A Review on Microdevices for Isolating Circulating Tumor Cells
Abstract
:1. Introduction
2. CTC Isolation Principles
2.1. Biochemical Approach
2.2. Biophysical Approach
3. Microdevices Adopting Single Approach
3.1. Single Biochemical Approach
3.2. Single Biophysical Approach
3.2.1. Filtration
3.2.2. Hydrodynamics
3.2.3. Dielectrophoresis
4. Microdevices Adopting Multiple Approach
4.1. Multiple Biochemical Approach
4.2. Multiple Biophysical Approach
4.3. Combined Biochemical and Biophysical Approach
5. Discussion and Conclusions
Funding
Conflicts of Interest
References
- Kapoor, A.; Kalwar, A.; Kumar, N.; Maharia, S.; Nirban, R.K.; Kumar, H.S. Detection of bone metastasis in nasopharyngeal carcinoma by bone scintigraphy: A retrospective study in perspective of limited resource settings. Clin. Cancer Investig. J. 2015, 4, 17–21. [Google Scholar] [CrossRef]
- Noguti, J.; De Moura, C.F.G.; De Jesus, G.P.P.; Da Slva, V.H.P.; Hossaka, T.A.; Oshima, C.T.F.; Ribeiro, D.A. Metastasis from oral cancer: An overview. Cancer Genom. Proteom. 2012, 9, 329–335. [Google Scholar]
- Gupta, G.P.; Massague, J. Cancer metastasis: Building a framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaks, V.; Koopman, C.D.; Werb, Z. Circulating tumor cells. Science 2013, 341, 1186–1188. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 1869, 14, 146–149. [Google Scholar]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [Green Version]
- de Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.W.M.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Kelly, K.M.; Dean, J.; Comulada, W.S.; Lee, S.J. Breast cancer detection using automated whole breast ultrasound and mammography in radiographically dense breasts. Eur. Radiol. 2010, 20, 734–742. [Google Scholar] [CrossRef] [Green Version]
- Maheswaran, S.; Haber, D.A. Circulating tumor cells: A window into cancer biology and metastasis. Curr. Opin. Genet. Dev. 2010, 20, 96–99. [Google Scholar] [CrossRef] [Green Version]
- Paterlini-Brechot, P.; Benali, N.L. Circulating tumor cells (CTC) detection: Clinical impact and future directions. Cancer Lett. 2007, 253, 180–204. [Google Scholar] [CrossRef] [PubMed]
- Fabisiewicz, A.; Grzybowska, E. CTC clusters in cancer progression and metastasis. Med. Oncol. 2017, 34, 12. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.C.; Proudhon, C.; Pierga, J.Y. Circulating tumor cells in breast cancer. Mol. Oncol. 2016, 10, 418–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulasinghe, A.; Schmidt, H.; Perry, C.; Whitfield, B.; Kenny, L.; Nelson, C.; Warkiani, M.E.; Punyadeera, C. A collective route of head and neck cancer metastasis. Sci. Rep. 2018, 8, 746. [Google Scholar] [CrossRef] [Green Version]
- Gorin, M.A.; Verdone, J.E.; van der Toom, E.; Bivalacqua, T.J.; Allaf, M.E.; Pienta, K.J. Circulating tumour cells as biomarkers of prostate, bladder, and kidney cancer. Nat. Rev. Urol. 2017, 14, 90–97. [Google Scholar] [CrossRef]
- Wei, R.R.; Sun, D.N.; Yang, H.; Yan, J.; Zhang, X.; Zheng, X.L.; Fu, X.H.; Geng, M.Y.; Huang, X.; Ding, J. CTC clusters induced by heparanase enhance breast cancer metastasis. Acta Pharmacol. Sin. 2018, 39, 1326–1337. [Google Scholar] [CrossRef] [Green Version]
- van de Stolpe, A.; Pantel, K.; Sleijfer, S.; Terstappen, L.W.; den Toonder, J.M.J. Circulating tumor cell isolation and diagnostics: Toward routine clinical use. Cancer Res. 2011, 71, 5955–5960. [Google Scholar] [CrossRef] [Green Version]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [Green Version]
- Talasaz, A.H.; Powell, A.A.; Huber, D.E.; Berbee, J.G.; Roh, K.H.; Yu, W.; Xiao, W.; Davis, M.M.; Pease, R.F.; Mindrinos, M.N.; et al. Isolating highly enriched populations of circulating epithelial cells and other rara cells from blood using a magnetic sweeper device. PNAS 2009, 106, 3970–3975. [Google Scholar] [CrossRef] [Green Version]
- Earhart, C.M.; Hughes, C.E.; Gaster, R.S.; Ooi, C.C.; Wilson, R.J.; Zhou, L.Y.; Humke, E.W.; Xu, L.; Wong, D.J.; Willingham, S.B.; et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip 2014, 14, 78–88. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Lin, H.; Liu, J.Q.; Balic, M.; Datar, R.; Cote, R.J.; Tai, Y.C. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J. Chromatogr. A 2007, 1162, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Lin, H.K.; Lu, B.; Williams, A.; Datar, R.; Cote, R.J.; Tai, Y.C. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed. Microdevices 2011, 13, 203–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.J.; Yobas, L.; Lee, G.Y.H.; Ong, C.N.; Lim, C.T. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed. Microdevices 2009, 11, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, R.; Gertler, R.; Friederichs, J.; Fuehrer, K.; Dahm, M.; Phelps, R.; Thorban, S.; Nekarda, H.; Siewert, J.R. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry 2002, 49, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, A.A.S.; Hou, H.W.; Li, L.D.; Lim, C.T.; Han, J. Pinched flow coupled shear-modulated inertial microfluidics for high-throughout rare blood cell separation. Lab Chip 2011, 11, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Jafferji, I.; Garza, M.; Melnikova, V.O.; Hasegawa, D.K.; Pethig, R.; Davis, D.W. ApoStreamTM, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012, 6, 024133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, A.D.; Mattison, J.; Western, L.T.; Powderly, J.D.; Greene, B.T.; King, M.R. Microtube device for selectin-mediated capture of viable circulating tumor cells from blood. Clin. Chem. 2012, 58, 846–853. [Google Scholar] [CrossRef]
- Moon, H.S.; Kwon, K.; Kim, S.I.; Han, H.; Sohn, J.; Lee, S.; Jung, H.I. Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP). Lab Chip 2011, 11, 1118–1125. [Google Scholar] [CrossRef]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef] [Green Version]
- Gleghorn, J.P.; Pratt, E.D.; Denning, D.; Liu, H.; Bander, N.H.; Tagawa, S.T.; Nanus, D.M.; Giannakakou, P.A.; Kirby, B.J. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip 2010, 10, 27–29. [Google Scholar] [CrossRef]
- Yi, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Rose, L.; Zollner, T.M.; Kraha, T.; von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Miller, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224. [Google Scholar] [CrossRef] [Green Version]
- Alix-Panabières, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Ang, R.R.; Duffy, S.P.; Bazov, J.; Chi, K.N.; Black, P.C.; Ma, H. Morphological differences between circulating tumor cells from prostate cancer patients and cultured prostate cancer cells. PLoS ONE 2014, 9, e85264. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-Zeni, N.; Mewes, S.; Niestroj, R.; Gasiorowski, L.; Murawa, D.; Nowaczyk, P.; Tomasi, T.; Weber, E.; Dworacki, G.; Morgenthaler, N.G.; et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int. J. Oncol. 2012, 41, 1241–1250. [Google Scholar]
- Chen, S.; El-Heliebi, A.; Tauber, G.; Langsenlehner, T.; Potscher, M.; Kashofer, K.; Czyz, Z.T.; Polzer, B.; Riethdorf, S.; Kuske, A.; et al. Catch and release: Rare cell analysis from a functionalized medical wire. Sci. Rep. 2017, 7, 43424. [Google Scholar] [CrossRef] [Green Version]
- Gorges, T.M.; Penkalla, N.; Schalk, T.; Joosse, S.A.; Riethdorf, S.; Tucholski, J.; Lucke, K.; Wikman, H.; Jackson, S.; Brychta, N.; et al. Enumeration and molecular characterization of tumor cells in lung cancer patients using a novel in vivo device for capturing circulating tumor cells. Clin. Cancer Res. 2016, 22, 2197–2206. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Stratton, Z.S.; Dao, M.; Ritz, J.; Huang, T.J. Probing circulating tumor cells in microfluidics. Lab Chip 2013, 13, 602–609. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, H.; Murray, M.; Turner, J.N.; Caggana, M. Isolation of tumor cells using size and deformation. J. Chromatogr. A 2009, 1216, 8289–8295. [Google Scholar] [CrossRef]
- Yusa, A.; Toneri, M.; Masuda, T.; Ito, S.; Yamamoto, S.; Okochi, M.; Kondo, N.; Iwata, H.; Yatabe, Y.; Ichinosawa, Y.; et al. Development of a new rapid isolation device for circulating tumor cells (CTCs) using 3D palladium filter and its application for genetic analysis. PLoS ONE 2014, 9, e88821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, H.W.; Warkiano, M.E.; Khoo, B.L.; Li, Z.R.; Soo, R.A.; Tan, D.S.W.; Lim, W.T.; Han, J.; Bhagat, A.A.S.; Lim, C.T. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci. Rep. 2013, 3, 1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, A.A.; Talasaz, A.H.; Zhang, H.; Coram, M.A.; Reddy, A.; Deng, G.; Telli, M.L.; Advani, R.H.; Carlson, R.W.; Mollick, J.A.; et al. Single cell profiling of circulating tumor cells: Transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE 2012, 7, e33788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, C.E.; Park, J.M.; Moon, H.S.; Joung, J.G.; Son, D.S.; Jeon, H.J.; Kim, Y.J.; Han, K.Y.; Sun, J.M.; Park, K.; et al. Vertical magnetic separation of circulating tumor cells for somatic genomic-alteration analysis in lung cancer patients. Sci. Rep. 2016, 6, 37392. [Google Scholar] [CrossRef] [Green Version]
- Gascoyne, R.R.C.; Noshari, J.; Anderson, T.J.; Becker, F.F. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis 2009, 30, 1388–1398. [Google Scholar] [CrossRef] [Green Version]
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 2015, 12, 685–691. [Google Scholar] [CrossRef]
- Li, H.; Chen, J.; Du, W.; Xia, Y.; Wang, D.; Zhao, G.; Chu, J. The optimization of a microfluidic CTC filtering chip by simulation. Micromachines 2017, 8, 79. [Google Scholar] [CrossRef] [Green Version]
- Lei, K.F. Microfluidic systems for diagnostic applications: A review. JALA 2012, 17, 330–347. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.S.A.; Lei, K.F. Advances in miniaturized instruments for genomics. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Lei, K.F. Review on impedance detection of cellular responses in micro/nano environment. Micromachines 2014, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Gerhardt, T.; Woo, S.; Ma, H. Chromatographic behaviour of single cells in a microchannel with dynamic geometry. Lab Chip 2011, 11, 2731–2737. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.J.; Lakshmi, R.L.; Chen, P.; Lim, W.T.; Yobas, L.; Lim, C.T. Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosens. Bioelectron. 2010, 26, 1701–1705. [Google Scholar] [CrossRef] [PubMed]
- Marrinucci, D.; Bethel, K.; Bruce, R.H.; Curry, D.N.; Hsieh, B.; Humphrey, M.; Krivacic, T.R.; Kroener, J.; Kroener, L.; Ladanyi, A.; et al. Case study of the morphologic variation of circulating tumor cells. Human Pathol. 2007, 38, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Kahn, H.J.; Presta, A.; Yang, L.Y.; Blondal, J.; Trudeau, M.; Lickley, L.; Holloway, C.; McCready, D.R.; Maclean, D.; Marks, A. Enumeration of circulating tumor cells in the blood of breast cancer patients after filtration enrichment: Correlation with disease stage. Breast Cancer Res. Treat. 2004, 86, 237–247. [Google Scholar] [CrossRef]
- Coumans, F.A.W.; van Dalum, G.; Beck, M.; Terstappen, L.W.M.M. Filter characteristics influrencing circulating tumor cell enrichment from whole blood. PLoS ONE 2013, 8, e61770. [Google Scholar]
- Gascoyne, P.R.; Shim, S.; Noshari, J.; Becker, F.F.; Stemke-Hale, K. Correlations between the dielectric properties and exterior morphology of cells revealed by dielectrophoretic field-flow fractionation. Electrophoresis 2013, 34, 1042–1050. [Google Scholar] [CrossRef]
- Pariset, E.; Pudda, C.; Boizot, F.; Verplanck, N.; Revol-Cavalier, F.; Berthier, J.; Thuaire, A.; Agache, V. Purification of complex samples: Implementation of a modular and reconfigurable droplet-based microfluidic platform with cascaded deterministic lateral displacement separation modules. PLoS ONE 2018, 13, e0197629. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, S.; Yuan, D.; Alici, G.; Nguyen, N.T.; Warkiani, M.E.; Li, W. Fundamentals and applications of inertial microfluidics: A review. Lab Chip 2016, 16, 10–34. [Google Scholar] [CrossRef] [Green Version]
- Gou, Y.; Jia, Y.; Wang, P.; Sun, C. Progress of inertial microfluidics in principle and application. Sensors 2018, 18, 1762. [Google Scholar] [CrossRef] [Green Version]
- Chudasama, D.Y.; Freydina, D.V.; Fridin, M.B.; Leung, M.; Fernandez, A.M.; Rice, A.; Nicholson, A.G.; Karteris, E.; Anikin, V.; Lim, E. Inertia based microfluidic capture and characterization of circulating tumor cells for the diagnosis of lung cancer. Ann. Transl. Med. 2016, 4, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Zhang, Z. An intertia-deformability hybrid circulating tumor cell chip: Design, clinical test, and numerical analysis. J. Med. Devices 2018, 12, 041004. [Google Scholar] [CrossRef]
- Khoo, B.L.; Warkiani, M.E.; Tan, D.S.W.; Bhagat, A.A.S.; Lrwin, D.; Lau, D.P.; Lim, A.S.T.; Lim, K.H.; Krisna, S.S.; Lim, W.T.; et al. Clinical validation of an ultra high-throughput spiral microdluidics for the detection and enrichment of viable circulating tumor cells. PLoS ONE 2014, 9, e99409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Ishikawa, T.; Numayama-Tsuruta, K.; Imai, Y.; Ueno, H.; Matsuki, N.; Yamaguchi, T. Separation of cancer cells from a red blood cell suspension using inertial force. Lab Chip 2012, 12, 4336–4343. [Google Scholar] [CrossRef]
- Pethig, R. Review article—dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics 2010, 4, 022811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morimoto, A.; Mogami, T.; Watanabe, M.; Iijima, K.; Akiyama, Y.; Katayama, K.; Futami, T.; Yamamoto, N.; Sawada, T.; Koizumi, F.; et al. High-density dielectrophoretic microwell array for detection, capture, and single-cell analysis of rare tumor cells in peripheral blood. PLoS ONE 2015, 10, e0130418. [Google Scholar] [CrossRef] [PubMed]
- Becker, F.F.; Wang, X.B.; Huang, Y.; Pethig, R.; Vykoukal, J.; Gascoyne, P.R. Separation of human breast cancer cells from blood by differential dielectric affinity. PNAS 1995, 92, 860–864. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Wang, X.B.; Becker, F.F.; Gascoyne, P.R. Introducing dielectrophoresis as a new force field for field-flow fractionation. Biophys. J. 1997, 73, 1118–1129. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, P.; Kinders, R.J.; Kummar, S.; Gupta, V.; Hasegawa, D.; Menachery, A.; Lawrence, S.M.; Wang, L.; Ferry-Galow, K.; Davis, D.; et al. Antibody-independent capture of circulating tumor cells of non-epithelial origin with the ApoStream® system. PLoS ONE 2017, 12, e0175414. [Google Scholar] [CrossRef] [Green Version]
- Myung, J.H.; Launiere, C.A.; Eddington, D.T.; Hong, S. Enhanced tumor cell isolation by a biomimetic combination of E-selectin and anti-EpCAM: Implications for the effective separation of circulating tumor cells (CTCs). Langmuir 2010, 25, 8589–8596. [Google Scholar] [CrossRef] [Green Version]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. PNAS 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Liu, K.; Liu, J.; Yu, Z.T.F.; Xu, X.; Zhao, L.; Lee, T.; Lee, E.K.; Reiss, J.; Lee, Y.K.; et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew. Chem. Int. Ed. 2011, 50, 3084–3088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krivacic, R.T.; Ladanyi, A.; Curry, D.N.; Hsieh, H.B.; Kuhn, P.; Bergsrud, D.E.; Kepros, J.F.; Barbera, T.; Ho, M.Y.; Chen, L.B.; et al. A rare-cell detector for cancer. PNAS 2004, 101, 10501–10504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Issadore, D.; Chung, J.; Shao, H.; Liong, M.; Ghazani, A.A.; Castro, C.M.; Weissleder, R. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci. Transl. Med. 2012, 4, 141ra92. [Google Scholar] [CrossRef] [Green Version]
- Chiu, T.K.; Lei, K.F.; Hsieh, C.H.; Hsiao, H.B.; Wang, H.M.; Wu, M.H. Development of a microfluidic-based optical sensing device for label-free detection of circulating tumor cells (CTCs) through their lactic acid metabolism. Sensors 2015, 15, 6789–6806. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kozminsky, M.; Nagrath, S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. ACS Nano 2014, 8, 1995–2017. [Google Scholar] [CrossRef]
- Hoshino, K.; Huang, Y.Y.; Lane, N.; Huebschman, M.; Uhr, J.W.; Frenkel, E.P.; Zhang, X. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip 2011, 11, 3449–3457. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Hoshino, K.; Chen, P.; Wu, C.H.; Lane, N.; Huebschman, M.; Liu, H.; Sokolov, K.; Uhr, J.W.; Frenkel, E.P.; et al. Immunomagnetic nanoscreening of circulating tumor cells with a motion controlled microfluidic system. Biomed. Microdevices 2013, 15, 673–681. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Hong, G.; Wang, H.; Welsher, K.; Tabakman, S.M.; Sherlock, S.P.; Robinson, J.T.; Liang, Y.; Dai, H. Graphite-coated magnetic nanoparticle microarray for few-cells enrichment and detection. ACS Nano 2012, 6, 1094–1101. [Google Scholar] [CrossRef]
- Fachin, F.; Chen, G.D.; Toner, M.; Wardle, B.L. Integration of bulk nanoporous elements in microfluidic devices with application to biomedical diagnostics. J. MEMS 2011, 20, 1428–1438. [Google Scholar] [CrossRef]
- Chen, G.D.; Fachin, F.; Fernandez-Suarez, M.; Wardle, B.L.; Toner, M. Nanoporous elements in microfluidics for multiscale manipulation of bioparticles. Small 2011, 7, 1061–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, S.; Zhao, H.; Zhao, L.; Shen, Q.; Wei, K.S.; Suh, D.Y.; Nakao, A.; Garcia, M.A.; Song, M.; Lee, T.; et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv. Mater. 2013, 25, 1547–1551. [Google Scholar] [CrossRef] [Green Version]
- Sheng, W.; Chen, T.; Tan, W.; Fan, Z.H. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano 2013, 7, 7067–7076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekine, J.; Luo, S.C.; Wang, S.; Zhu, B.; Tseng, H.R.; Yu, H.H. Functionalized conducting polymer nanodots for enhanced cell capturing: The synergistic effect of capture agents and nanostructures. Adv. Mater. 2011, 23, 4788–4792. [Google Scholar] [CrossRef]
- Myung, S.; Solanki, A.; Kim, C.; Park, J.; Kim, K.S.; Lee, K.B. Graphene-encapsulated nanoparticle-based biosensor for the selective detection of cancer biomarkers. Adv. Mater. 2011, 23, 2221–2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, H.J.; Kim, T.H.; Zhang, Z.; Azizi, E.; Pham, T.M.; Paoletti, C.; Lin, J.; Ramnath, N.; Wicha, M.S.; Hayes, D.F.; et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 2013, 8, 735–741. [Google Scholar] [CrossRef] [PubMed]
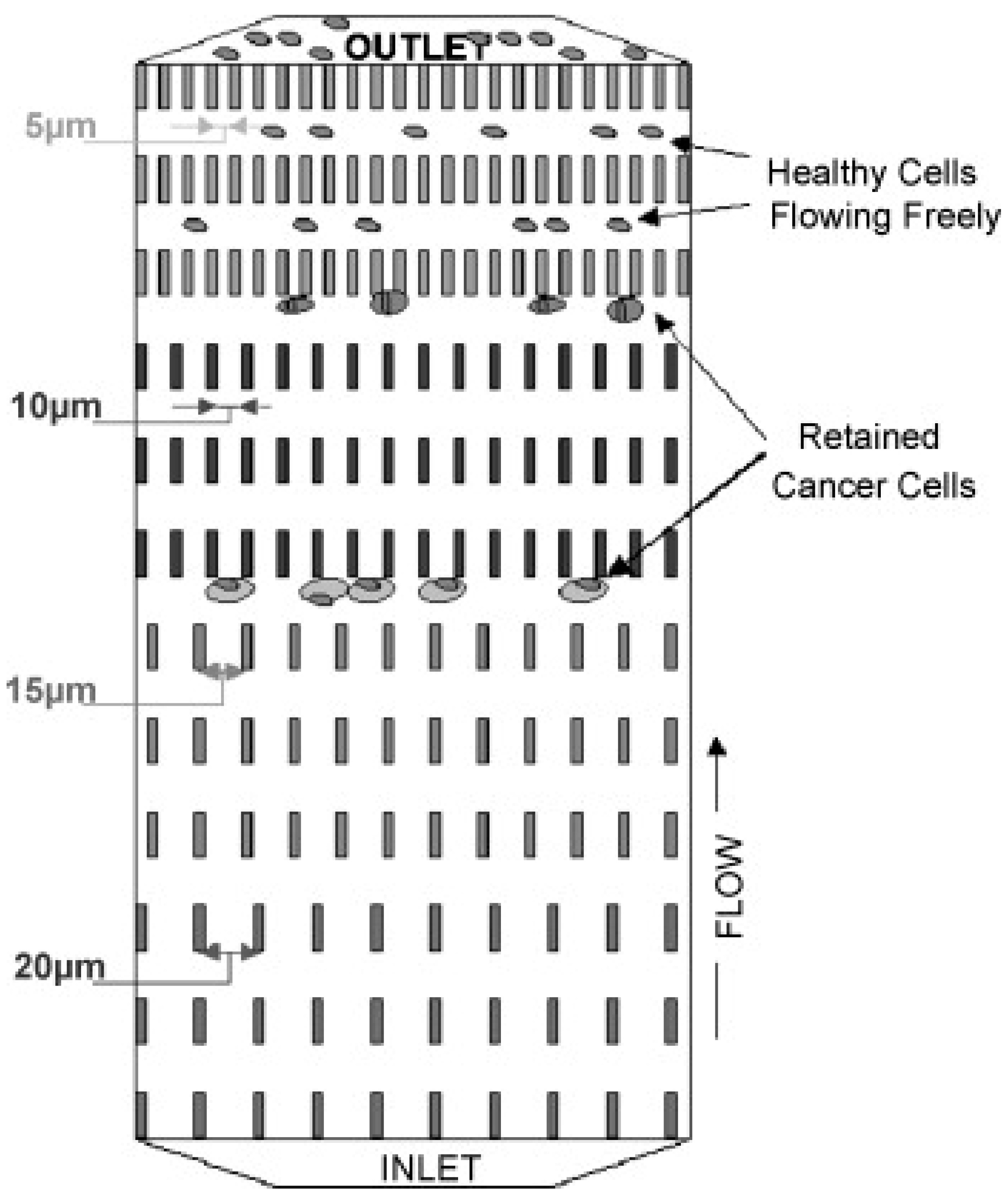



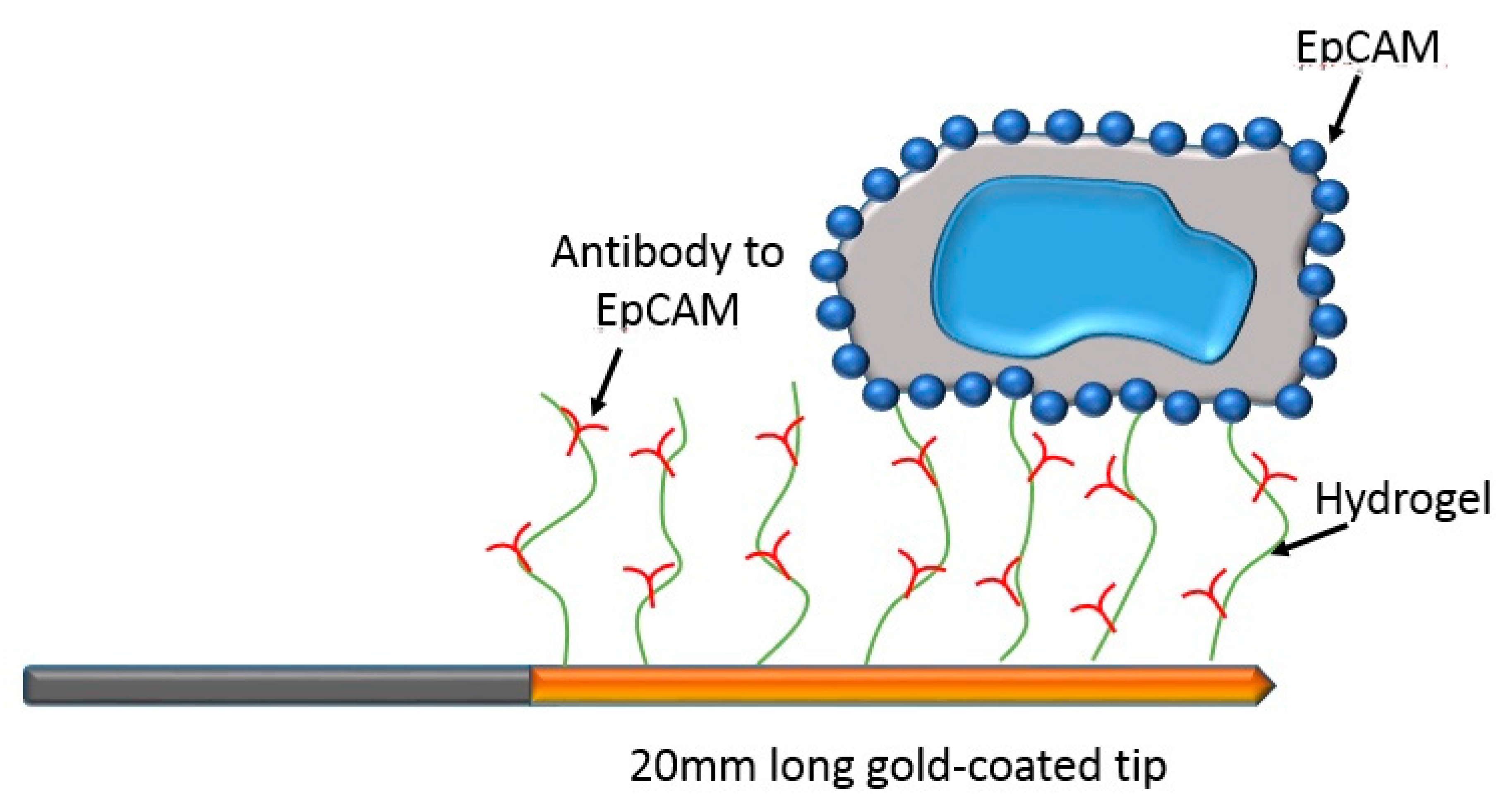
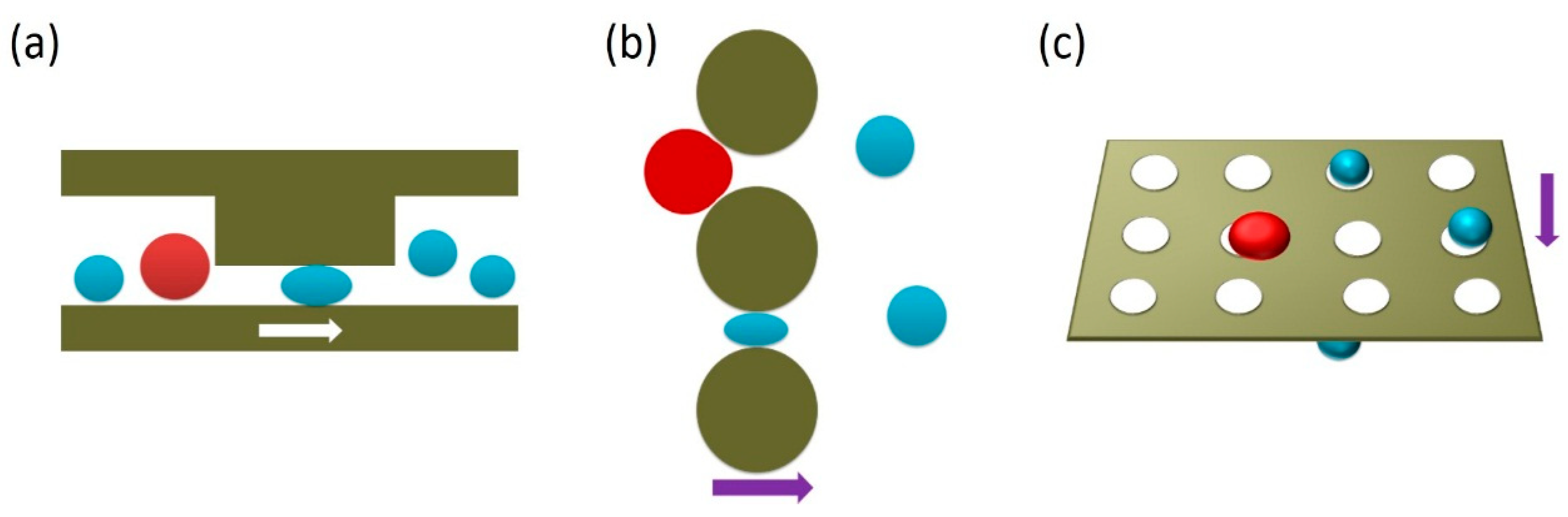






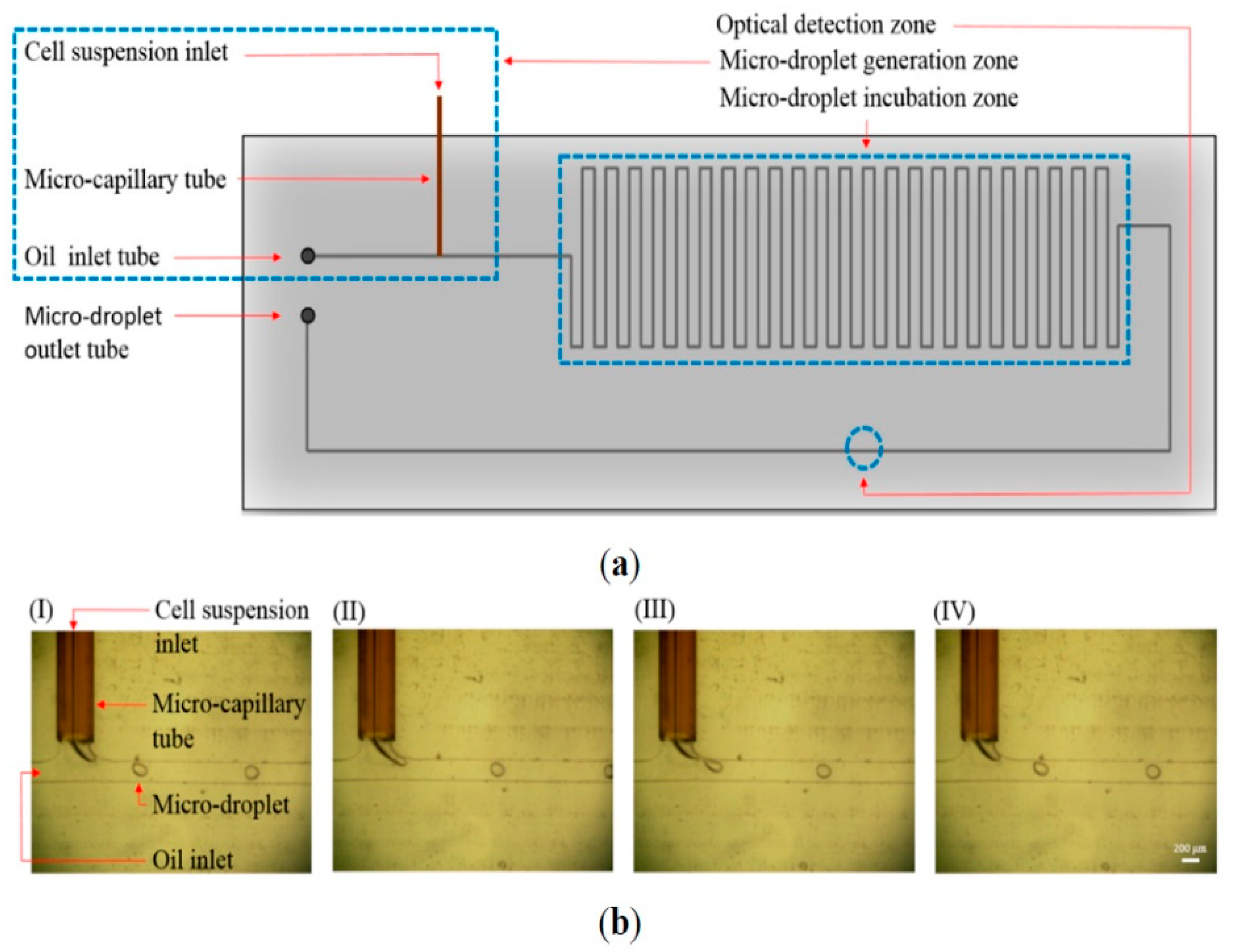
| Category | Isolation Principle |
|---|---|
| Microdevices adopting single approach | |
| Biochemical approach |
|
| Biophysical approach |
|
| Microdevices adopting multiple approach | |
| Multiple biochemical approach |
|
| Multiple biophysical approach |
|
| Combined biochemical and biophysical approach |
|
| Isolation Principle | Capture Efficiency | Isolation Purity | Throughput | Ref. |
|---|---|---|---|---|
| CTC-specific marker | 62 ± 7% | 51 ± 18% | 9 mL/h | [19] |
| 91.4% | 17.7 ± 9.3% | 10 mL/h | [20] | |
| Filtration | 89 ± 9.5% | - | <10 min | [21] |
| 86.5 ± 5.3% | - | 12–20 mL/h | [22] | |
| Hydrodynamics | >80% | - | 0.7 mL/h | [23] |
| ~85% | - | - | [24] | |
| ~80% | - | 1.2 mL/h | [25] | |
| Dielectrophoresis | ~70% | - | 7.5 mL/h | [26] |
| Selectin and CTC-specific marker | ~50% | 66 ± 3.9% | 4.8 mL/h | [27] |
| Microfluidic flow control and dielectrophoresis | >90% | 162-fold | 126 μL/min | [28] |
| Microstructures functionalized by CTC-specific markers | ≥60% | ~50% | 2.5 mL/h | [29] |
| 85 ± 5% | 68 ± 6% | 1 mL/h | [30] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, K.F. A Review on Microdevices for Isolating Circulating Tumor Cells. Micromachines 2020, 11, 531. https://doi.org/10.3390/mi11050531
Lei KF. A Review on Microdevices for Isolating Circulating Tumor Cells. Micromachines. 2020; 11(5):531. https://doi.org/10.3390/mi11050531
Chicago/Turabian StyleLei, Kin Fong. 2020. "A Review on Microdevices for Isolating Circulating Tumor Cells" Micromachines 11, no. 5: 531. https://doi.org/10.3390/mi11050531





