2011, 21, 57–60. [Google Scholar] [CrossRef][Green Version]
- Kopple, J.D.; Kalantar-Zadeh, K.; Mehrotra, R. Risks of chronic metabolic acidosis in patients with chronic kidney disease. Kidney Int. Suppl. 2005, 67, S21–S27. [Google Scholar] [CrossRef]
- Majchrzak, K.M.; Pupim, L.B.; Flakoll, P.J.; Ikizler, T.A. Resistance exercise augments the acute anabolic effects of intradialytic oral nutritional supplementation. Nephrol. Dial. Transplant. 2008, 23, 1362–1369. [Google Scholar] [CrossRef] [PubMed]
- Workeneh, B.T.; Rondon-Berrios, H.; Zhang, L.; Hu, Z.; Ayehu, G.; Ferrando, A.; Kopple, J.D.; Wang, H.; Storer, T.; Fournier, M.; et al. Development of a diagnostic method for detecting increased muscle protein degradation in patients with catabolic conditions. J. Am. Soc. Nephrol. 2006, 17, 3233–3239. [Google Scholar] [CrossRef] [PubMed]
- Watson, E.L.; Kosmadakis, G.C.; Smith, A.C.; Viana, J.L.; Brown, J.R.; Molyneux, K.; Pawluczyk, I.Z.A.; Mulheran, M.; Bishop, N.C.; Shirreffs, S.; et al. Combined walking exercise and alkali therapy in patients with CKD4–5 regulates intramuscular free amino acid pools and ubiquitin E3 ligase expression. Eur. J. Appl. Physiol. 2013, 113, 2111–2124. [Google Scholar] [CrossRef] [PubMed]
- Jo, D.; Yoon, G.; Kim, O.Y.; Song, J. A new paradigm in sarcopenia: Cognitive impairment caused by imbalanced myokine secretion and vascular dysfunction. Biomed. Pharmacother. 2022, 147, 112636. [Google Scholar] [CrossRef] [PubMed]
- Mendes, S.; Leal, D.V.; Baker, L.A.; Ferreira, A.; Smith, A.C.; Viana, J.L. The Potential Modulatory Effects of Exercise on Skeletal Muscle Redox Status in Chronic Kidney Disease. Int. J. Mol. Sci. 2023, 24, 6017. [Google Scholar] [CrossRef]
- Kawao, N.; Kawaguchi, M.; Ohira, T.; Ehara, H.; Mizukami, Y.; Takafuji, Y.; Kaji, H. Renal failure suppresses muscle irisin expression, and irisin blunts cortical bone loss in mice. J. Cachex. Sarcopenia Muscle 2022, 13, 758–771. [Google Scholar] [CrossRef]
- Yang, M.; Luo, S.; Yang, J.; Chen, W.; He, L.; Liu, D.; Zhao, L.; Wang, X. Myokines: Novel therapeutic targets for diabetic nephropathy. Front. Endocrinol. 2022, 13, 1014581. [Google Scholar] [CrossRef]
- Mageswari, R.; Sridhar, M.G.; Nandeesha, H.; Parameshwaran, S.; Vinod, K.V. Irisin and Visfatin Predicts Severity of Diabetic Nephropathy. Indian J. Clin. Biochem. 2019, 34, 342–346. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vandecruys, M.; De Smet, S.; De Beir, J.; Renier, M.; Leunis, S.; Van Criekinge, H.; Glorieux, G.; Raes, J.; Vanden Wyngaert, K.; Nagler, E.; et al. Revitalizing the Gut Microbiome in Chronic Kidney Disease: A Comprehensive Exploration of the Therapeutic Potential of Physical Activity. Toxins 2024, 16, 242. https://doi.org/10.3390/toxins16060242
Vandecruys M, De Smet S, De Beir J, Renier M, Leunis S, Van Criekinge H, Glorieux G, Raes J, Vanden Wyngaert K, Nagler E, et al. Revitalizing the Gut Microbiome in Chronic Kidney Disease: A Comprehensive Exploration of the Therapeutic Potential of Physical Activity. Toxins. 2024; 16(6):242. https://doi.org/10.3390/toxins16060242
Chicago/Turabian StyleVandecruys, Marieke, Stefan De Smet, Jasmine De Beir, Marie Renier, Sofie Leunis, Hanne Van Criekinge, Griet Glorieux, Jeroen Raes, Karsten Vanden Wyngaert, Evi Nagler, and et al. 2024. "Revitalizing the Gut Microbiome in Chronic Kidney Disease: A Comprehensive Exploration of the Therapeutic Potential of Physical Activity" Toxins 16, no. 6: 242. https://doi.org/10.3390/toxins16060242
APA StyleVandecruys, M., De Smet, S., De Beir, J., Renier, M., Leunis, S., Van Criekinge, H., Glorieux, G., Raes, J., Vanden Wyngaert, K., Nagler, E., Calders, P., Monbaliu, D., Cornelissen, V., Evenepoel, P., & Van Craenenbroeck, A. H. (2024). Revitalizing the Gut Microbiome in Chronic Kidney Disease: A Comprehensive Exploration of the Therapeutic Potential of Physical Activity. Toxins, 16(6), 242. https://doi.org/10.3390/toxins16060242







