The Usual Suspects 2019: of Chips, Droplets, Synthesis, and Artificial Cells
Abstract
:1. Introduction
2. Cell-Based Protein Synthesis—A Ménage à Trois of Droplets, Digital Microfluidics, and Cells
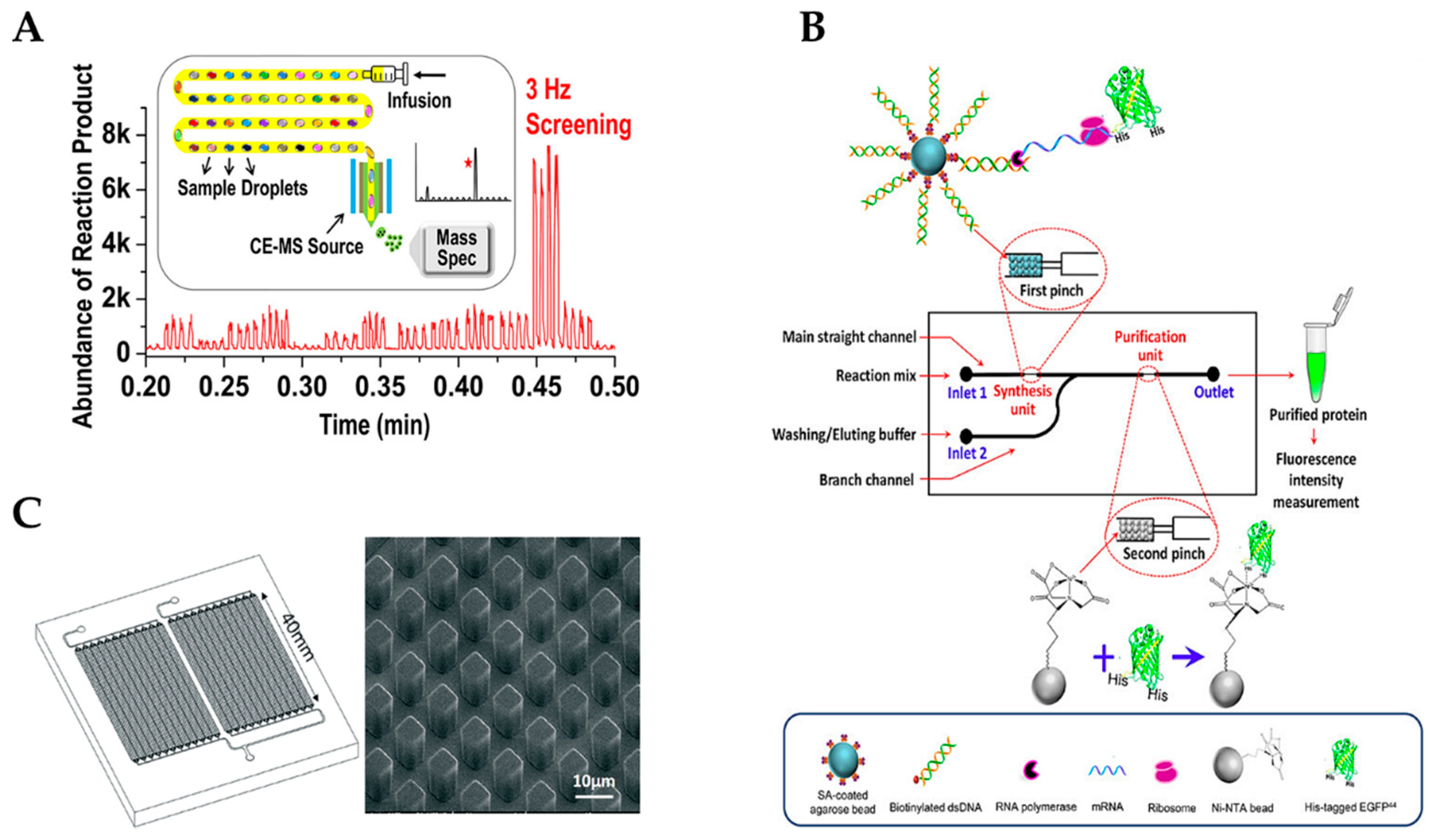
3. Microfluidic Devices for Cell-Free Protein Expression
4. Microfluidics and Artificial Cells
5. Conclusions and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ehmoser-Sinner, E.-K.; Tan, C.-W.D. Lessons on Synthetic Bioarchitectures—Interaction of Living Matter with Synthetic Structural Analogues; Springer International Publishing: Cham, Switzerland, 2018; Volume 1. [Google Scholar]
- Malinova, V.; Nallani, M.; Meier, W.P.; Sinner, E.K. Synthetic biology, inspired by synthetic chemistry. FEBS Lett. 2012, 586, 2146–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gulati, S.; Rouilly, V.; Niu, X.; Chappell, J.; Kitney, R.I.; Edel, J.B.; Freemont, P.S.; deMello, A.J. Opportunities for microfluidic technologies in synthetic biology. J. R. Soc. Interface 2009, 6, S493–S506. [Google Scholar] [CrossRef] [Green Version]
- Heinemann, M.; Panke, S. Synthetic biology—putting engineering into biology. Bioinformatics 2006, 22, 2790–2799. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Han, P.; You, C.; Zhang, Y.P.J. An in vitro synthetic biology platform for emerging industrial biomanufacturing: Bottom-up pathway design. Synth. Syst. Biotechnol. 2018, 3, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Linshiz, G.; Jensen, E.; Stawski, N.; Bi, C.; Elsbree, N.; Jiao, H.; Kim, J.; Mathies, R.; Keasling, J.D.; Hillson, N.J. End-to-end automated microfluidic platform for synthetic biology: from design to functional analysis. J. Biol. Eng. 2016, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Gach, P.C.; Iwai, K.; Kim, P.W.; Hillson, N.J.; Singh, A.K. Droplet microfluidics for synthetic biology. Lab Chip 2017, 17, 3388–3400. [Google Scholar] [CrossRef] [Green Version]
- Benner, S.A.; Sismour, A.M. Synthetic biology. Nat. Rev. Genet. 2005, 6, 533–543. [Google Scholar] [CrossRef]
- Jessop-Fabre, M.M.; Sonnenschein, N. Improving reproducibility in synthetic biology. Front. Bioeng. Biotechnol. 2019, 7, 18. [Google Scholar] [CrossRef]
- Szita, N.; Polizzi, K.; Jaccard, N.; Baganz, F. Microfluidic approaches for systems and synthetic biology. Curr. Opin. Biotechnol. 2010, 21, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Wartmann, D.; Rothbauer, M.; Kuten, O.; Barresi, C.; Visus, C.; Felzmann, T.; Ertl, P. Automated, miniaturized, and integrated quality control-on-chip (QC-on-a-chip) for cell-based cancer therapy applications. Front. Mater. 2015, 2, 60. [Google Scholar] [CrossRef]
- Rothbauer, M.; Zirath, H.; Ertl, P. Recent advances in microfluidic technologies for cell-to-cell interaction studies. Lab Chip 2018, 18, 249–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothbauer, M.; Wartmann, D.; Charwat, V.; Ertl, P. Recent advances and future applications of microfluidic live-cell microarrays. Biotechnol. Adv. 2015, 33, 948–961. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Gao, M.; Wen, L.; He, C.; Chen, Y.; Liu, C.; Fu, X.; Huang, S. Applications of microfluidics in quantitative biology. Biotechnol. J. 2018, 13, e1700170. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Huang, J.H.; Westerhof, T.M.; Lombardo, J.A.; Henrikson, K.M.; Pennell, M.; Pourfard, P.P.; Nelson, E.L.; Nath, P.; Haun, J.B. Microfluidic channel optimization to improve hydrodynamic dissociation of cell aggregates and tissue. Sci. Rep. 2018, 8, 2774. [Google Scholar] [CrossRef]
- Seemann, R.; Brinkmann, M.; Pfohl, T.; Herminghaus, S. Droplet based microfluidics. Rep. Prog. Phys. 2012, 75, 016601. [Google Scholar] [CrossRef]
- Göpfrich, K.; Platzman, I.; Spatz, J.P. Mastering complexity: Towards bottom-up construction of multifunctional eukaryotic synthetic cells. Trends Biotechnol. 2018, 36, 938–951. [Google Scholar] [CrossRef] [PubMed]
- Postiglione, L.; Napolitano, S.; Pedone, E.; Rocca, D.L.; Aulicino, F.; Santorelli, M.; Tumaini, B.; Marucci, L.; di Bernardo, D. Regulation of gene expression and signaling pathway activity in mammalian cells by automated microfluidics feedback control. ACS Synth. Biol. 2018, 7, 2558–2565. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, M.; Jug, F.; Julou, T.; Deshpande, S.; Pfohl, T.; Silander, O.K.; Myers, G.; van Nimwegen, E. Monitoring single-cell gene regulation under dynamically controllable conditions with integrated microfluidics and software. Nat. Commun. 2018, 9, 212. [Google Scholar] [CrossRef] [Green Version]
- Kolnik, M.; Tsimring, L.S.; Hasty, J. Vacuum-assisted cell loading enables shear-free mammalian microfluidic culture. Lab Chip 2012, 12, 4732–4737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Boehm, C.R.; Hibberd, J.M.; Abell, C.; Haseloff, J.; Burgess, S.J.; Reyna-Llorens, I. Droplet-based microfluidic analysis and screening of single plant cells. PLoS ONE 2018, 13, e0196810. [Google Scholar] [CrossRef]
- Siltanen, C.A.; Cole, R.H.; Poust, S.; Chao, L.; Tyerman, J.; Kaufmann-Malaga, B.; Ubersax, J.; Gartner, Z.J.; Abate, A.R. An oil-free picodrop bioassay platform for synthetic biology. Sci. Rep. 2018, 8, 7913. [Google Scholar] [CrossRef] [PubMed]
- Jebrail, M.J.; Bartsch, M.S.; Patel, K.D. Digital microfluidics: A versatile tool for applications in chemistry, biology and medicine. Lab Chip 2012, 12, 2452–2463. [Google Scholar] [CrossRef] [PubMed]
- Husser, M.C.; Vo, P.Q.N.; Sinha, H.; Ahmadi, F.; Shih, S.C.C. An automated induction microfluidics system for synthetic biology. ACS Synth. Biol. 2018, 7, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, F.; Samlali, K.; Vo, P.Q.N.; Shih, S.C.C. An integrated droplet-digital microfluidic system for on-demand droplet creation, mixing, incubation, and sorting. Lab Chip 2019, 19, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Aufinger, L.; Simmel, F.C. Artificial gel-based organelles for spatial organization of cell-free gene expression reactions. Angewandte Chem. Int. Ed. 2018, 57, 17245–17248. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.W.; Majewska, N.I.; Chen, C.X.; Albanetti, T.E.; Jimenez, R.B.C.; Schmelzer, A.E.; Jewett, M.C.; Roy, V. Development of a CHO-based cell-free platform for synthesis of active monoclonal antibodies. ACS Synth. Biol. 2017, 6, 1370–1379. [Google Scholar] [CrossRef]
- Garamella, J.; Marshall, R.; Rustad, M.; Noireaux, V. The all E. coli TX-TL toolbox 2.0: A platform for cell-free synthetic biology. ACS Synth. Biol. 2016, 5, 344–355. [Google Scholar] [CrossRef]
- Endo, Y.; Sawasaki, T. Cell-free expression systems for eukaryotic protein production. Curr. Opin. Biotechnol. 2006, 17, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Damiati, S.; Mhanna, R.; Kodzius, R.; Ehmoser, E.K. Cell-free approaches in synthetic biology utilizing microfluidics. Genes 2018, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, H.; Kanamori, T.; Shimizu, Y.; Ueda, T. A highly controllable reconstituted cell-free system—A breakthrough in protein synthesis research. Curr. Pharm. Biotechnol. 2010, 11, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Spirin, A.S.; Baranov, V.I.; Ryabova, L.A.; Ovodov, S.Y.; Alakhov, Y.B. A continuous cell-free translation system capable of producing polypeptides in high yield. Science 1988, 242, 1162–1164. [Google Scholar] [CrossRef]
- Park, N.; Um, S.H.; Funabashi, H.; Xu, J.; Luo, D. A cell-free protein-producing gel. Nat. Mater. 2009, 8, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Kahn, J.S.; Ruiz, R.C.; Sureka, S.; Peng, S.; Derrien, T.L.; An, D.; Luo, D. DNA microgels as a platform for cell-free protein expression and display. Biomacromolecules 2016, 17, 2019–2026. [Google Scholar] [CrossRef]
- Thiele, J.; Ma, Y.; Foschepoth, D.; Hansen, M.M.; Steffen, C.; Heus, H.A.; Huck, W.T. DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation. Lab Chip 2014, 14, 2651–2656. [Google Scholar] [CrossRef]
- Jiao, Y.; Liu, Y.; Luo, D.; Huck, W.T.S.; Yang, D. Microfluidic-assisted fabrication of clay microgels for cell-free protein synthesis. ACS Appl. Mater. Interfaces 2018, 10, 29308–29313. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Majumder, S.; Emery, N.J.; Liu, A.P. Simultaneous monitoring of transcription and translation in mammalian cell-free expression in bulk and in cell-sized droplets. Synth. Biol. 2018, 3, ysy005. [Google Scholar] [CrossRef] [PubMed]
- Niederholtmeyer, H.; Sun, Z.Z.; Hori, Y.; Yeung, E.; Verpoorte, A.; Murray, R.M.; Maerkl, S.J. Rapid cell-free forward engineering of novel genetic ring oscillators. Elife 2015, 4, e09771. [Google Scholar] [CrossRef] [PubMed]
- Stricker, J.; Cookson, S.; Bennett, M.R.; Mather, W.H.; Tsimring, L.S.; Hasty, J. A fast, robust and tunable synthetic gene oscillator. Nature 2008, 456, 516–519. [Google Scholar] [CrossRef]
- Rosier, B.J.; de Greef, T.F. How to make an oscillator. Elife 2015, 4, e12260. [Google Scholar] [CrossRef]
- Uriu, K. Genetic oscillators in development. Dev. Growth Differ. 2016, 58, 16–30. [Google Scholar] [CrossRef] [Green Version]
- Goldbeter, A. Dissipative structures in biological systems: Bistability, oscillations, spatial patterns and waves. Philos. Trans. A Math. Phys. Eng. Sci. 2018, 376. [Google Scholar] [CrossRef] [PubMed]
- Purcell, O.; Savery, N.J.; Grierson, C.S.; di Bernardo, M. A comparative analysis of synthetic genetic oscillators. J. R. Soc. Interface 2010, 7, 1503–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novák, B.; Tyson, J.J. Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 2008, 9, 981–991. [Google Scholar] [CrossRef]
- Yelleswarapu, M.; van der Linden, A.J.; van Sluijs, B.; Pieters, P.A.; Dubuc, E.; de Greef, T.F.A.; Huck, W.T.S. Sigma factor-mediated tuning of bacterial cell-free synthetic genetic oscillators. ACS Synth. Biol. 2018, 7, 2879–2887. [Google Scholar] [CrossRef] [PubMed]
- Levskaya, A.; Weiner, O.D.; Lim, W.A.; Voigt, C.A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009, 461, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Dobson, J. Remote control of cellular behaviour with magnetic nanoparticles. Nat. Nanotechnol. 2008, 3, 139–143. [Google Scholar] [CrossRef]
- Tschirhart, T.; Kim, E.; McKay, R.; Ueda, H.; Wu, H.C.; Pottash, A.E.; Zargar, A.; Negrete, A.; Shiloach, J.; Payne, G.F.; et al. Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling. Nat. Commun. 2017, 8, 14030. [Google Scholar] [CrossRef]
- Efrat, Y.; Tayar, A.M.; Daube, S.S.; Levy, M.; Bar-Ziv, R.H. Electric-field manipulation of a compartmentalized cell-free gene expression reaction. ACS Synth. Biol. 2018, 7, 1829–1833. [Google Scholar] [CrossRef] [PubMed]
- Shojaeian, M.; Lehr, F.C.O.; Göringer, H.U.; Hardt, S. On-demand production of femtoliter drops in microchannels and their use as biological reaction compartments. Anal. Chem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Quertinmont, L.T.; Orru, R.; Lutz, S. RApid Parallel Protein EvaluatoR (RAPPER), from gene to enzyme function in one day. Chem. Commun. 2015, 51, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, A.B.; Govindarajan, S.; Lutz, S. Improved biocatalysts from a synthetic circular permutation library of the flavin-dependent oxidoreductase old yellow enzyme. J. Am. Chem. Soc. 2013, 135, 14425–14432. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, X.W.; Farasat, I.; Guetschow, E.D.; Welch, C.J.; Kennedy, R.T.; Sun, S.; Moore, J.C. Enabling biocatalysis by high-throughput protein engineering using droplet microfluidics coupled to mass spectrometry. ACS Omega 2018, 3, 1498–1508. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Wang, S. Application of microfluidic chip technology in pharmaceutical analysis: A review. J. Pharm. Anal. 2019. [Google Scholar] [CrossRef]
- Tsao, C.W. Polymer microfluidics: Simple, low-cost fabrication process bridging academic lab research to commercialized production. Micromachines 2016, 7, 225. [Google Scholar] [CrossRef] [PubMed]
- Holtze, C.; Weisse, S.A.; Vranceanu, M. Commercial value and challenges of drop-based microfluidic screening platforms—An opinion. Micromachines 2017, 8, 193. [Google Scholar] [CrossRef]
- Gale, B.K.; Jafek, A.R.; Lambert, C.J.; Goenner, B.L.; Moghimifam, H.; Nze, U.C.; Kamarap, S.K. A review of current methods in microfluidic device fabrication and future commercialization prospects. Inventions 2018, 3, 60. [Google Scholar] [CrossRef]
- Xiao, X.; Zhou, Y.; Sun, Y.; Wang, Q.; Liu, J.; Huang, J.; Zhu, X.; Yang, X.; Wang, K. Integration of cell-free protein synthesis and purification in one microfluidic chip for on-demand production of recombinant protein. Biomicrofluidics 2018, 12, 054102. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.D.M.; Gamage, S.S.T.; Jackson, J.M.; Witek, M.A.; Park, D.S.; Murphy, M.C.; Godwin, A.K.; Soper, S.A. Microfluidic-based solid phase extraction of cell free DNA. Lab Chip 2018, 18, 3459–3470. [Google Scholar] [CrossRef]
- Yewdall, N.A.; Mason, A.F.; van Hest, J.C.M. The hallmarks of living systems: Towards creating artificial cells. Interface Focus 2018, 8, 20180023. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M.; Karimi, N.; Safaei, M. Application of various types of liposomes in drug delivery systems. Adv. Pharm. Bull. 2017, 7, 3–9. [Google Scholar] [CrossRef]
- Buddingh, B.C.; van Hest, J.C.M. Artificial cells: Synthetic compartments with life-like functionality and adaptivity. Acc. Chem. Res. 2017, 50, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xin, X.; Liu, T.; Wang, S.; Yang, Y.; Luan, X.; Xu, G.; Yuan, S. Ionic self-assembly of a giant vesicle as a smart microcarrier and microreactor. Langmuir 2016, 32, 9548–9556. [Google Scholar] [CrossRef]
- Trantidou, T.; Friddin, M.S.; Salehi-Reyhani, A.; Ces, O.; Elani, Y. Droplet microfluidics for the construction of compartmentalised model membranes. Lab Chip 2018, 18, 2488–2509. [Google Scholar] [CrossRef]
- Jorgensen, I.L.; Kemmer, G.C.; Pomorski, T.G. Membrane protein reconstitution into giant unilamellar vesicles: A review on current techniques. Eur. Biophys. J. 2017, 46, 103–119. [Google Scholar] [CrossRef]
- Elani, Y.; Trantidou, T.; Wylie, D.; Dekker, L.; Polizzi, K.; Law, R.V.; Ces, O. Constructing vesicle-based artificial cells with embedded living cells as organelle-like modules. Sci. Rep. 2018, 8, 4564. [Google Scholar] [CrossRef] [PubMed]
- Trantidou, T.; Dekker, L.; Polizzi, K.; Ces, O.; Elani, Y. Functionalizing cell-mimetic giant vesicles with encapsulated bacterial biosensors. Interface Focus 2018, 8, 20180024. [Google Scholar] [CrossRef]
- Weiss, M.; Frohnmayer, J.P.; Benk, L.T.; Haller, B.; Janiesch, J.W.; Heitkamp, T.; Borsch, M.; Lira, R.B.; Dimova, R.; Lipowsky, R.; et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater. 2018, 17, 89–96. [Google Scholar] [CrossRef]
- Deshpande, S.; Spoelstra, W.K.; van Doorn, M.; Kerssemakers, J.; Dekker, C. Mechanical division of cell-sized liposomes. ACS Nano 2018, 12, 2560–2568. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, S.; Caspi, Y.; Meijering, A.E.; Dekker, C. Octanol-assisted liposome assembly on chip. Nat. Commun. 2016, 7, 10447. [Google Scholar] [CrossRef] [Green Version]
- Yandrapalli, N.; Robinson, T. Ultra-high capacity microfluidic trapping of giant vesicles for high-troughput membrane studies. Lab Chip 2019, 19, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Chen, Y.Y.; Gonzalez, R.; Peterson, T.C.; Zhao, H.; Baltz, R.H. Synthetic biology advances and applications in the biotechnology industry: A perspective. J. Ind. Microbiol. Biotechnol. 2018, 45, 449–461. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eilenberger, C.; Spitz, S.; Bachmann, B.E.M.; Ehmoser, E.K.; Ertl, P.; Rothbauer, M. The Usual Suspects 2019: of Chips, Droplets, Synthesis, and Artificial Cells. Micromachines 2019, 10, 285. https://doi.org/10.3390/mi10050285
Eilenberger C, Spitz S, Bachmann BEM, Ehmoser EK, Ertl P, Rothbauer M. The Usual Suspects 2019: of Chips, Droplets, Synthesis, and Artificial Cells. Micromachines. 2019; 10(5):285. https://doi.org/10.3390/mi10050285
Chicago/Turabian StyleEilenberger, Christoph, Sarah Spitz, Barbara Eva Maria Bachmann, Eva Kathrin Ehmoser, Peter Ertl, and Mario Rothbauer. 2019. "The Usual Suspects 2019: of Chips, Droplets, Synthesis, and Artificial Cells" Micromachines 10, no. 5: 285. https://doi.org/10.3390/mi10050285