A Micropatterning Strategy to Study Nuclear Mechanotransduction in Cells
Abstract
:1. Introduction
2. Materials and Methods
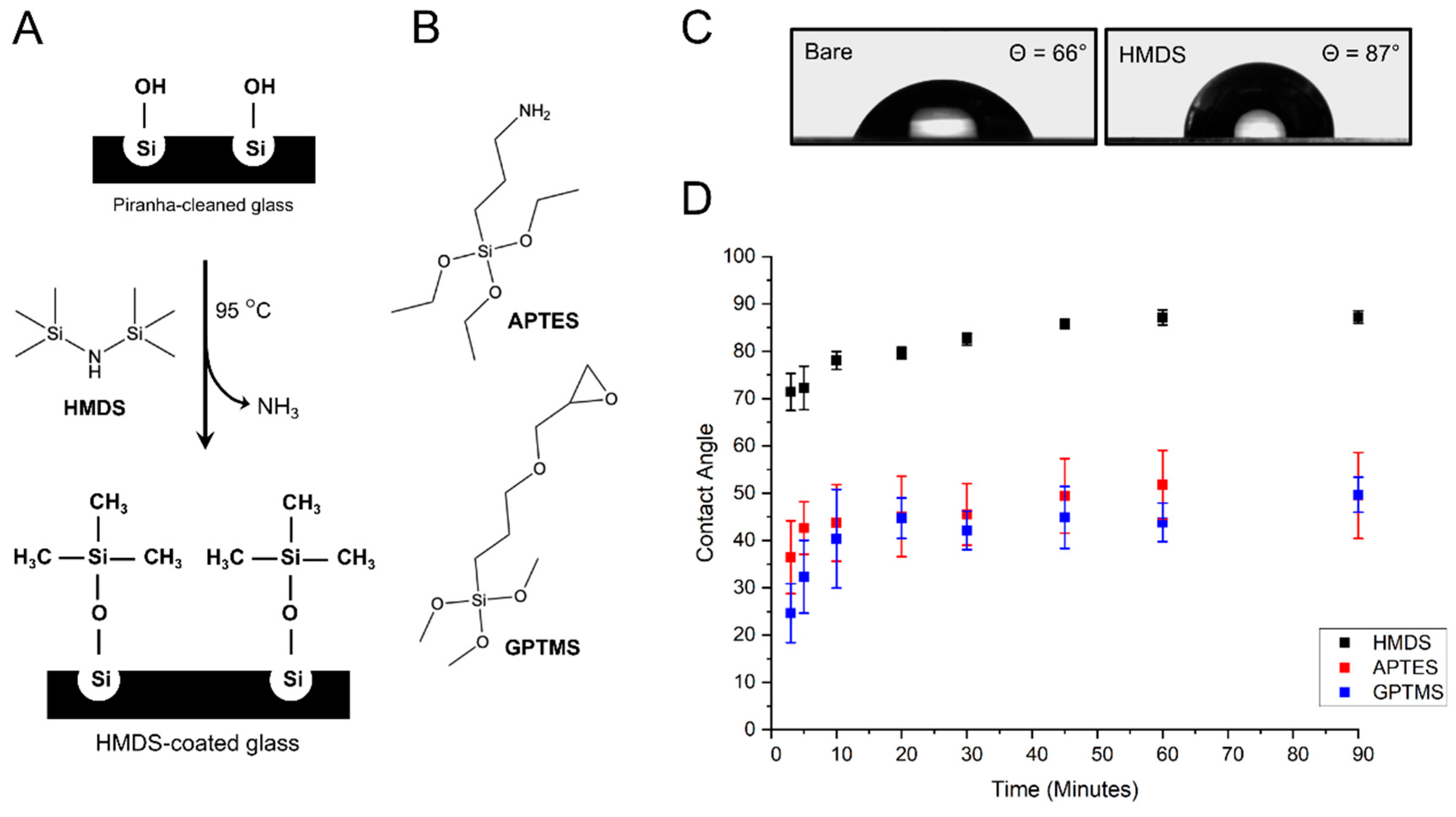
2.1. Surface Modification of Glass Coverslips
2.2. Fabrication of PDMS stamps
2.3. Microcontact Printing
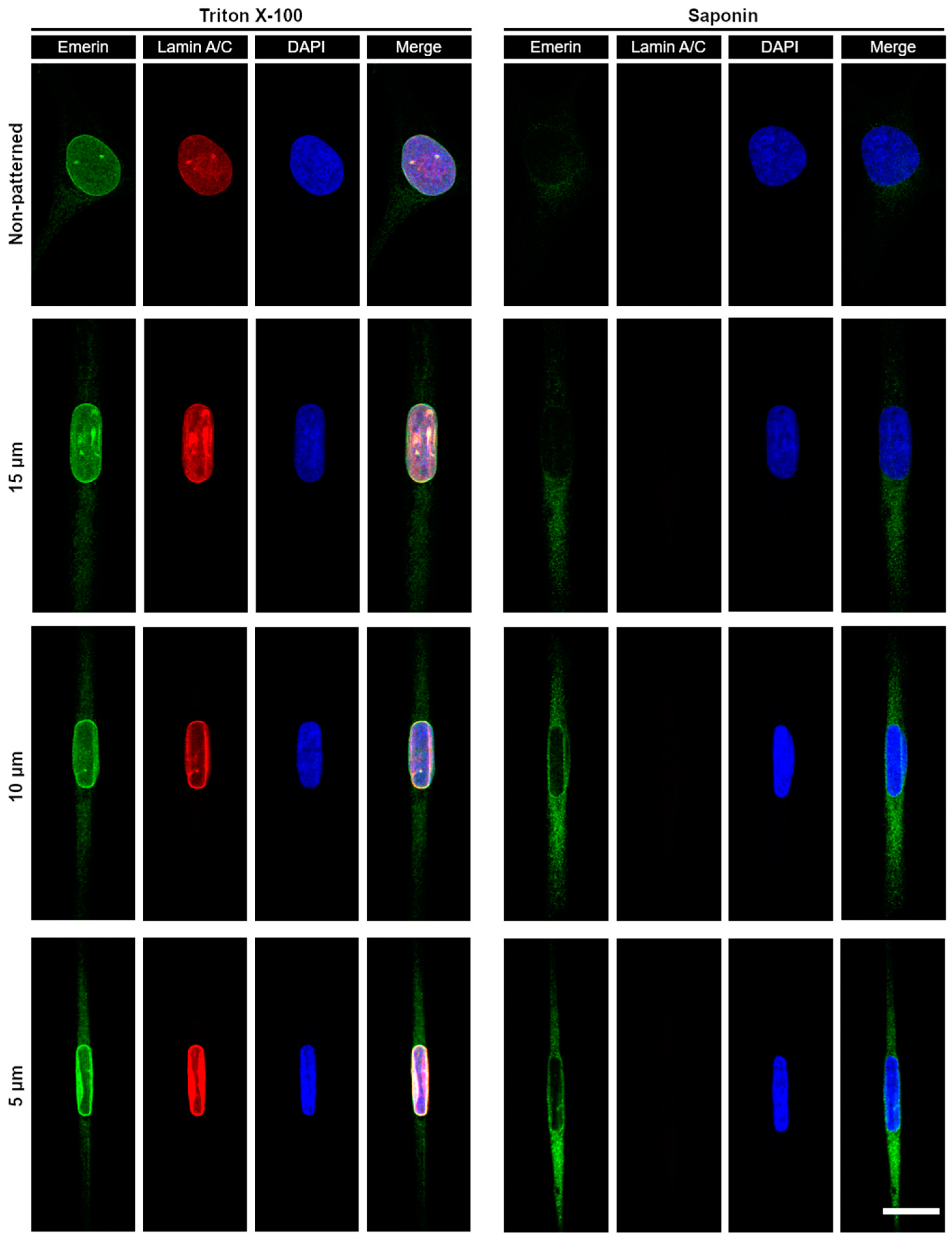
2.4. Cell Culture and Expression of Emerin Fusion
2.5. Immunofluorescence
2.6. Microscopy Imaging
2.7. Image Analysis and Statistics
3. Results
3.1. Vapor-Phase Surface Silanization of Coverslip Surface
3.2. Cell Micropatterning
3.3. Micropatterning to Study Emerin Mechanotransduction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thery, M. Micropatterning as a tool to decipher cell morphogenesis and functions. J. Cell Sci. 2010, 123, 4201–4213. [Google Scholar] [CrossRef]
- Falconnet, D.; Csucs, G.; Grandin, H.M.; Textor, M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials 2006, 27, 3044–3063. [Google Scholar] [CrossRef]
- Martinez-Rivas, A.; González-Quijano, G.; Proa-Coronado, S.; Séverac, C.; Dague, E. Methods of micropatterning and manipulation of cells for biomedical applications. Micromachines 2017, 8, 347. [Google Scholar] [CrossRef] [PubMed]
- Théry, M.; Piel, M. Adhesive micropatterns for cells: A microcontact printing protocol. Cold Spring Harb. Protoc. 2009, 2009. [Google Scholar] [CrossRef] [PubMed]
- Azioune, A.; Carpi, N.; Tseng, Q.; Théry, M.; Piel, M. Protein micropatterns: A direct printing protocol using deep uvs. In Methods Cell Biol.; Cassimeris, L., Tran, P., Eds.; Academic Press: Cambridge, MA, USA, 2010; Volume 97, pp. 133–146. [Google Scholar]
- Tu, C.; Huang, B.; Zhou, J.; Liang, Y.; Tian, J.; Ji, L.; Liang, X.; Ye, X. A microfluidic chip for cell patterning utilizing paired microwells and protein patterns. Micromachines 2016, 8, 1. [Google Scholar] [CrossRef]
- Sahni, G.; Yuan, J.; Toh, Y.C. Stencil micropatterning of human pluripotent stem cells for probing spatial organization of differentiation fates. J. Vis. Exp. 2016, 112, e54097. [Google Scholar] [CrossRef] [PubMed]
- Quist, A.P.; Oscarsson, S. Micropatterned surfaces: Techniques and applications in cell biology. Expert Opin. Drug Discov. 2010, 5, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Desai, R.A.; Gao, L.; Raghavan, S.; Liu, W.F.; Chen, C.S. Cell polarity triggered by cell-cell adhesion via e-cadherin. J. Cell Sci. 2009, 122, 905–911. [Google Scholar] [CrossRef]
- Labouesse, C.; Verkhovsky Alexander, B.; Meister, J.J.; Gabella, C.; Vianay, B. Cell shape dynamics reveal balance of elasticity and contractility in peripheral arcs. Biophys. J. 2015, 108, 2437–2447. [Google Scholar] [CrossRef]
- Versaevel, M.; Grevesse, T.; Gabriele, S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun. 2012, 3, 671. [Google Scholar] [CrossRef]
- Pan, C.J.; Qin, H.; Nie, Y.D.; Ding, H.Y. Control of osteoblast cells adhesion and spreading by microcontact printing of extracellular matrix protein patterns. Colloids Surf. B Biointerfaces 2013, 104, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Alom Ruiz, S.; Chen, C.S. Microcontact printing: A tool to pattern. Soft Matter 2007, 3, 168–177. [Google Scholar] [CrossRef]
- Kumar, A.; Whitesides, G.M. Features of gold having micrometer to centimeter dimensions can be formed through a combination of stamping with an elastomeric stamp and an alkanethiol ink followed by chemical etching. Appl. Phys. Lett. 1993, 63, 2002–2004. [Google Scholar] [CrossRef]
- Wang, D.Y.; Huang, Y.C.; Chiang, H.; Wo, A.M.; Huang, Y.Y. Microcontact printing of laminin on oxygen plasma activated substrates for the alignment and growth of schwann cells. J. Biomed. Mater. Res. Part B Appl. Biomater. 2007, 80, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Qi, J.; Kam, L.C. Microcontact printing of proteins for cell biology. J. Vis. Exp. 2008, 22, e1065. [Google Scholar] [CrossRef]
- Altomare, L.; Riehle, M.; Gadegaard, N.; Tanzi, M.C.; Fare, S. Microcontact printing of fibronectin on a biodegradable polymeric surface for skeletal muscle cell orientation. Int. J. Artif. Organs 2010, 33, 535–543. [Google Scholar] [CrossRef]
- Collins, C.M.; Nee, K.A.; Holaska, J.M. The Nuclear Envelope Protein Emerin and Its Interacting Proteins; Els: New Lisbon, NJ, USA, 2016; pp. 1–9. [Google Scholar]
- Berk, J.M.; Simon, D.N.; Jenkins-Houk, C.R.; Westerbeck, J.W.; Gronning-Wang, L.M.; Carlson, C.R.; Wilson, K.L. The molecular basis of emerin-emerin and emerin-baf interactions. J. Cell Sci. 2014, 127, 3956–3969. [Google Scholar] [CrossRef]
- Rowat, A.C.; Lammerding, J.; Ipsen, J.H. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys. J. 2006, 91, 4649–4664. [Google Scholar] [CrossRef]
- Lammerding, J.; Hsiao, J.; Schulze, P.C.; Kozlov, S.; Stewart, C.L.; Lee, R.T. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 2005, 170, 781–791. [Google Scholar] [CrossRef]
- Méjat, A. Linc complexes in health and disease. Nucleus 2014, 1, 40–52. [Google Scholar] [CrossRef]
- Emery, A.E.; Dreifuss, F.E. Unusual type of benign x-linked muscular dystrophy. J. Neurol. Neurosurg. Psychiatry 1966, 29, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, L.C.; Jackson, J.A.; Elsas, L.J. Emery-dreifuss humeroperoneal muscular dystrophy: An x-linked myopathy with unusual contractures and bradycardia. Ann. Neurol. 1981, 10, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Fairley, E.A.; Kendrick-Jones, J.; Ellis, J.A. The emery-dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J. Cell Sci. 1999, 112 (Pt 15), 2571–2582. [Google Scholar]
- Emery, A.E. Emery-dreifuss muscular dystrophy—A 40 year retrospective. Neuromuscul. Disord. 2000, 10, 228–232. [Google Scholar] [CrossRef]
- Holaska, J.M.; Wilson, K.L. Multiple roles for emerin: Implications for emery-dreifuss muscular dystrophy. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2006, 288, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Guilluy, C.; Osborne, L.D.; Van Landeghem, L.; Sharek, L.; Superfine, R.; Garcia-Mata, R.; Burridge, K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 2014, 16, 376–381. [Google Scholar] [CrossRef]
- Fernandez, A.; Bautista, M.; Stanciauskas, R.; Chung, T.; Pinaud, F. Cell-shaping micropatterns for quantitative super-resolution microscopy imaging of membrane mechanosensing proteins. ACS Appl. Mater. Interfaces 2017, 9, 27575–27586. [Google Scholar] [CrossRef]
- Kawai, A.; Kawakami, J. Wetting analysis of hydrophobic substrate treated by hmds primer. J. Photopolym. Sci. Technol. 2007, 20, 815–816. [Google Scholar] [CrossRef]
- Tsukruk, V.V.; Luzinov, I.; Julthongpiput, D. Sticky molecular surfaces: Epoxysilane self-assembled monolayers. Langmuir 1999, 15, 3029–3032. [Google Scholar] [CrossRef]
- Luzinov, I.; Julthongpiput, D.; Liebmann-Vinson, A.; Cregger, T.; Foster, M.D.; Tsukruk, V.V. Epoxy-terminated self-assembled monolayers: Molecular glues for polymer layers. Langmuir 2000, 16, 504–516. [Google Scholar] [CrossRef]
- Kyaw, H.H.; Al-Harthi, S.H.; Sellai, A.; Dutta, J. Self-organization of gold nanoparticles on silanated surfaces. Beilstein J. Nanotechnol. 2015, 6, 2345–2353. [Google Scholar] [CrossRef] [PubMed]
- Acres, R.G.; Ellis, A.V.; Alvino, J.; Lenahan, C.E.; Khodakov, D.A.; Metha, G.F.; Andersson, G.G. Molecular structure of 3-aminopropyltriethoxysilane layers formed on silanol-terminated silicon surfaces. J. Phys. Chem. C 2012, 116, 6289–6297. [Google Scholar] [CrossRef]
- Zhu, M.; Lerum, M.Z.; Chen, W. How to prepare reproducible, homogeneous, and hydrolytically stable aminosilane-derived layers on silica. Langmuir 2011, 28, 416–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.L.; Liu, W.; Nelson, C.M.; Raghavan, S.; Chen, C.S. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 2004, 10, 865–872. [Google Scholar] [CrossRef]
- Koch, A.J.; Holaska, J.M. Emerin in health and disease. Semin. Cell Dev. Biol. 2014, 29, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Fidzianska, A.; Hausmanowa-Petrusewicz, I. Architectural abnormalities in muscle nuclei. Ultrastructural differences between x-linked and autosomal dominant forms of edmd. J. Neurol. Sci. 2003, 210, 47–51. [Google Scholar] [CrossRef]
- Le, H.Q.; Ghatak, S.; Yeung, C.Y.C.; Tellkamp, F.; Günschmann, C.; Dieterich, C.; Yeroslaviz, A.; Habermann, B.; Pombo, A.; Niessen, C.M.; et al. Mechanical regulation of transcription controls polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 2016, 18, 864–875. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bautista, M.; Fernandez, A.; Pinaud, F. A Micropatterning Strategy to Study Nuclear Mechanotransduction in Cells. Micromachines 2019, 10, 810. https://doi.org/10.3390/mi10120810
Bautista M, Fernandez A, Pinaud F. A Micropatterning Strategy to Study Nuclear Mechanotransduction in Cells. Micromachines. 2019; 10(12):810. https://doi.org/10.3390/mi10120810
Chicago/Turabian StyleBautista, Markville, Anthony Fernandez, and Fabien Pinaud. 2019. "A Micropatterning Strategy to Study Nuclear Mechanotransduction in Cells" Micromachines 10, no. 12: 810. https://doi.org/10.3390/mi10120810
APA StyleBautista, M., Fernandez, A., & Pinaud, F. (2019). A Micropatterning Strategy to Study Nuclear Mechanotransduction in Cells. Micromachines, 10(12), 810. https://doi.org/10.3390/mi10120810





