Identification of a Novel Aflatoxin B1-Degrading Strain, Bacillus halotolerans DDC-4, and Its Response Mechanisms to Aflatoxin B1
Abstract
:1. Introduction
2. Results
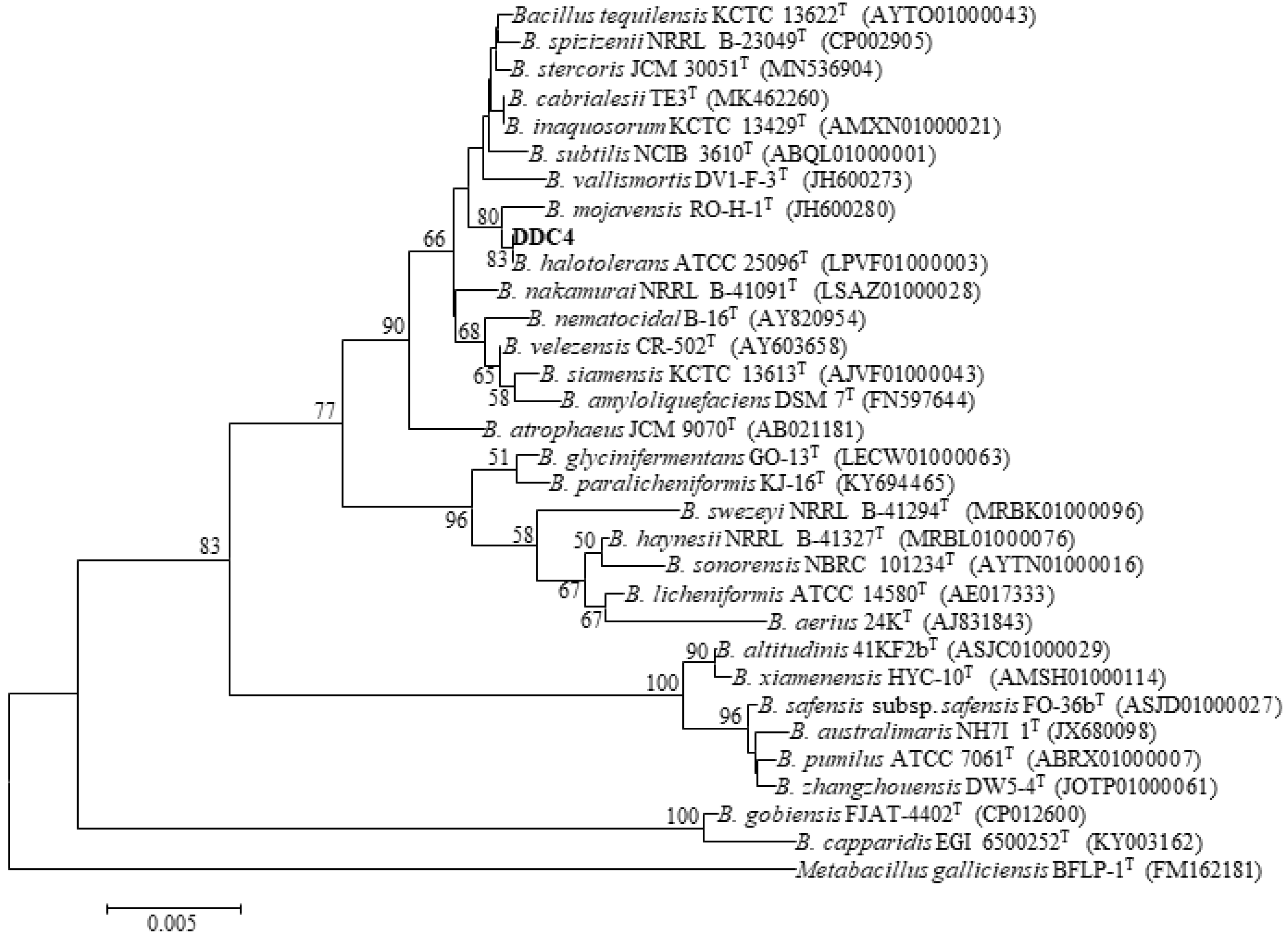
2.1. Isolation and Identification of AFB1-Degrading Strains
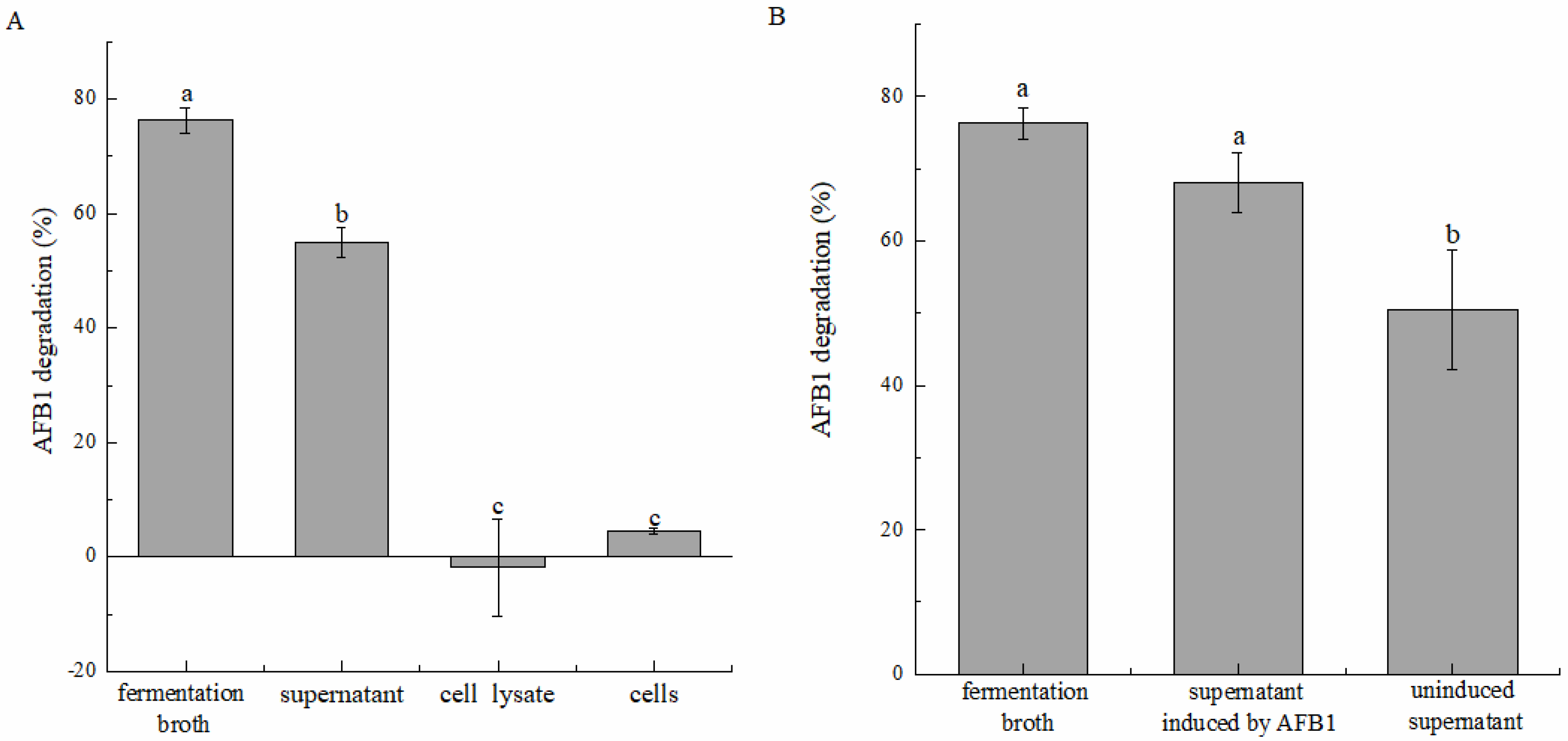
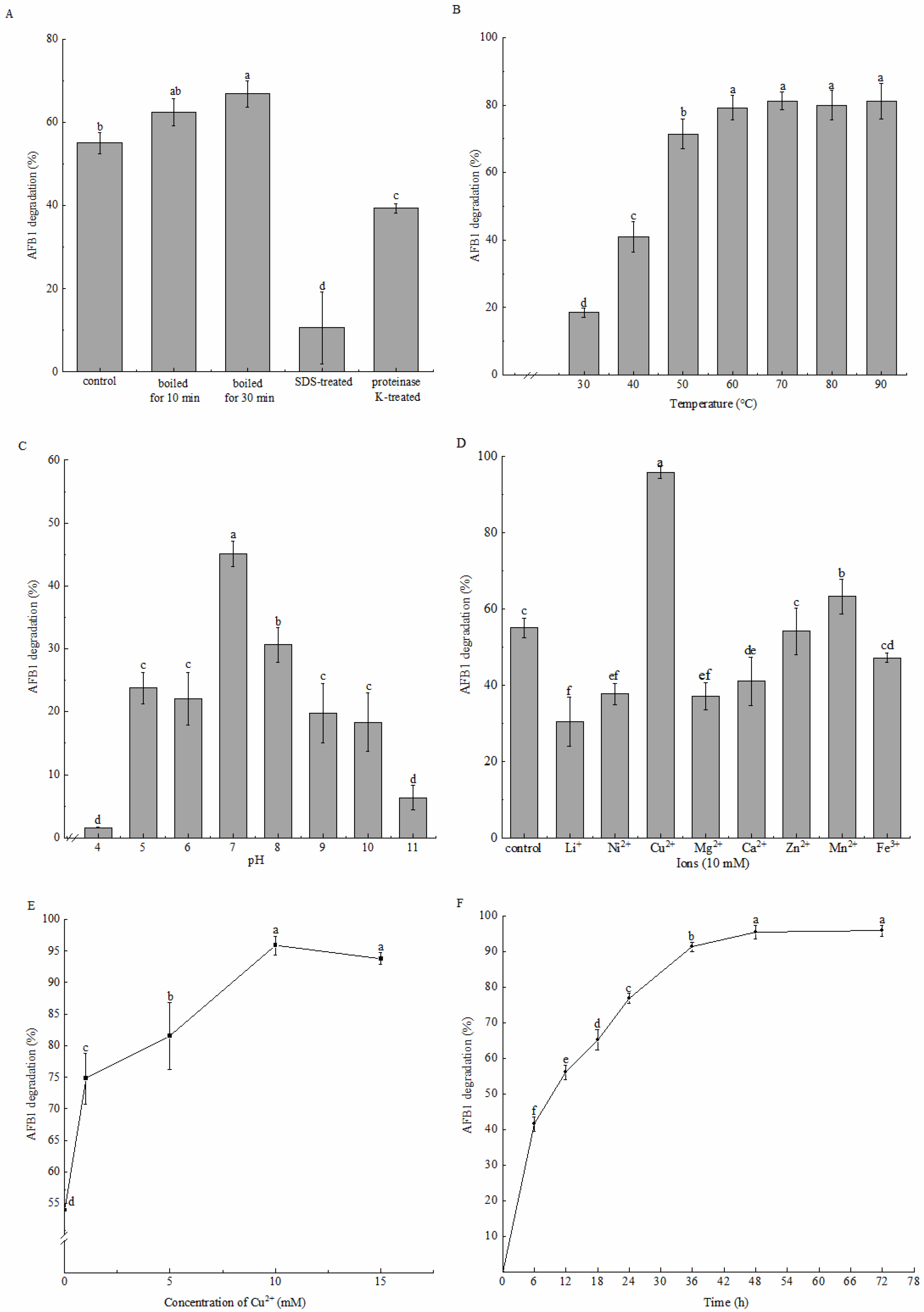
2.2. AFB1 Degradation by the Active Component of Strain DDC-4 and Its Characteristics
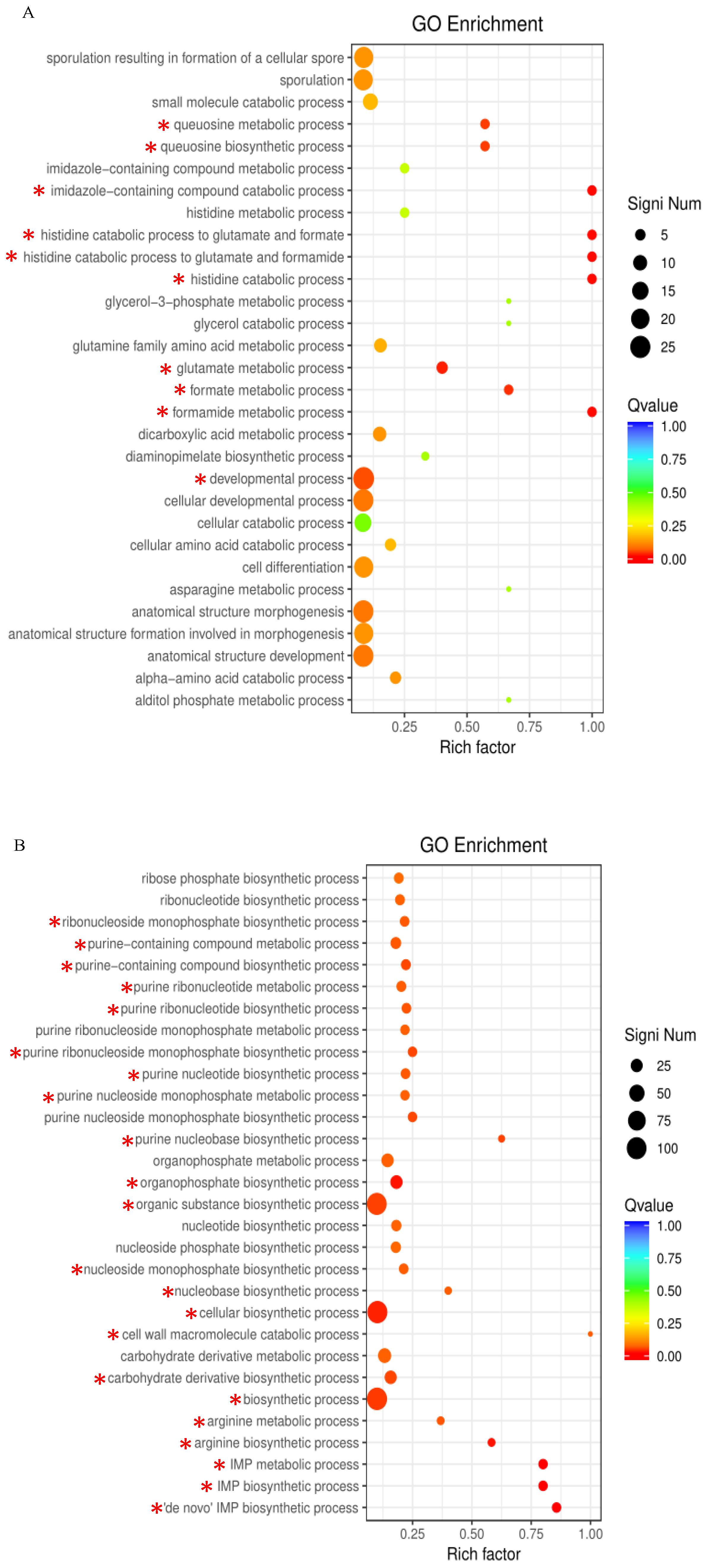
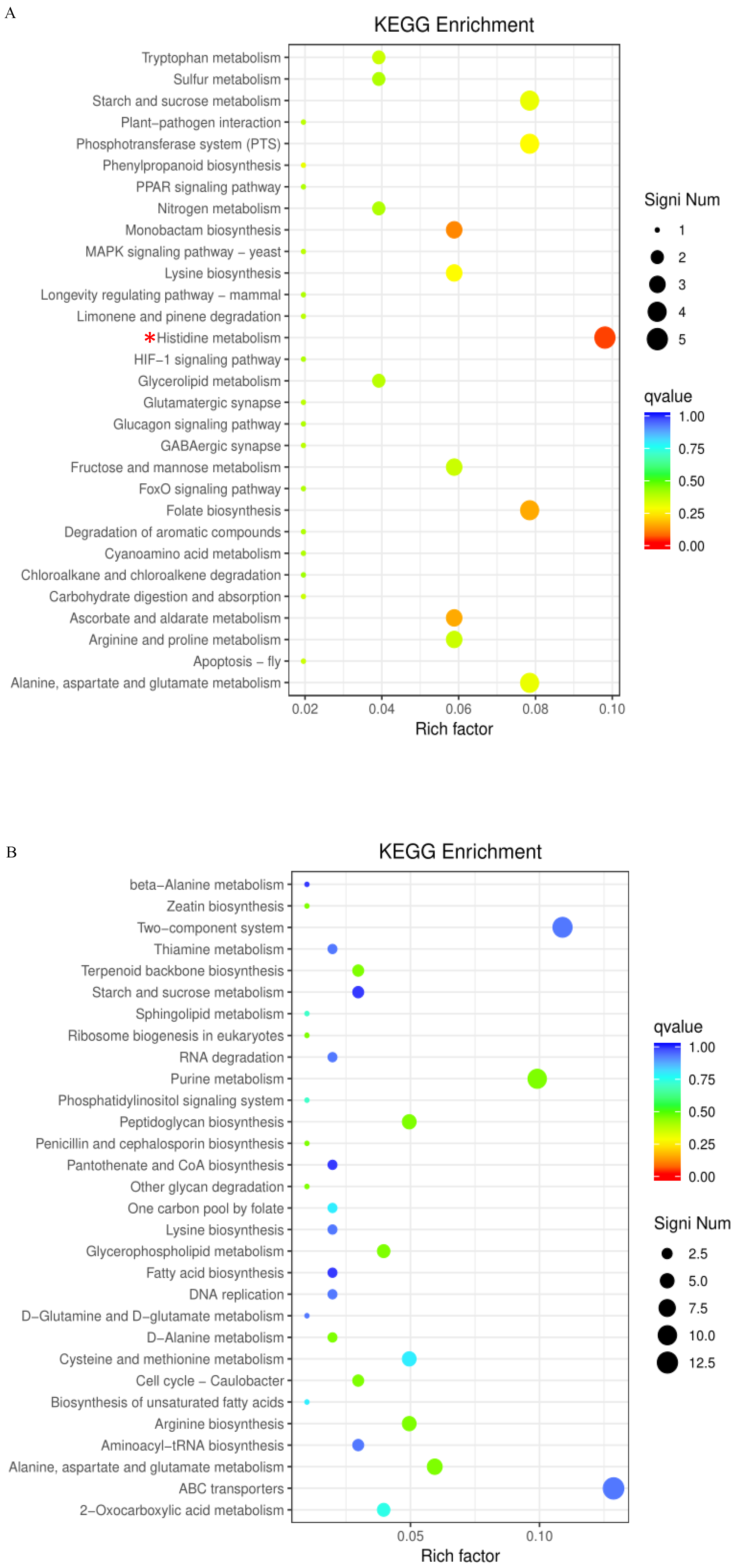
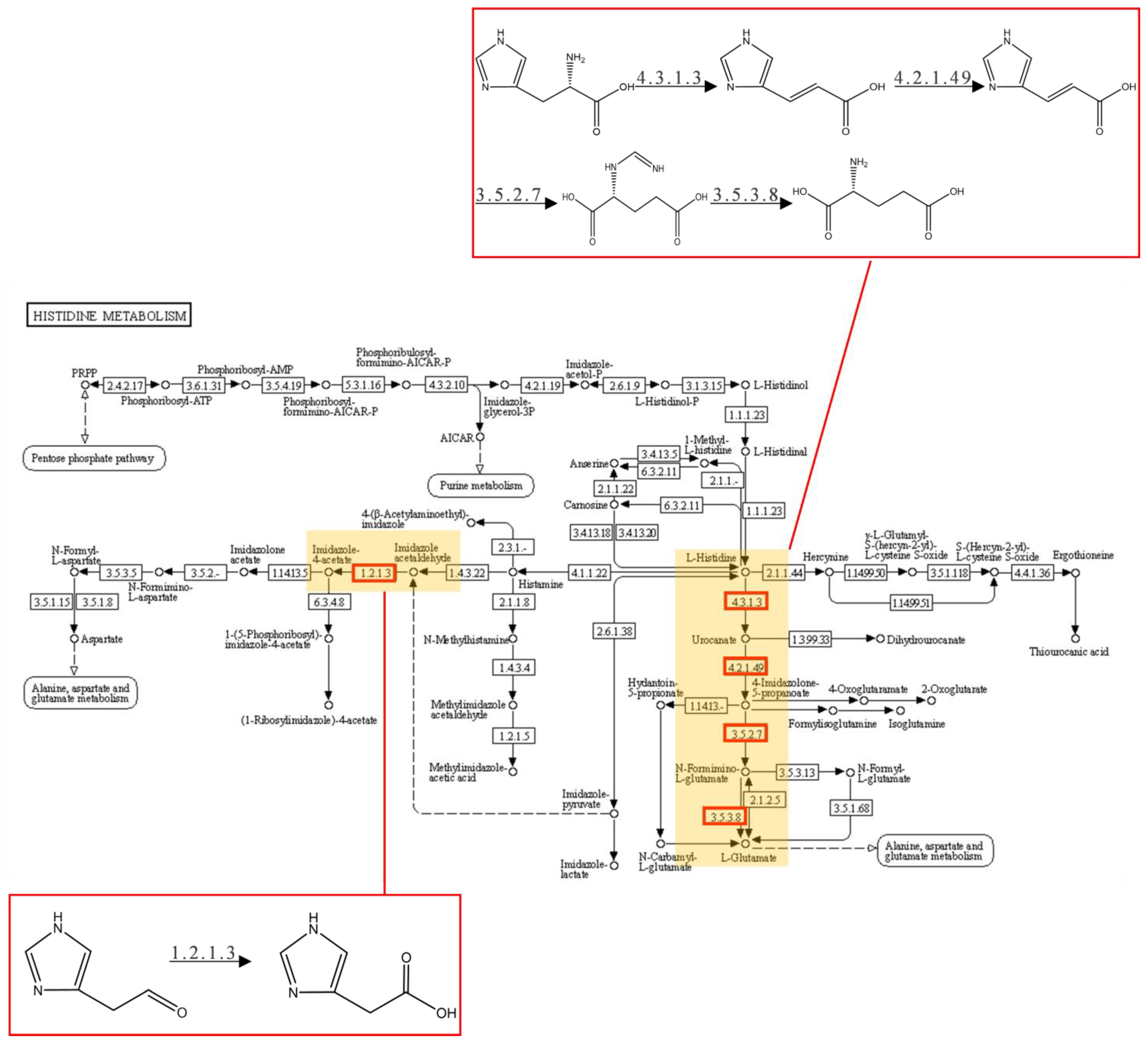
2.3. GO Term and KEGG Pathway Enrichment Analyses
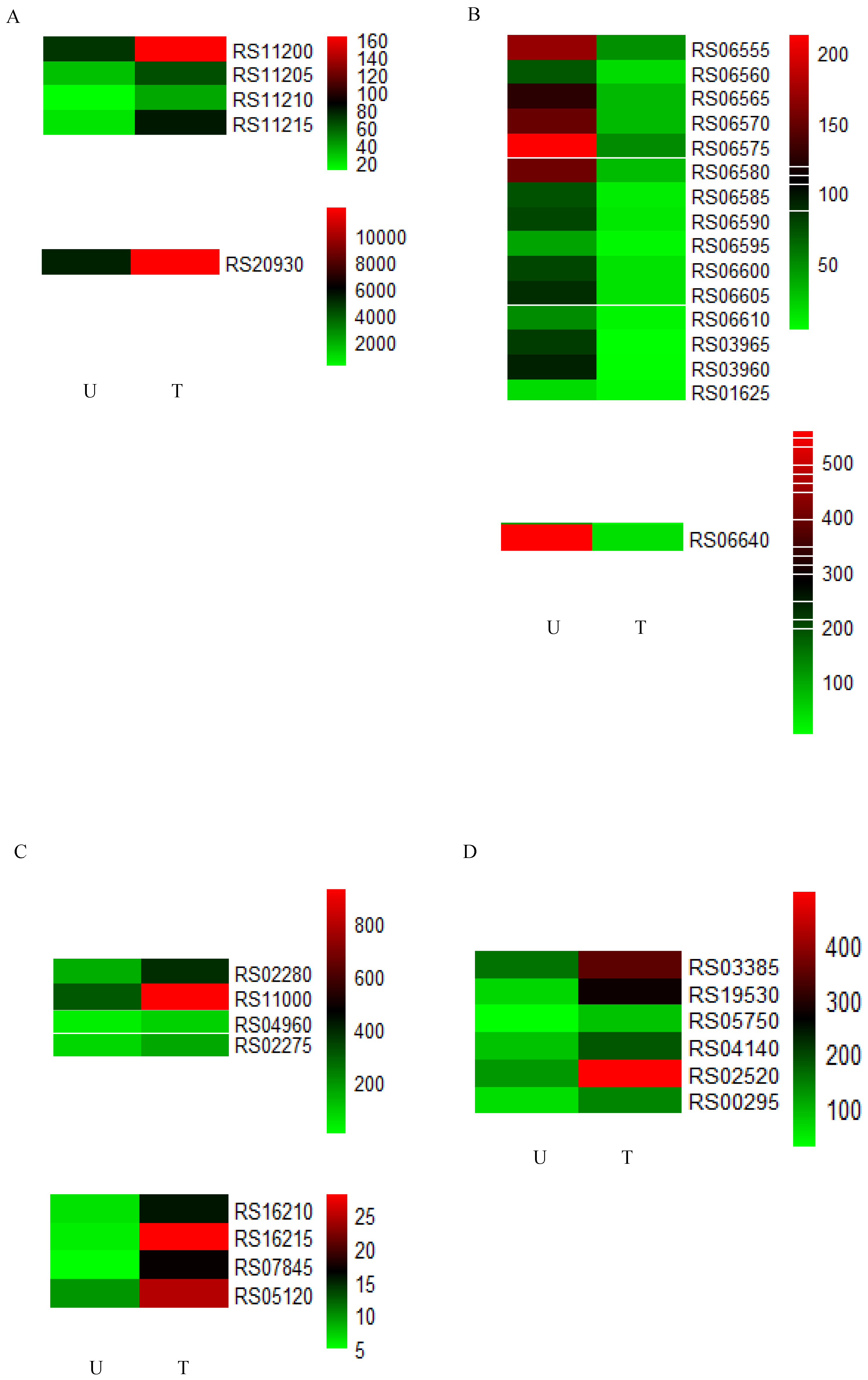
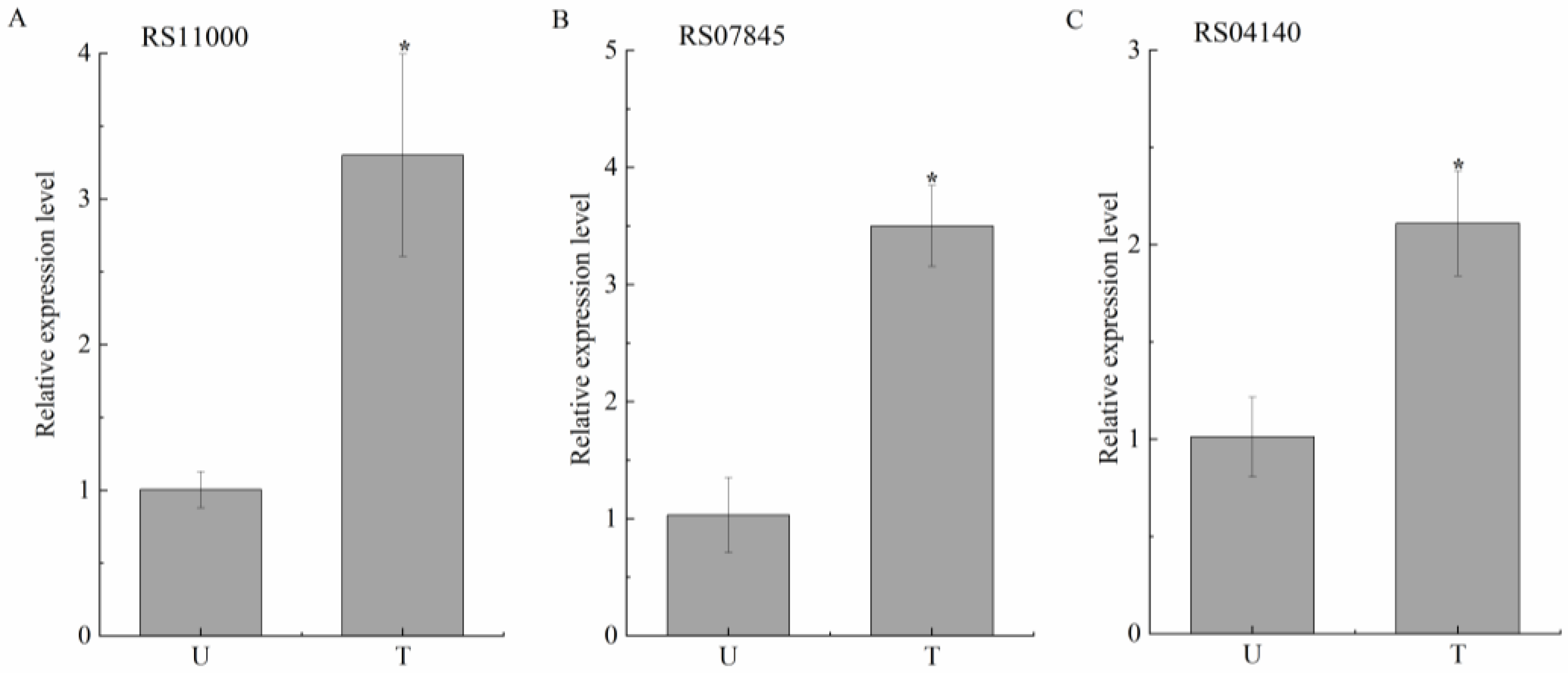
2.4. Identification and Expression Analysis of Potential Degrading Genes
3. Discussion
3.1. AFB1-Degrading Strains and Functional Genes
3.2. Toxicological Effect of AFB1 on Nucleic Acid Synthesis
4. Conclusions
5. Materials and Methods
5.1. Isolation of AFB1-Degrading Strains
5.2. Identification of Strain DDC-4
5.3. AFB1 Degradation by the Cell-Free Supernatant, Cell Suspension, and Cell Lysate
5.4. Induction Effect of Degradation by AFB1
5.5. Effects of Heat, SDS, and Proteinase K Treatments on AFB1 Degradation by the Cell-Free Supernatant
5.6. Effects of Incubation Conditions on AFB1 Degradation by the Cell-Free Supernatant
5.7. Application of the Cell-Free Supernatant to Remove AFB1 from the Moldy Maize Powder
5.8. RNA Extraction and Sequencing
5.9. GO Term and KEGG Pathway Enrichment Analyses
5.10. Quantitative Real-Time PCR
5.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, V.; Bahuguna, A.; Lee, J.S.; Sood, A.; Han, S.S.; Chun, H.S.; Kim, M. Degradation mechanism of aflatoxin B1 and aflatoxin G1 by salt tolerant Bacillus albus YUN5 isolated from ‘doenjang’, a traditional Korean food. Food Res. Int. 2023, 165, 112479. [Google Scholar] [CrossRef]
- Owolabi, I.O.; Karoonuthaisiri, N.; Elliott, C.T.; Petchkongkaew, A. A 10-year analysis of RASFF notifications for my-cotoxins in nuts. Trend in key mycotoxins and impacted countries. Food Res. Int. 2023, 172, 112915. [Google Scholar] [CrossRef]
- Focker, M.; van Asselt, E.D.; Berendsen, B.J.A.; van de Schans, M.G.M.; van Leeuwen, S.P.J.; Visser, S.M.; Van der Fels-Klerx, H.J. Review of food safety hazards in circular food systems in Europe. Food Res. Int. 2022, 158, 111505. [Google Scholar] [CrossRef]
- Ahmad, M.; Han, Z.; Kong, Q. Aflatoxin in peanuts and maize: An overview on occurrence, regulations, prevention, and control methods. World Mycotoxin J. 2023, 16, 99–114. [Google Scholar] [CrossRef]
- Guo, Y.P.; Zhao, L.H.; Ma, Q.G.; Ji, C. Novel strategies for degradation of aflatoxins in food and feed: A review. Food Res. Int. 2021, 140, 109878. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Bahuguna, A.; Ramalingam, S.; Dhakal, G.; Shim, J.J.; Kim, M. Recent technological advances in mechanism, toxicity, and food perspectives of enzyme-mediated aflatoxin degradation. Crit. Rev. Food Sci. 2022, 62, 5395–5412. [Google Scholar] [CrossRef]
- Farzaneh, M.; Shi, Z.Q.; Ghassempour, A.; Sedaghat, N.; Ahmadzadeh, M.; Mirabolfathy, M.; Javan-Nikkhah, M. Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control 2012, 23, 100–106. [Google Scholar] [CrossRef]
- Samuel, M.S.; Sivaramakrishna, A.; Mehta, A. Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeter. Biodegr. 2014, 86, 202–209. [Google Scholar] [CrossRef]
- Taylor, M.C.; Jackson, C.J.; Tattersall, D.B.; French, N.; Peat, T.S.; Newman, J.; Briggs, L.J.; Lapalikar, G.V.; Campbell, P.M.; Scott, C.; et al. Identification and characterization of two families of F420H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010, 78, 561–575. [Google Scholar] [CrossRef]
- Prettl, Z.; Dési, E.; Lepossa, A.; Kriszt, B.; Kukolya, J.; Nagy, E. Biological degradation of aflatoxin B1 by a Rhodococcus pyridinivorans strain in by-product of bioethanol. Anim. Feed. Sci. Tech. 2017, 224, 104–114. [Google Scholar] [CrossRef]
- Xu, D.; Wang, H.X.; Zhang, Y.Z.; Yang, Z.D.; Sun, X.L. Inhibition of non-toxigenic Aspergillus niger FS10 isolated from Chinese fermented soybean on growth and aflatoxin B1 production by Aspergillus flavus. Food Control 2013, 32, 359–365. [Google Scholar] [CrossRef]
- Zhou, G.H.; Chen, Y.J.; Kong, Q.; Ma, Y.X.; Liu, Y. Detoxification of aflatoxin B1 by Zygosaccharomyces rouxii with solid state fermentation in peanut meal. Toxins 2017, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.T.; Xie, C.F.; Yao, D.S.; Zhou, C.Z.; Liu, J.S. Crystal structures of aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme. Biochem. Bioph Res. Commun. 2017, 494, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Suresh, G.; Cabezudo, I.; Pulicharla, R.; Cuprys, A.; Rouissi, T.; Brar, S.K. Biodegradation of aflatoxin B1 with cell-free extracts of Trametes versicolor and Bacillus subtilis. Res. Vet. Sci. 2020, 133, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Alberts, J.F.; Gelderblom, W.C.A.; Botha, A.; van Zyl, W.H. Degradation of aflatoxin B1 by fungal laccase enzymes. Int. J. Food Microbiol. 2009, 135, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Ogata, M.; Hirai, H.; Kawagishi, H. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 2011, 314, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Ranjith, A. Metabolites and degradation pathways of microbial detoxification of aflatoxins: A review. Mycotoxin Res. 2024, 40, 71–83. [Google Scholar] [CrossRef]
- Cao, H.; Liu, D.L.; Mo, X.M.; Xie, C.F.; Yao, D.S. A fungal enzyme with the ability of aflatoxin B1 conversion: Purification and ESI-MS/MS identification. Microbiol. Res. 2011, 166, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, R.; Ng, T.B.; Lai, Y.; Yang, J.; Ye, X. A new laccase of Lac 2 from the white rot fungus Cerrena unicolor 6884 and Lac 2-mediated degradation of aflatoxin B1. Toxins 2020, 12, 476. [Google Scholar] [CrossRef]
- Guo, Y.P.; Qin, X.J.; Tang, Y.; Ma, Q.G.; Zhang, J.Y.; Zhao, L.H. CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 2020, 325, 126877. [Google Scholar] [CrossRef]
- Loi, M.; Renaud, J.B.; Rosini, E.; Pollegioni, L.; Vignali, E.; Haidukowski, M.; Sumarah, M.W.; Logrieco, A.F.; Mulè, G. Enzymatic transformation of aflatoxin B1 by Rh_DypB peroxidase and characterization of the reaction products. Chemosphere 2020, 250, 126296. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, J.; Jin, Y.; Wu, C.; Shen, D.; Zhang, S.; Zhou, R. Aflatoxin B1 degradation by salt tolerant Tetragenococcus halophilus CGMCC 3792. Food Chem. Toxicol. 2018, 121, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.R.; Chen, Y.X.; Zhang, L.Q.; Wu, J.Z.; Zeng, X.H.; Shi, X.G.; Liu, L.M.; Chen, J.F. A comprehensive review on enzymatic biodegradation of polyethylene terephthalate. Environ. Res. 2024, 240, 117427. [Google Scholar] [CrossRef]
- Xu, L.P.; Sun, X.L.; Wan, X.H.; Li, H.; Yan, F.B.; Han, R.L.; Li, H.; Li, Z.J.; Tian, Y.D.; Liu, X.J.; et al. Identification of a Bacillus amyloliquefaciens H6 thioesterase involved in zearalenone detoxification by transcriptomic analysis. J. Agric. Food Chem. 2020, 68, 10071–10080. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Lu, H.; Li, Q.S.; Zhou, Y.; Zhou, J.T. Comparative genome and transcriptome of Rhodococcus pyridinivorans GF3 for analyzing the detoxification mechanism of anthraquinone compounds. Ecotox Environ. Safe 2022, 237, 113545. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.L.; Dhanasekaran, S.; Yang, Q.Y.; Ngea, G.L.N.; Godana, E.A.; Zhang, H.Y. Degradation and stress response mechanism of Cryptococcus podzolicus Y3 on ochratoxin A at the transcriptional level. LWT-Food Sci. Technol. 2022, 157, 113061. [Google Scholar] [CrossRef]
- Hormisch, D.; Brost, I.; Kohring, G.W.; Giffhorn, F.; Kroppenstedt, R.M.; Stackebrandt, E.; Farber, P.; Holzapfel, W.H. Mycobacterium fluoranthenivorans sp. nov., a fluoranthene and aflatoxin B1 degrading bacterium from contaminated soil of a former coal gas plant. Syst. Appl. Microbiol. 2004, 27, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.-X.; Ma, Q.-G.; Niu, T.-G. Aflatoxin B1 degradation by Stenotrophomonas Maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.R.; Vipin, A.V.; Hariprasad, P.; Appaiah, K.A.A.; Venkateswaran, G. Biological detoxification of aflatoxin B1 by Bacillus licheniformis CFR1. Food Control 2017, 71, 234–241. [Google Scholar] [CrossRef]
- Shu, X.; Wang, Y.T.; Zhou, Q.; Li, M.H.; Hu, H.; Ma, Y.H.; Chen, X.; Ni, J.; Zhao, W.W.; Huang, S.W.; et al. Biological degradation of aflatoxin B1 by cell-free extracts of Bacillus velezensis DY3108 with broad pH stability and excellent thermostability. Toxins 2018, 10, 330. [Google Scholar] [CrossRef]
- Chen, G.J.; Fang, Q.A.; Liao, Z.L.; Xu, C.W.; Liang, Z.B.; Liu, T.; Zhong, Q.P.; Wang, L.; Fang, X.; Wang, J. Detoxification of aflatoxin B1 by a potential probiotic Bacillus amyloliquefaciens WF2020. Front. Microbiol. 2022, 13, 891091. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ahmed, M.F.E.; Sangare, L.; Zhao, Y.J.; Selvaraj, J.N.; Xing, F.G.; Wang, Y.; Yang, H.P.; Liu, Y. Novel aflatoxin-degrading enzyme from Bacillus shackletonii L7. Toxins 2017, 9, 36. [Google Scholar] [CrossRef]
- Sangare, L.; Zhao, Y.J.; Folly, Y.M.E.; Chang, J.H.; Li, J.H.; Selvaraj, J.N.; Xing, F.G.; Zhou, L.; Wang, Y.; Liu, Y. Aflatoxin B1 degradation by a Pseudomonas Strain. Toxins 2014, 6, 3028–3040. [Google Scholar] [CrossRef]
- Qiu, T.Y.; Wang, H.M.; Yang, Y.; Yu, J.; Ji, J.; Sun, J.D.; Zhang, S.; Sun, X.L. Exploration of biodegradation mechanism by AFB1-degrading strain Aspergillus niger FS10 and its metabolic feedback. Food Control 2021, 121, 107609. [Google Scholar] [CrossRef]
- Pereyra, M.L.G.; Martínez, M.P.; Cavaglieri, L.R. Presence of aii homologue genes encoding for N-Acyl homoserine lactone-degrading enzyme in aflatoxin B1-decontaminating Bacillus strains with potential use as feed additives. Food Chem. Toxicol. 2019, 124, 316–323. [Google Scholar] [CrossRef]
- Xia, X.S.; Zhang, Y.; Li, M.Y.; Garba, B.; Zhang, Q.; Wang, Y.; Zhang, H.Y.; Li, P.W. Isolation and characterization of a Bacillus subtilis strain with aflatoxin B1 biodegradation capability. Food Control 2017, 75, 92–98. [Google Scholar] [CrossRef]
- Afsharmanesh, H.; Perez-Garcia, A.; Zeriouh, H.; Ahmadzadeh, M.; Romero, D. Aflatoxin degradation by Bacillus subtilis UTB1 is based on production of an oxidoreductase involved in bacilysin biosynthesis. Food Control 2018, 94, 48–55. [Google Scholar] [CrossRef]
- Wang, L.; Huang, W.; Sha, Y.; Yin, H.C.; Liang, Y.; Wang, X.; Shen, Y.; Wu, X.Q.; Wu, D.P.; Wang, J.S. Co-cultivation of two Bacillus strains for improved cell growth and enzyme production to enhance the degradation of aflatoxin B1. Toxins 2021, 13, 435. [Google Scholar] [CrossRef]
- Qian, Z.; Fields, C.J.; Yu, Y.; Lutz, S. Recent progress in engineering alpha/beta hydrolase-fold family members. Biotechnol. J. 2007, 2, 192–200. [Google Scholar] [CrossRef]
- Xing, M.Y.; Chen, Y.; Li, B.Q.; Tian, S.P. Characterization of a short-chain dehydrogenase/reductase and its function in patulin biodegradation in apple juice. Food Chem. 2021, 348, 129046. [Google Scholar] [CrossRef]
- Bernhardt, J.R.; V Lker, U.; V Lker, A.; Antelmann, H.; Schmid, R.; Mach, H.; Hecker, M. Specific and general stress proteins in Bacillus subtilis—A two-deimensional protein electrophoresis study. Microbiology 1997, 143, 999–1017. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, M.; Peng, B.-Z. Transcriptomic analysis reveals the metabolic mechanism of patulin by Saccharomyces cerevisiae during fermentation. LWT-Food Sci. Technol. 2021, 149, 111808. [Google Scholar] [CrossRef]
- Butler, W.H.; Neal, G.E. Mode of action and human health aspects of aflatoxin carcinogenesis. Ann. Nutr. L’alimentation 1977, 31, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Bedard, L.L.; Massey, T.E. Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 2006, 241, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Edwards, U.; Rogall, T.; Blocker, H.; Emde, M.; Bottger, E.C. Isolation and direct complete nucleotide determination of entire genes-Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Micr 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]


| Items | DDC-4 |
|---|---|
| Gram stain | Gram-positive rod |
| Moveability | + |
| Voges-Proskauer | + |
| Oxidation of | |
| glucose | + |
| arabinose | + |
| xylose | + |
| mannitol | + |
| Hydrolysis of | |
| gelatin | + |
| starch | + |
| casein | + |
| Growth on | |
| citrate | + |
| lysozyme | − |
| 5 °C | − |
| 10 °C | − |
| 30 °C | + |
| 40 °C | + |
| 50 °C | + |
| 55 °C | − |
| 65 °C | − |
| NaCl (2%) | + |
| NaCl (5%) | + |
| NaCl (7%) | + |
| NaCl (10%) | + |
| pH 5.7 | + |
| Phenylalanine dehydrolase | − |
| Catalase activity | + |
| Nitrate reduction | + |
| Malonate | + |
| Indole | − |
| Sample Name | The Content of AFB1 (μg/kg) |
|---|---|
| Initial | 6.76 ± 0.85 a |
| Sterilization | 6.39 ± 0.43 a |
| Treatment | 2.96 ± 0.92 b |
| Gene id | Gene Name | Gene Description |
|---|---|---|
| RS06555 | PurD | phosphoribosylamine-glycine ligase |
| RS06560 | PurH | IMP cyclohydrolase |
| RS06565 | PurN | phosphoribosylglycinamide formyltransferase |
| RS06570 | RS06570 | phosphoribosylformylglycinamidine cyclo-ligase |
| RS06575 | RS06575 | amidophosphoribosyltransferase |
| RS06580 | PurL | phosphoribosylformylglycinamidine synthase subunit PurL |
| RS06585 | PurQ | phosphoribosylformylglycinamidine synthase subunit PurQ |
| RS06590 | PurS | phosphoribosylformylglycinamidine synthase subunit PurS |
| RS06595 | RS06595 | phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| RS06600 | PurB | adenylosuccinate lyase |
| RS06605 | PurK | 5-(carboxyamino)imidazole ribonucleotide synthase |
| RS06610 | PurE | 5-(carboxyamino)imidazole ribonucleotide mutase |
| Gene id | Gene Name | Gene Description |
|---|---|---|
| RS16210 | RS16210 | cytochrome ubiquinol oxidase subunit II |
| RS16215 | RS16215 | cytochrome ubiquinol oxidase subunit I |
| RS02280 | AhpA | biofilm-specific peroxidase AhpA |
| RS11000 | RS11000 | aldo/keto reductase |
| RS07845 | RS07845 | SDR family oxidoreductase |
| RS04960 | RS04960 | NAD(P)H-dependent oxidoreductase |
| RS05120 | RS05120 | NAD(P)H-dependent oxidoreductase |
| RS02275 | YkuV | thiol-disulfide oxidoreductase YkuV |
| RS03385 | RS03385 | NUDIX hydrolase |
| RS19530 | RS19530 | alpha/beta hydrolase (haloalkane dehalogenase) |
| RS05750 | RS05750 | amidohydrolase |
| RS04140 | RS04140 | alpha/beta hydrolase (arylesterase) |
| RS02520 | RS02520 | glycoside hydrolase family 18 protein |
| RS00295 | RS00295 | poly-gamma-glutamate hydrolase family protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Zhang, H.; Zhao, Y.; Hao, X.; Liu, Y.; Li, S.; Wu, R. Identification of a Novel Aflatoxin B1-Degrading Strain, Bacillus halotolerans DDC-4, and Its Response Mechanisms to Aflatoxin B1. Toxins 2024, 16, 256. https://doi.org/10.3390/toxins16060256
Guo J, Zhang H, Zhao Y, Hao X, Liu Y, Li S, Wu R. Identification of a Novel Aflatoxin B1-Degrading Strain, Bacillus halotolerans DDC-4, and Its Response Mechanisms to Aflatoxin B1. Toxins. 2024; 16(6):256. https://doi.org/10.3390/toxins16060256
Chicago/Turabian StyleGuo, Jia, Hanlu Zhang, Yixuan Zhao, Xiaoxu Hao, Yu Liu, Suhong Li, and Rina Wu. 2024. "Identification of a Novel Aflatoxin B1-Degrading Strain, Bacillus halotolerans DDC-4, and Its Response Mechanisms to Aflatoxin B1" Toxins 16, no. 6: 256. https://doi.org/10.3390/toxins16060256





