Low-Cost Battery-Powered and User-Friendly Real-Time Quantitative PCR System for the Detection of Multigene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
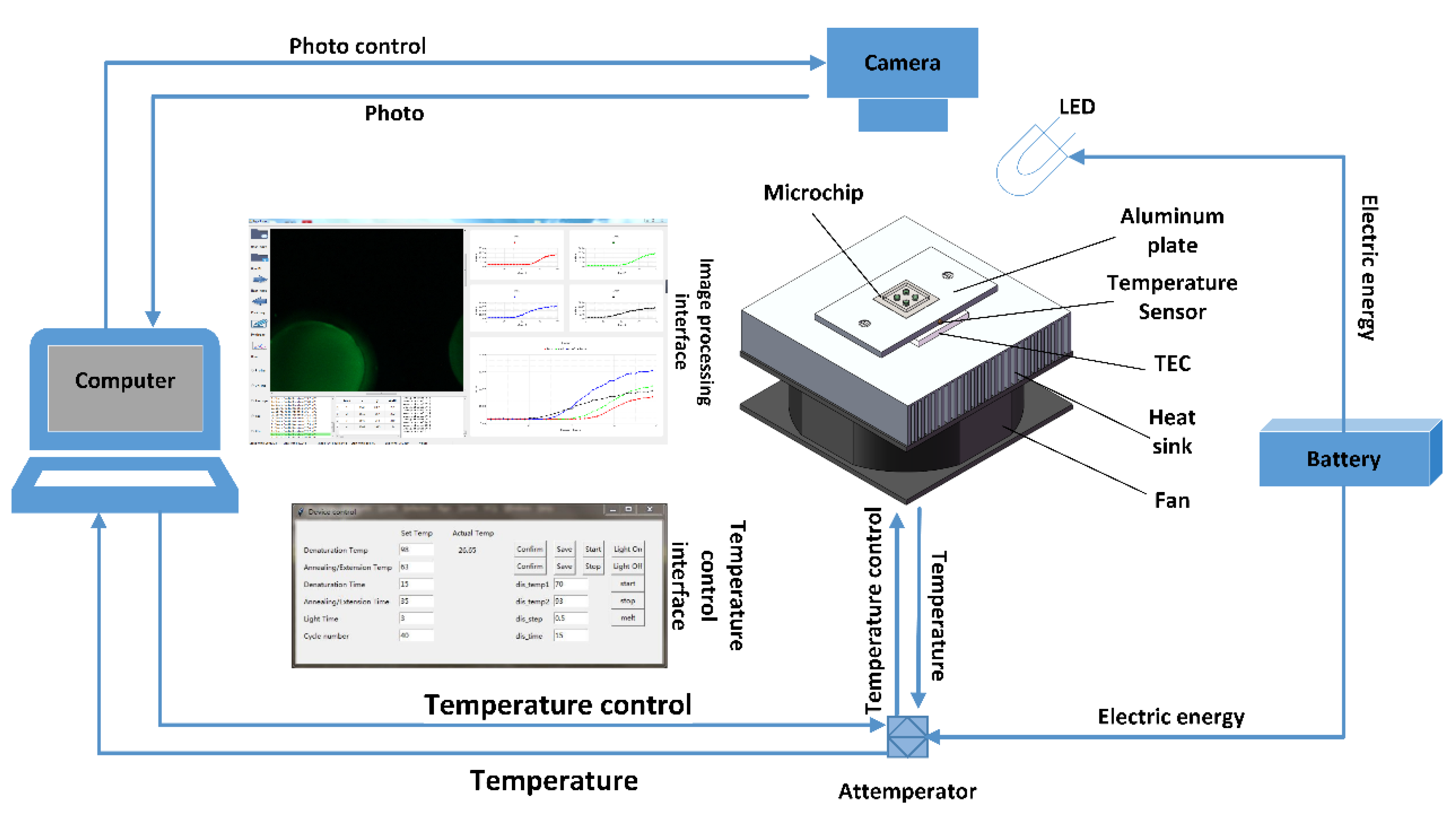
2.2. Instrument Build
2.3. Microchip Fabrication
2.4. PCR Amplification
3. Results and Discussion
3.1. Bubble Elimination
3.2. Temperature Cycling
3.3. Fluorescence Imaging
3.4. PCR Amplification
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Welsh, J.; McClelland, M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990, 18, 7213–7218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koshland, D.E. The molecule of the year. Science 1992, 258, 1861–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saiki, R.K.; Scharf, S.; Faloona, F.; Mullis, K.B.; Horn, G.T.; Erlich, H.A.; Arnheim, N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle-cell anemia. Science 1985, 230, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Stramer, S.L.; Wend, U.; Candotti, D.; Foster, G.A.; Hollinger, F.B.; Dodd, R.Y.; Allain, J.P.; Gerlich, W. Nucleic Acid Testing to Detect HBV Infection in Blood Donors. N. Engl. J. Med. 2011, 364, 236–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrimonti, C.; Bottari, B.; Sardaro, M.L.S.; Marmiroli, N. Application of real-time PCR (qPCR) for characterization of microbial populations and type of milk in dairy food products. Crit. Rev. Food Sci. Nutr. 2019, 59, 423–442. [Google Scholar] [CrossRef]
- Xu, X.; Li, X.; Chen, Q.; Yuan, J. Detection of Treponema denticola in Chronic Periodontitis by Quantitative Real-Time Polymerase Chain Reaction. J. Nanosci. Nanotechnol. 2020, 20, 1463–1469. [Google Scholar] [CrossRef]
- Bharucha, T.; Sengvilaipaseuth, O.; Vongsouvath, M.; Vongsouvath, M.; Davong, V.; Panyanouvong, P.; Piorkowski, G.; Garson, J.A.; Newton, P.N.; de Lamballerie, X.; et al. Development of an improved RT-qPCR Assay for detection of Japanese encephalitis virus (JEV) RNA including a systematic review and comprehensive comparison with published methods. PLoS ONE 2018, 13, 18. [Google Scholar] [CrossRef]
- Balcazar, W.; Rondon, J.; Rengifo, M.; Ball, M.M.; Melfo, A.; Gomez, W.; Andres Yarzabal, L. Bioprospecting glacial ice for plant growth promoting bacteria. Microbiol. Res. 2015, 177, 1–7. [Google Scholar] [CrossRef]
- Piaggio, A.J.; Engeman, R.M.; Hopken, M.W.; Humphrey, J.S.; Keacher, K.L.; Bruce, W.E.; Avery, M.L. Detecting an elusive invasive species: A diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Mol. Ecol. Resour. 2014, 14, 374–380. [Google Scholar] [CrossRef] [Green Version]
- Moignard, V.; Macaulay, I.C.; Swiers, G.; Buettner, F.; Schuette, J.; Calero-Nieto, F.J.; Kinston, S.; Joshi, A.; Hannah, R.; Theis, F.J.; et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat. Cell Biol. 2013, 15, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Freire, V.; Ebert, A.D.; Kalisky, T.; Quake, S.R.; Wu, J.C. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat. Protoc. 2012, 7, 829–838. [Google Scholar] [CrossRef] [Green Version]
- Son, J.H.; Cho, B.; Hong, S.; Lee, S.H.; Hoxha, O.; Haack, A.J.; Lee, L.P. Ultrafast photonic PCR. Light-Sci. Appl. 2015, 4. [Google Scholar] [CrossRef] [Green Version]
- Hein, I.; Flekna, G.; Krassnig, M.; Wagner, M. Real-time PCR for the detection of Salmonella spp. in food: An alternative approach to a conventional PCR system suggested by the FOOD-PCR project. J. Microbiol. Methods 2006, 66, 538–547. [Google Scholar] [CrossRef]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Higuchi, R.; Dollinger, G.; Walsh, P.S.; Griffith, R. SIMULTANEOUS AMPLIFICATION AND DETECTION OF SPECIFIC DNA-SEQUENCES. Bio-Technol. 1992, 10, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Walker, N.J. A technique whose time has come. Science 2002, 296, 557. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. Quantification Strategies in Real-time Polymerase Chain Reaction; Caister Academic Press: Norfolk, UK, 2012; pp. 53–61. [Google Scholar]
- Navarro, E.; Serrano-Heras, G.; Castano, M.J.; Solera, J. Real-time PCR detection chemistry. Clin. Chim. Acta 2015, 439, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Putignani, L.; Mancinelli, L.; Del Chierico, F.; Menichella, D.; Adlerstein, D.; Angelici, M.C.; Marangi, M.; Berrilli, F.; Caffara, M.; di Regalbono, D.A.F.; et al. Investigation of Toxoplasma gondii presence in farmed shellfish by nested-PCR and real-time PCR fluorescent amplicon generation assay (FLAG). Exp. Parasitol. 2011, 127, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Demeke, T.; Jenkins, G.R. Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal. Bioanal. Chem. 2010, 396, 1977–1990. [Google Scholar] [CrossRef]
- Li, X.; Wu, W.; Manz, A. Thermal gradient for fluorometric optimization of droplet PCR in virtual reaction chambers. Microchim. Acta 2017, 184, 3433–3439. [Google Scholar] [CrossRef]
- Shi, B.; Li, Y.; Wu, D.; Wu, W. A handheld continuous-flow real-time fluorescence qPCR system with a PVC microreactor. Analyst 2020, 145, 2767–2773. [Google Scholar] [CrossRef]
- Shi, B.; He, G.; Wu, W. A PCR microreactor machinery with passive micropump and battery-powered heater for thermo-cycled amplifications of clinical-level and multiplexed DNA targets. Microchim. Acta 2018, 185. [Google Scholar] [CrossRef]
- Wu, W.; Kang, K.-T.; Lee, N.Y. Bubble-free on-chip continuous-flow polymerase chain reaction: Concept and application. Analyst 2011, 136, 2287–2293. [Google Scholar] [CrossRef]
- Ahrberg, C.D.; Manz, A.; Neuzil, P. Palm-Sized Device for Point-of-Care Ebola Detection. Anal. Chem. 2016, 88, 4803–4807. [Google Scholar] [CrossRef]
- Ahrberg, C.D.; Ilic, B.R.; Manz, A.; Neuzil, P. Handheld real-time PCR device. Lab Chip 2016, 16, 586–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Li, B.; Wu, W. Application of automatic feedback photographing by portable smartphone in PCR. Sens. Actuators B Chem. 2019, 298. [Google Scholar] [CrossRef]
- Wu, W.; Wu, J.; Kim, J.H.; Lee, N.Y. Instantaneous room temperature bonding of a wide range of non-silicon substrates with poly(dimethylsiloxane) (PDMS) elastomer mediated by a mercaptosilane. Lab Chip 2015, 15, 2819–2825. [Google Scholar] [CrossRef]
- Lee, D.-J.; Kim, J.-D.; Kim, Y.-S.; Song, H.-J.; Park, C.-Y. Evaluation-independent system for DNA section amplification. Biomed. Eng. Online 2018, 17. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.; Datta, A.; Berg, J.M.; Gangopadhyay, S. Studies on surface wettability of poly(dimethyl) siloxane (PDMS) and glass under oxygen-plasma treatment and correlation with bond strength. J. Microelectromech. Syst. 2005, 14, 590–597. [Google Scholar] [CrossRef]
- Virlogeux, V.; Li, M.; Tsang, T.K.; Feng, L.; Fang, V.J.; Jiang, H.; Wu, P.; Zheng, J.; Lau, E.H.Y.; Cao, Y.; et al. Estimating the Distribution of the Incubation Periods of Human Avian Influenza A(H7N9) Virus Infections. Am. J. Epidemiol. 2015, 182, 723–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, L.-Q.; Li, X.-L.; Liu, K.; Li, Y.-J.; Yao, H.-W.; Liang, S.; Yang, Y.; Feng, Z.-J.; Gray, G.C.; Cao, W.-C. Mapping Spread and Risk of Avian Influenza A (H7N9) in China. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, J.; Jiang, Y.; Shi, B.; Wu, D.; Wu, W. Low-Cost Battery-Powered and User-Friendly Real-Time Quantitative PCR System for the Detection of Multigene. Micromachines 2020, 11, 435. https://doi.org/10.3390/mi11040435
An J, Jiang Y, Shi B, Wu D, Wu W. Low-Cost Battery-Powered and User-Friendly Real-Time Quantitative PCR System for the Detection of Multigene. Micromachines. 2020; 11(4):435. https://doi.org/10.3390/mi11040435
Chicago/Turabian StyleAn, Junru, Yangyang Jiang, Bing Shi, Di Wu, and Wenming Wu. 2020. "Low-Cost Battery-Powered and User-Friendly Real-Time Quantitative PCR System for the Detection of Multigene" Micromachines 11, no. 4: 435. https://doi.org/10.3390/mi11040435
APA StyleAn, J., Jiang, Y., Shi, B., Wu, D., & Wu, W. (2020). Low-Cost Battery-Powered and User-Friendly Real-Time Quantitative PCR System for the Detection of Multigene. Micromachines, 11(4), 435. https://doi.org/10.3390/mi11040435





