3D Printed Monolithic Microreactors for Real-Time Detection of Klebsiella pneumoniae and the Resistance Gene blaNDM-1 by Recombinase Polymerase Amplification
Abstract
:1. Introduction
- Optically clear
- Non-inhibitory to RPA chemistry without the need for surface pre-treatment
- Low autofluorescence
- Low fluorescence drift
2. Experimental
2.1. 3D Printed Monolithic Microreactors
2.2. Post Curing
2.3. Mobile Temperature-Controlled Fluorescence Reader (mTFR) Instrument
2.4. DNA Standards
2.4.1. Klebsiella Pneumoniae Genomic DNA
2.4.2. blaNDM-1 Resistance Gene
2.5. Recombinase Polymerase Amplification (RPA)
2.5.1. Reaction Setup
2.5.2. RPA Primers and exo Probes
2.6. Fluorescence Data Processing
3. Results and Discussion
3.1. Influence of Post Curing on Autofluorescence
3.2. Influence of Aging on Autofluorescence
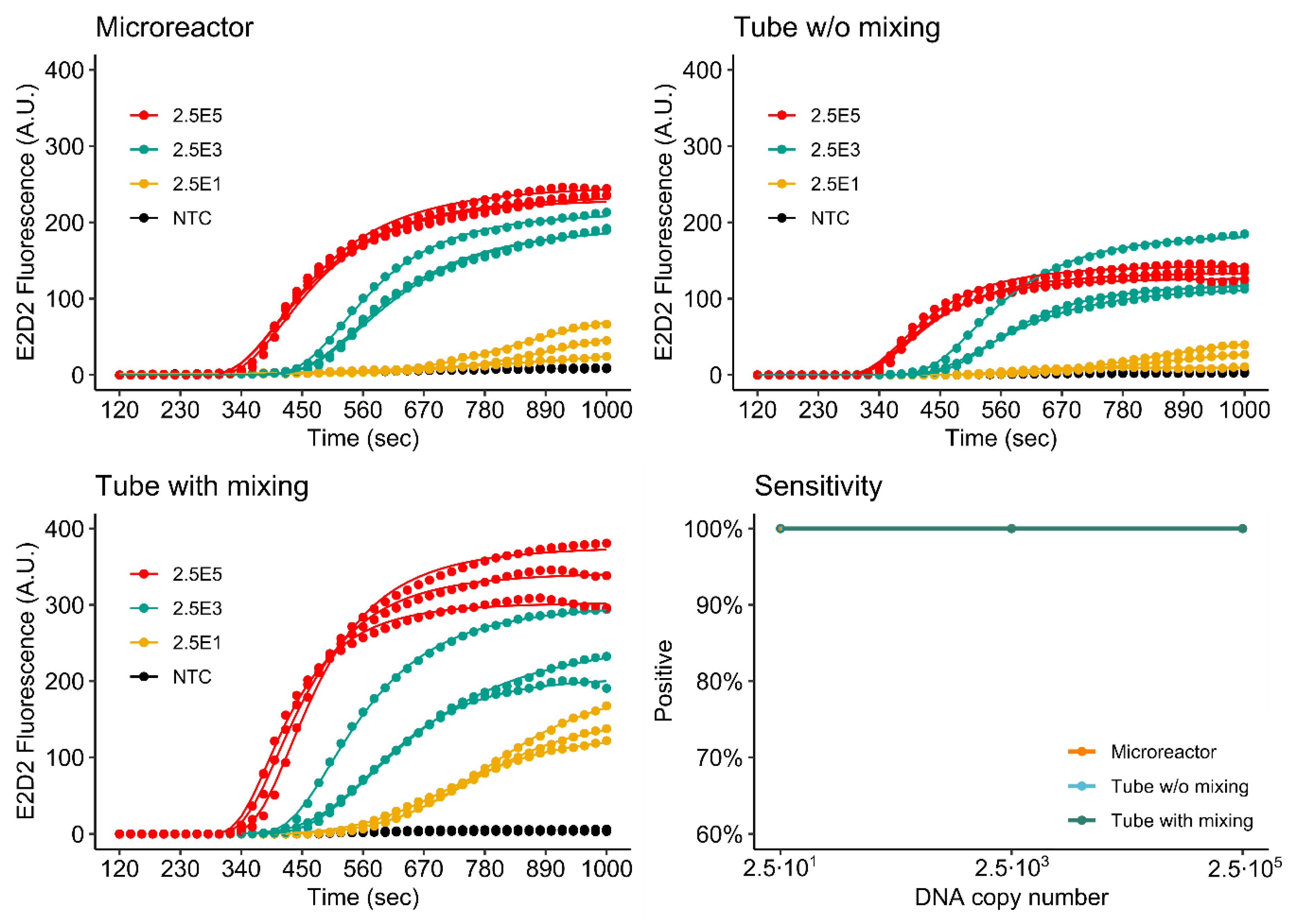
3.3. Influence of 3D Printing Materials on RPA Kinetics
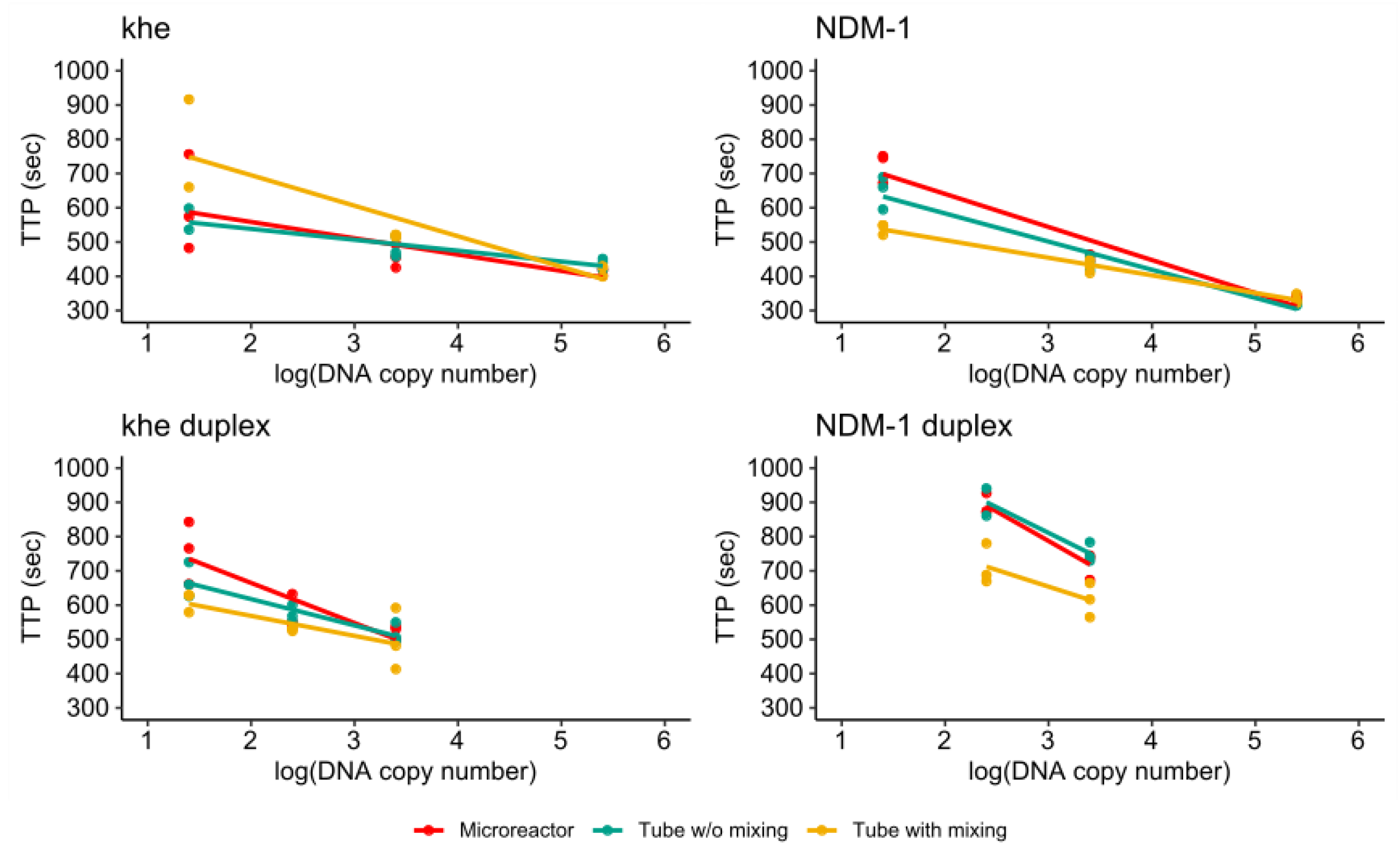
3.4. RPA of the khe and blaNDM-1 Genes
3.4.1. Singleplex Assay
3.4.2. Duplex Assay
3.4.3. Comparison of Single- and Duplex Assays
4. Summary and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| (rt)RPA | (real-time)Recombinase Polymerase Amplification |
| mTFR | Mobile temperature-controlled fluorescence reader |
| NXSG | NextDent Surgical Guide |
| SFSG | SolFlex Surgical Guide |
| NXOC | NextDent Ortho Clear |
| BSA | Bovine Serum Albumin |
| PEG | Polyethylene glycol |
| TTP | Time-to-positive |
| DLP | Digital Light Processing |
| PID | Proportional-integral-derivative |
| IPA | Isopropyl alcohol |
| SLA | Stereolithography |
| FDM | Fused Deposition Modeling |
| khe | Klebsiella haemolysin |
| blaNDM-1 | New Delhi metallo-β-lactamase 1 |
| NTC | No template control |
| AMR | Antimicrobial Resistance |
| FAM | Carboxyfluorescein |
| LC610 | LightCycler Red 610 |
| KPN | Klebsiella pneumoniae |
| HTR | Horizontal Gene Transfer |
| UTI | Urinary Tract Infection |
| µTAS | Micro Total Analysis Systems |
| Mg(OAc)2 | Magnesium acetate |
References
- Chaurasia, S.; Sivanandan, S.; Agarwal, R.; Ellis, S.; Sharland, M.; Sankar, M.J. Neonatal sepsis in South Asia: Huge burden and spiralling antimicrobial resistance. BMJ 2019, 364, k5314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe (2017); European Centre for Disease Prevention and Control: Stockholm, Sweden, 2018; pp. 1–71.
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.-W.; Walsh, T.R. Characterization of a New Metallo-β-Lactamase Gene, blaNDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [Green Version]
- Wailan, A.M.; Paterson, D.L. The spread and acquisition of NDM-1: A multifactorial problem. Expert Rev. Anti-Infect. Ther. 2013, 12, 91–115. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Carrër, A.; Toleman, M.A.; Walsh, T.R. How to Detect NDM-1 Producers. J. Clin. Microbiol. 2011, 49, 718–721. [Google Scholar] [CrossRef] [Green Version]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J.; Von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2019, 144, 31–67. [Google Scholar] [CrossRef] [Green Version]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Isothermal Recombinase Polymerase Amplification Assay Applied to the Detection of Group B Streptococci in Vaginal/Anal Samples. Clin. Chem. 2014, 60, 660–666. [Google Scholar] [CrossRef] [Green Version]
- Gumaa, M.; Cao, X.; Li, Z.; Lou, Z.; Zhang, N.-Z.; Zhang, Z.; Zhou, J.; Fu, B. Establishment of a recombinase polymerase amplification (RPA) assay for the detection of Brucella spp. Infection. Mol. Cell. Probes 2019, 47, 101434. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, J.-L. Development of a multiplex real-time recombinase polymerase amplification (RPA) assay for rapid quantitative detection of Campylobacter coli and jejuni from eggs and chicken products. Food Control 2017, 73, 1247–1255. [Google Scholar] [CrossRef] [Green Version]
- Higgins, O.; Clancy, E.; Forrest, M.; Piepenburg, O.; Cormican, M.; Boo, T.W.; O’Sullivan, N.; McGuinness, C.; Cafferty, D.; Cunney, R.; et al. Duplex recombinase polymerase amplification assays incorporating competitive internal controls for bacterial meningitis detection. Anal. Biochem. 2018, 546, 10–16. [Google Scholar] [CrossRef]
- Raja, B.; Goux, H.J.; Marapadaga, A.; Rajagopalan, S.; Kourentzi, K.; Willson, R.C. Development of a panel of recombinase polymerase amplification assays for detection of common bacterial urinary tract infection pathogens. J. Appl. Microbiol. 2017, 123, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Euler, M.; Wang, Y.; Nentwich, O.; Piepenburg, O.; Hufert, F.T.; Weidmann, M. Recombinase polymerase amplification assay for rapid detection of Rift Valley fever virus. J. Clin. Virol. 2012, 54, 308–312. [Google Scholar] [CrossRef] [PubMed]
- El Wahed, A.A.; Patel, P.; Heidenreich, D.; Hufert, F.T.; Weidmann, M. Reverse Transcription Recombinase Polymerase Amplification Assay for the Detection of Middle East Respiratory Syndrome Coronavirus. PLoS Curr. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- El Wahed, A.A.; Patel, P.; Faye, O.; Thaloengsok, S.; Heidenreich, D.; Matangkasombut, P.; Manopwisedjaroen, K.; Sakuntabhai, A.; Sall, A.A.; Hufert, F.T.; et al. Recombinase Polymerase Amplification Assay for Rapid Diagnostics of Dengue Infection. PLoS ONE 2015, 10, e0129682. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, M.A.; El-Deeb, A.; El-Tholoth, M.; Hoffmann, D.; Czerny, C.-P.; Hufert, F.; Weidmann, M.; Wahed, A. Recombinase polymerase amplification assay for rapid detection of lumpy skin disease virus. BMC Vet. Res. 2016, 12, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davi, S.D.; Kissenkötter, J.; Faye, M.; Böhlken-Fascher, S.; Stahl-Hennig, C.; Faye, O.; Faye, O.; Sall, A.A.; Weidmann, M.; Ademowo, O.G.; et al. Recombinase polymerase amplification assay for rapid detection of Monkeypox virus. Diagn. Microbiol. Infect. Dis. 2019, 95, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Behrmann, O.; Bachmann, I.; Spiegel, M.; Schramm, M.; El Wahed, A.A.; Dobler, G.; Dame, G.; Hufert, F.T. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (exo-IQ). Clin. Chem. 2020. [Google Scholar] [CrossRef]
- Tian, A.-L.; Elsheikha, H.; Zhou, D.-H.; Wu, Y.-D.; Chen, M.-X.; Wang, M.; Chen, D.; Zhang, X.-C.; Zhu, X.-Q. A novel recombinase polymerase amplification (RPA) assay for the rapid isothermal detection of Neospora caninum in aborted bovine fetuses. Vet. Parasitol. 2018, 258, 24–29. [Google Scholar] [CrossRef]
- Lai, M.-Y.; Ooi, C.-H.; Lau, Y.-L. Rapid Detection of Plasmodium knowlesi by Isothermal Recombinase Polymerase Amplification Assay. Am. J. Trop. Med. Hyg. 2017, 97, 1597–1599. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Coello, M.; Shelite, T.; Castellanos-González, A.; Saldarriaga, O.; Rivero, R.; Ortega-Pacheco, A.; Acevedo-Arcique, C.; Amaya-Guardia, K.; Garg, N.; Melby, P.; et al. Efficacy of Recombinase Polymerase Amplification to Diagnose Trypanosoma cruzi Infection in Dogs with Cardiac Alterations from an Endemic Area of Mexico. Vector-Borne Zoonotic Dis. 2018, 18, 417–423. [Google Scholar] [CrossRef]
- Wee, E.J.H.; Trau, M. Simple Isothermal Strategy for Multiplexed, Rapid, Sensitive, and Accurate miRNA Detection. ACS Sens. 2016, 1, 670–675. [Google Scholar] [CrossRef]
- Lutz, S.; Weber, P.; Focke, M.; Faltin, B.; Hoffmann, J.; Müller, C.; Mark, D.; Roth, G.; Munday, P.; Armes, N.; et al. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA). Lab Chip 2010, 10, 887–893. [Google Scholar] [CrossRef]
- Kunze, A.; Dilcher, M.; El Wahed, A.A.; Hufert, F.; Niessner, R.; Seidel, M. On-Chip Isothermal Nucleic Acid Amplification on Flow-Based Chemiluminescence Microarray Analysis Platform for the Detection of Viruses and Bacteria. Anal. Chem. 2015, 88, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Yeh, E.-C.; Fu, C.-C.; Hu, L.; Thakur, R.; Feng, J.; Lee, L.P. Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv. 2017, 3, e1501645. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.E.; Lee, T.Y.; Koo, B.; Sung, H.; Kim, S.-H.; Shin, Y. Rapid virus diagnostic system using bio-optical sensor and microfluidic sample processing. Sens. Actuators B Chem. 2018, 255, 2399–2406. [Google Scholar] [CrossRef]
- Yamanaka, E.S.; Tortajada-Genaro, L.A.; Maquieira, A. Low-cost genotyping method based on allele-specific recombinase polymerase amplification and colorimetric microarray detection. Microchim. Acta 2017, 184, 1453–1462. [Google Scholar] [CrossRef]
- Piruska, A.; Nikcevic, I.; Lee, S.H.; Ahn, C.; Heineman, W.R.; Limbach, P.A.; Seliskar, C.J. The autofluorescence of plastic materials and chips measured under laser irradiation. Lab Chip 2005, 5, 1348–1354. [Google Scholar] [CrossRef]
- Jurischka, C.; Dinter, F.; Efimova, A.; Weiss, R.; Schiebel, J.; Schulz, C.; Fayziev, B.; Schierack, P.; Fischer, T.; Rödiger, S. An explorative study of polymers for 3D printing of bioanalytical test systems. Clin. Hemorheol. Microcirc. 2020, 1–28. [Google Scholar] [CrossRef]
- Kadimisetty, K.; Song, J.; Doto, A.M.; Hwang, Y.; Peng, J.; Mauk, M.G.; Bushman, F.D.; Gross, R.; Jarvis, J.N.; Liu, C. Fully 3D printed integrated reactor array for point-of-care molecular diagnostics. Biosens. Bioelectron. 2018, 109, 156–163. [Google Scholar] [CrossRef]
- Bagheri, A.; Jin, J. Photopolymerization in 3D Printing. ACS Appl. Polym. Mater. 2019, 1, 593–611. [Google Scholar] [CrossRef] [Green Version]
- Gong, H.; Bickham, B.P.; Woolley, A.T.; Nordin, G.P. Custom 3D printer and resin for 18 μm × 20 μm microfluidic flow channels. Lab Chip 2017, 17, 2899–2909. [Google Scholar] [CrossRef]
- Yin-Ching, C.; Jer-Horng, S.; Ching-Nan, L.; Ming-Chung, C. Cloning of a gene encoding a unique haemolysin from Klebsiella pneumoniae and its potential use as a species-specific gene probe. Microb. Pathog. 2002, 33, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Higgins, M.; Ravenhall, M.; Ward, D.; Phelan, J.; Ibrahim, A.; Forrest, M.S.; Clark, T.G.; Campino, S. PrimedRPA: Primer design for recombinase polymerase amplification assays. Bioinformatics 2019, 35, 682–684. [Google Scholar] [CrossRef] [Green Version]
- TwistDx Ltd. TwistAmp DNA Amplification Kits Assay Design Manual. 2019. Available online: https://www.twistdx.co.uk/docs/default-source/RPA-assay-design/twistamp-assay-design-manual-v2-5.pdf (accessed on 16 June 2020).
- Ritz, C.; Spiess, A.-N. qpcR: An R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics 2008, 24, 1549–1551. [Google Scholar] [CrossRef] [PubMed]

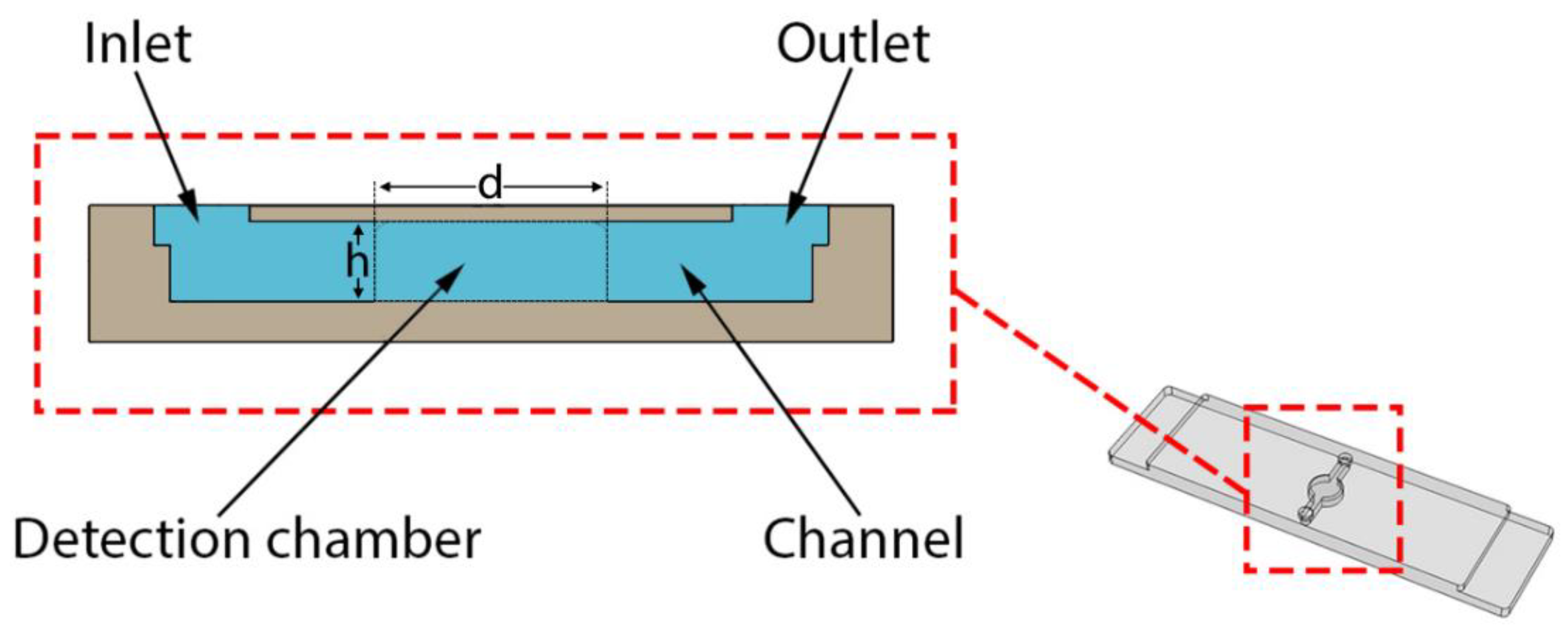
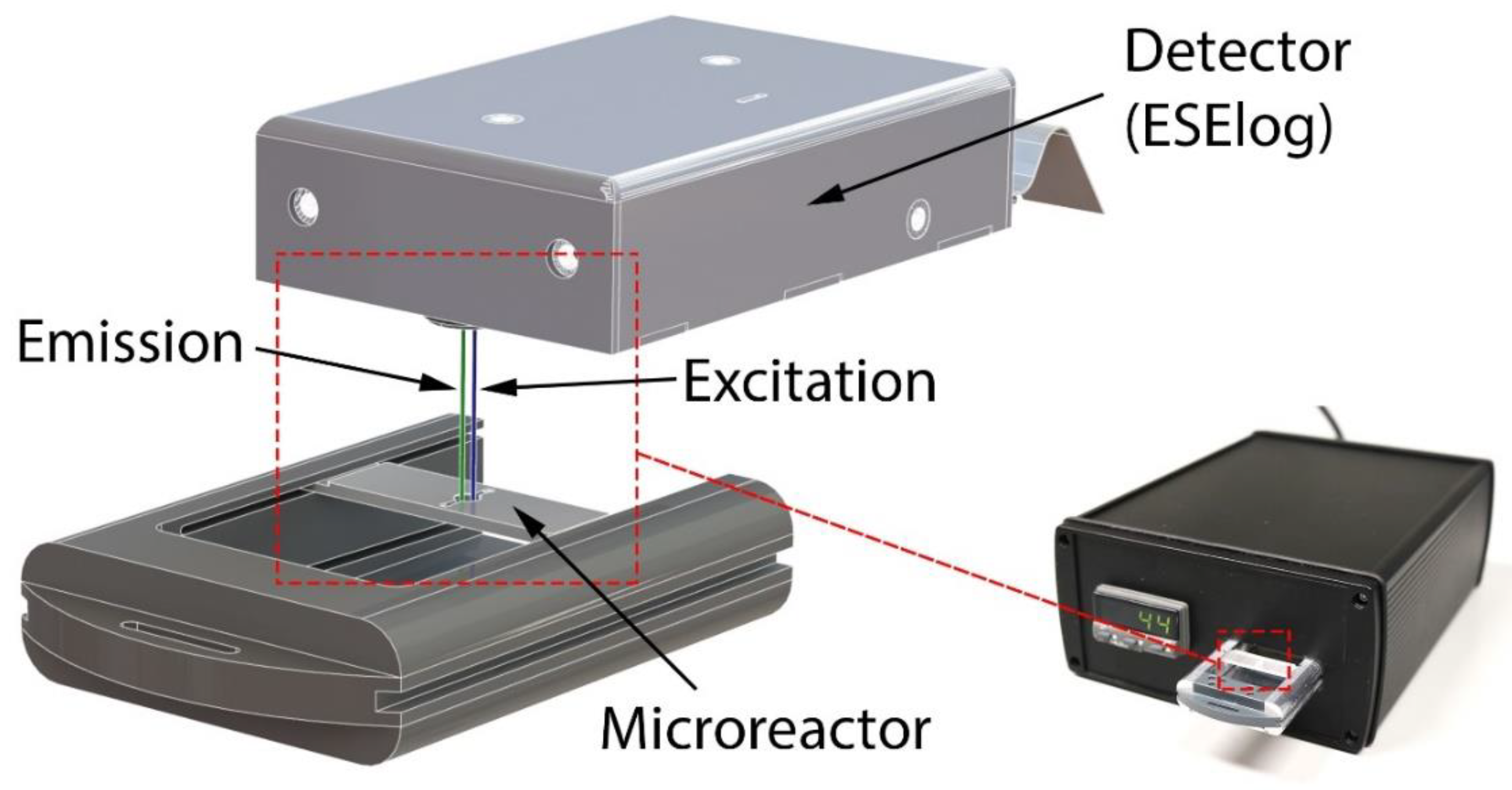








| Material | Manufacturer | Abbreviation |
|---|---|---|
| NextDent Surgical Guide | NextDent B.V., Soesterberg, The Netherlands | NXSG |
| NextDent Ortho Clear | NextDent B.V., Soesterberg, The Netherlands | NXOC |
| SolFlex Surgical Guide | VOCO GmbH, Cuxhaven, Germany | SFSG |
| Protocol | Illumination (min) | Temperature |
|---|---|---|
| Uncured | - | - |
| RT | 60 | Ambient (~23 °C) |
| 60 °C | 60 | 60 °C |
| Name | Sequence 5′-3′ |
|---|---|
| khe Forward | ACACTTTTCTCAATAACACCGAGCAGGAGGTTC |
| khe Reverse | CGCATAGTGCGCCGCGCTTCGCCCCTTCCCCGG |
| khe exo probe | CGCTCAATCCAGGCTATGCCGCGACGCGCCAGGA(dT-BHQ1)C(dspacer)(dT-FAM)TGGGTTGACCATCC-PH |
| blaNDM-1 Forward | GACCAGACCGCCCAGATCCTCAACTGGATCAAGCA |
| blaNDM-1 Reverse | CTGGTTCGACAACGCATTGGCATAAGTCGCAA |
| blaNDM-1 exo Probe | CCCCGCCGCATGCAGCGCGTCCATACCGCCCA(dT-BHQ2)(dspacer)(dT-LC610)TGTCCTGATGCGCG-PH |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behrmann, O.; Hügle, M.; Eckardt, F.; Bachmann, I.; Heller, C.; Schramm, M.; Turner, C.; Hufert, F.T.; Dame, G. 3D Printed Monolithic Microreactors for Real-Time Detection of Klebsiella pneumoniae and the Resistance Gene blaNDM-1 by Recombinase Polymerase Amplification. Micromachines 2020, 11, 595. https://doi.org/10.3390/mi11060595
Behrmann O, Hügle M, Eckardt F, Bachmann I, Heller C, Schramm M, Turner C, Hufert FT, Dame G. 3D Printed Monolithic Microreactors for Real-Time Detection of Klebsiella pneumoniae and the Resistance Gene blaNDM-1 by Recombinase Polymerase Amplification. Micromachines. 2020; 11(6):595. https://doi.org/10.3390/mi11060595
Chicago/Turabian StyleBehrmann, Ole, Matthias Hügle, Franz Eckardt, Iris Bachmann, Cecilia Heller, Marina Schramm, Carrie Turner, Frank T. Hufert, and Gregory Dame. 2020. "3D Printed Monolithic Microreactors for Real-Time Detection of Klebsiella pneumoniae and the Resistance Gene blaNDM-1 by Recombinase Polymerase Amplification" Micromachines 11, no. 6: 595. https://doi.org/10.3390/mi11060595
APA StyleBehrmann, O., Hügle, M., Eckardt, F., Bachmann, I., Heller, C., Schramm, M., Turner, C., Hufert, F. T., & Dame, G. (2020). 3D Printed Monolithic Microreactors for Real-Time Detection of Klebsiella pneumoniae and the Resistance Gene blaNDM-1 by Recombinase Polymerase Amplification. Micromachines, 11(6), 595. https://doi.org/10.3390/mi11060595





