Molecular Dynamics Insight into the Lipid II Recognition by Type A Lantibiotics: Nisin, Epidermin, and Gallidermin
Abstract
:1. Introduction

- Discover the general lipid II recognition pharmacophore in the three related peptides: nisin1–12, epidermin1–12, and gallidermin1–12. This was done by the MD simulations in water in presence and absence of the dimethyl pyrophosphate (DMPPi), which mimics the lipid II binding site, revealing the mutual adaptation of the peptides and their target.
- Reveal sequence features of gallidermin and epidermin, which offer superior biological activity (in some cases) as compared to nisin.
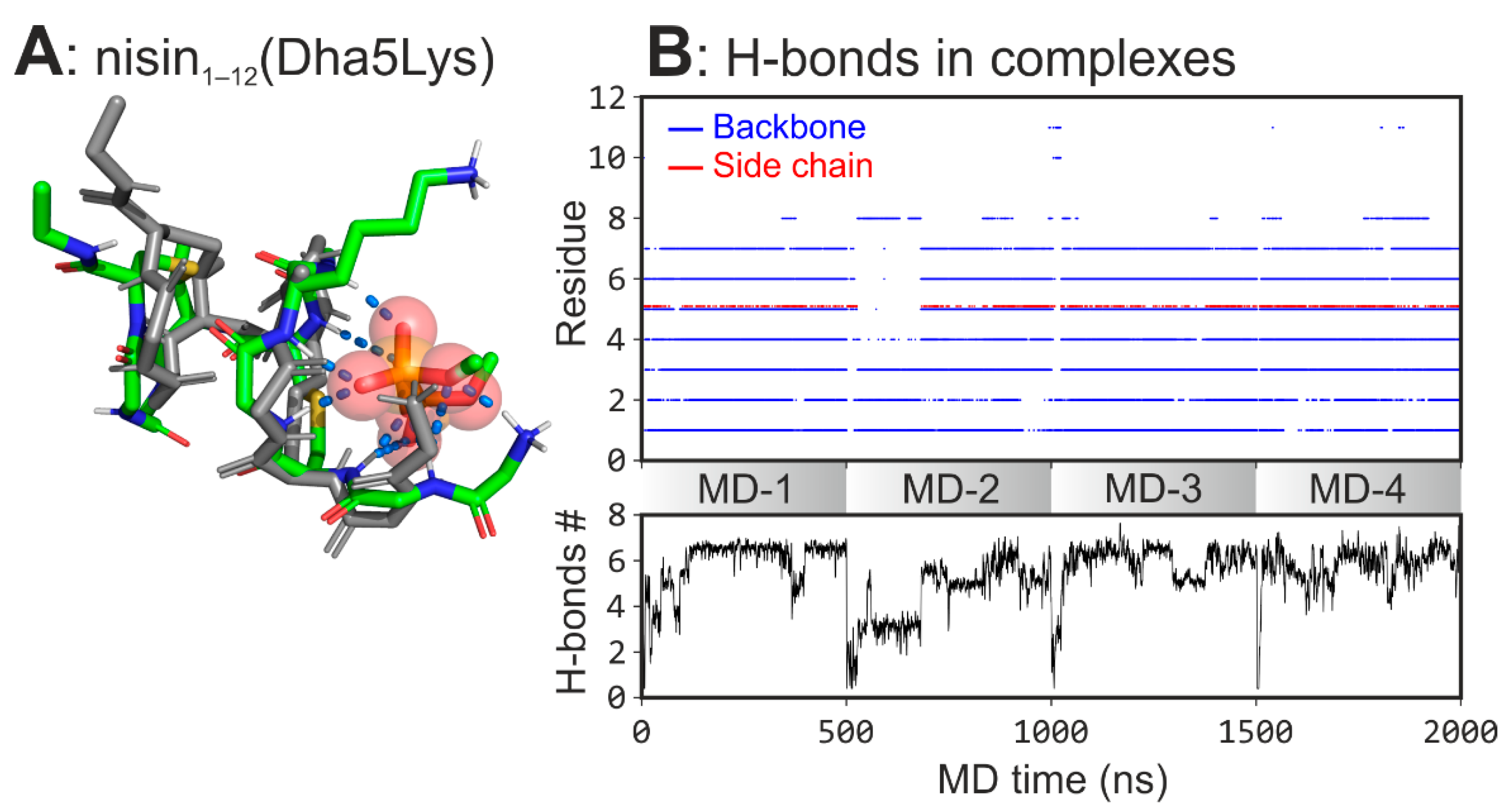
- Verify if this feature upon transfer to the nisin backbone increases its activity (in silico).
2. Materials and Methods
Molecular Dynamics Simulations
3. Results
3.1. Nisin, Epidermin, and Gallidermin Reveal Similar Binding Motif
3.2. The Source of Epidermin and Gallidermin Advantage over Nisin
3.3. Single Mutation May Improve Nisin Binding Ability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Luepke, K.H.; Suda, K.J.; Boucher, H.; Russo, R.L.; Bonney, M.W.; Hunt, T.D.; Mohr, J.F., III. Past, Present, and Future of Antibacterial Economics: Increasing Bacterial Resistance, Limited Antibiotic Pipeline, and Societal Implications. Pharmacotherapy 2017, 37, 71–84. [Google Scholar] [CrossRef]
- Jackson, N.; Czaplewski, L.; Piddock, L.J.V. Discovery and Development of New Antibacterial Drugs: Learning from Experience? J. Antimicrob. Chemother. 2018, 73, 1452–1459. [Google Scholar] [CrossRef] [Green Version]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Bierbaum, G.; Sahl, H.-G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef]
- Chatterjee, C.; Paul, M.; Xie, L.; van der Donk, W.A. Biosynthesis and Mode of Action of Lantibiotics. Chem. Rev. 2005, 105, 633–684. [Google Scholar] [CrossRef]
- McAuliffe, O. Lantibiotics: Structure, Biosynthesis and Mode of Action. FEMS Microbiol. Rev. 2001, 25, 285–308. [Google Scholar] [CrossRef] [Green Version]
- Jack, R.W.; Sahl, H.G. Unique Peptide Modifications Involved in the Biosynthesis of Lantibiotics. Trends Biotechnol. 1995, 13, 269–278. [Google Scholar] [CrossRef]
- Breukink, E.; de Kruijff, B. Lipid II as a Target for Antibiotics. Nat. Rev. Drug Discov. 2006, 5, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, Q.; Breukink, E. Non-Lipid II Targeting Lantibiotics. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183244. [Google Scholar] [CrossRef] [PubMed]
- Garneau, S.; Martin, N.I.; Vederas, J.C. Two-Peptide Bacteriocins Produced by Lactic Acid Bacteria. Biochimie 2002, 84, 577–592. [Google Scholar] [CrossRef]
- Oppedijk, S.F.; Martin, N.I.; Breukink, E. Hit ’Em Where It Hurts: The Growing and Structurally Diverse Family of Peptides That Target Lipid-II. Biochim. Biophys. Acta 2016, 1858, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Barbour, A.; Wescombe, P.; Smith, L. Evolution of Lantibiotic Salivaricins: New Weapons to Fight Infectious Diseases. Trends Microbiol. 2020, 28, 578–593. [Google Scholar] [CrossRef] [PubMed]
- Shenkarev, Z.O.; Finkina, E.I.; Nurmukhamedova, E.K.; Balandin, S.V.; Mineev, K.S.; Nadezhdin, K.D.; Yakimenko, Z.A.; Tagaev, A.A.; Temirov, Y.V.; Arseniev, A.S.; et al. Isolation, Structure Elucidation, and Synergistic Antibacterial Activity of a Novel Two-Component Lantibiotic Lichenicidin from Bacillus Licheniformis VK21. Biochemistry 2010, 49, 6462–6472. [Google Scholar] [CrossRef]
- Mattick, A.T.R.; Hirsch, A. Further Observations on an Inhibitory Substance (nisin) from Lactic Streptococci. Lancet 1947, 2, 5–8. [Google Scholar] [CrossRef]
- Scherer, K.M.; Spille, J.-H.; Sahl, H.-G.; Grein, F.; Kubitscheck, U. The Lantibiotic Nisin Induces Lipid II Aggregation, Causing Membrane Instability and Vesicle Budding. Biophys. J. 2015, 108, 1114–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiedemann, I.; Breukink, E.; van Kraaij, C.; Kuipers, O.P.; Bierbaum, G.; de Kruijff, B.; Sahl, H.G. Specific Binding of Nisin to the Peptidoglycan Precursor Lipid II Combines Pore Formation and Inhibition of Cell Wall Biosynthesis for Potent Antibiotic Activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef] [Green Version]
- Brötz, H.; Josten, M.; Wiedemann, I.; Schneider, U.; Götz, F.; Bierbaum, G.; Sahl, H.G. Role of Lipid-Bound Peptidoglycan Precursors in the Formation of Pores by Nisin, Epidermin and Other Lantibiotics. Mol. Microbiol. 1998, 30, 317–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breukink, E.; Wiedemann, I.; van Kraaij, C.; Kuipers, O.P.; Sahl, H.-G.; de Kruijff, B. Use of the Cell Wall Precursor Lipid II by a Pore-Forming Peptide Antibiotic. Science 1999, 286, 2361–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasper, H.E.; de Kruijff, B.; Breukink, E. Assembly and Stability of Nisin-Lipid II Pores. Biochemistry 2004, 43, 11567–11575. [Google Scholar] [CrossRef]
- Hsu, S.-T.; Breukink, E.; de Kruijff, B.; Kaptein, R.; Bonvin, A.M.J.J.; van Nuland, N.A.J. Mapping the Targeted Membrane Pore Formation Mechanism by Solution NMR: The Nisin Z and Lipid II Interaction in SDS Micelles. Biochemistry 2002, 41, 7670–7676. [Google Scholar] [CrossRef] [Green Version]
- Hsu, S.-T.D.; Breukink, E.; Tischenko, E.; Lutters, M.A.G.; de Kruijff, B.; Kaptein, R.; Bonvin, A.M.J.J.; van Nuland, N.A.J. The Nisin-Lipid II Complex Reveals a Pyrophosphate Cage That Provides a Blueprint for Novel Antibiotics. Nat. Struct. Mol. Biol. 2004, 11, 963–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medeiros-Silva, J.; Jekhmane, S.; Paioni, A.L.; Gawarecka, K.; Baldus, M.; Swiezewska, E.; Breukink, E.; Weingarth, M. High-Resolution NMR Studies of Antibiotics in Cellular Membranes. Nat. Commun. 2018, 9, 3963. [Google Scholar] [CrossRef] [Green Version]
- Allgaier, H.; Jung, G.; Werner, R.G.; Schneider, U.; Zähner, H. Epidermin: Sequencing of a Heterodetic Tetracyclic 21-Peptide Amide Antibiotic. Eur. J. Biochem. 1986, 160, 9–22. [Google Scholar] [CrossRef]
- Kellner, R.; Jung, G.; Hörner, T.; Zähner, H.; Schnell, N.; Entian, K.D.; Götz, F. Gallidermin: A New Lanthionine-Containing Polypeptide Antibiotic. Eur. J. Biochem. 1988, 177, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Ulm, H.; Reder-Christ, K.; Sahl, H.-G.; Schneider, T. Interaction of Type A Lantibiotics with Undecaprenol-Bound Cell Envelope Precursors. Microb. Drug Resist. 2012, 18, 261–270. [Google Scholar] [CrossRef]
- Bonelli, R.R.; Schneider, T.; Sahl, H.-G.; Wiedemann, I. Insights into in Vivo Activities of Lantibiotics from Gallidermin and Epidermin Mode-of-Action Studies. Antimicrob. Agents Chemother. 2006, 50, 1449–1457. [Google Scholar] [CrossRef] [Green Version]
- Panina, I.; Krylov, N.; Nolde, D.; Efremov, R.; Chugunov, A. Environmental and Dynamic Effects Explain How Nisin Captures Membrane-Bound Lipid II. Sci. Rep. 2020, 10, 8821. [Google Scholar] [CrossRef]
- Chan, W.C.; Leyland, M.; Clark, J.; Dodd, H.M.; Lian, L.Y.; Gasson, M.J.; Bycroft, B.W.; Roberts, G.C. Structure-Activity Relationships in the Peptide Antibiotic Nisin: Antibacterial Activity of Fragments of Nisin. FEBS Lett. 1996, 390, 129–132. [Google Scholar] [CrossRef]
- Slootweg, J.C.; van Herwerden, E.F.; van Doremalen, M.F.M.; Breukink, E.; Liskamp, R.M.J.; Rijkers, D.T.S. Synthesis of Nisin AB Dicarba Analogs Using Ring-Closing Metathesis: Influence of sp(3) versus sp(2) Hybridization of the α-Carbon Atom of Residues Dehydrobutyrine-2 and Dehydroalanine-5 on the Lipid II Binding Affinity. Org. Biomol. Chem. 2015, 13, 5997–6009. [Google Scholar] [CrossRef]
- Salsbury, F.R., Jr.; Crowley, M.F.; Brooks, C.L., III. Modeling of the Metallo-Beta-Lactamase from B. Fragilis: Structural and Dynamic Effects of Inhibitor Binding. Proteins 2001, 44, 448–459. [Google Scholar] [CrossRef]
- Salsbury, F.R. Molecular Dynamics Simulations of Protein Dynamics and Their Relevance to Drug Discovery. Curr. Opin. Pharmacol. 2010, 10, 738–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panina, I.S.; Chugunov, A.O.; Efremov, R.G. Lipid II as a Target for Novel Antibiotics: Structural and Molecular Dynamics Studies. Russ. J. Bioorg. Chem. 2018, 44, 653–664. [Google Scholar] [CrossRef]
- Bakhtiary, A.; Cochrane, S.A.; Mercier, P.; McKay, R.T.; Miskolzie, M.; Sit, C.S.; Vederas, J.C. Insights into the Mechanism of Action of the Two-Peptide Lantibiotic Lacticin 3147. J. Am. Chem. Soc. 2017, 139, 17803–17810. [Google Scholar] [CrossRef] [Green Version]
- Dickman, R.; Danelius, E.; Mitchell, S.A.; Flemming Hansen, D.; Erdélyi, M.; Tabor, A.B. A Chemical Biology Approach to Understanding Molecular Recognition of Lipid II by Nisin(1–12): Synthesis and NMR Ensemble Analysis of Nisin(1–12) and Analogues. Chem. A Eur. J. 2019, 25, 14572–14582. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces, Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry, Jerusalem, Israel, 13–16 April 1981; Pullman, B., Ed.; Springer Netherlands: Dordrecht, The Netherlands, 1981; pp. 331–342. ISBN 9789401576581. [Google Scholar]
- Yang, H.; Du Bois, D.R.; Ziller, J.W.; Nowick, J.S. X-ray Crystallographic Structure of a Teixobactin Analogue Reveals Key Interactions of the Teixobactin Pharmacophore. Chem. Commun. 2017, 53, 2772–2775. [Google Scholar] [CrossRef] [Green Version]


| System Composition | MD Duration, ns |
|---|---|
| Peptides1–12 in solution | |
| Nisin1–12 (1)/Water(5694)/Cl−(1) | 5 × 500 |
| Epidermin1–12 (1)/Water(5687)/Cl−(1) | 5 × 500 |
| Gallidermin1–12 (1)/Water(5687)/Cl−(1) | 5 × 500 |
| Peptides1–12 with DMPPi in solution | |
| Nisin1–12 (1)/DMPPi(1)/Water(6707)/Na+(1) | 5 × 500 |
| Epidermin1–12 (1)/DMPPi(1)/Water(6706)/Na+(1) | 5 × 500 |
| Gallidermin1–12 (1)/DMPPi(1)/Water(6707)/Na+(1) | 5 × 500 |
| Nisin1–12(Dha5Lys)(1)/DMPPi(1)/Water (5690) | 4 × 500 |
| Nisin1–12 (1)/DMPPi(3)/Water(5671)/Na+(5) | 5 × 500 |
| Epidermin1–12 (1)/DMPPi(3)/Water(5661)/Na+(5) | 5 × 500 |
| Gallidermin1–12 (1)/DMPPi(3)/Water(5670)/Na+(5) | 5 × 500 |
| Nisin1–12(Dha5Lys)(1)/DMPPi(3)/Water(5673)/Na+(4) | 4 × 500 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panina, I.; Taldaev, A.; Efremov, R.; Chugunov, A. Molecular Dynamics Insight into the Lipid II Recognition by Type A Lantibiotics: Nisin, Epidermin, and Gallidermin. Micromachines 2021, 12, 1169. https://doi.org/10.3390/mi12101169
Panina I, Taldaev A, Efremov R, Chugunov A. Molecular Dynamics Insight into the Lipid II Recognition by Type A Lantibiotics: Nisin, Epidermin, and Gallidermin. Micromachines. 2021; 12(10):1169. https://doi.org/10.3390/mi12101169
Chicago/Turabian StylePanina, Irina, Amir Taldaev, Roman Efremov, and Anton Chugunov. 2021. "Molecular Dynamics Insight into the Lipid II Recognition by Type A Lantibiotics: Nisin, Epidermin, and Gallidermin" Micromachines 12, no. 10: 1169. https://doi.org/10.3390/mi12101169
APA StylePanina, I., Taldaev, A., Efremov, R., & Chugunov, A. (2021). Molecular Dynamics Insight into the Lipid II Recognition by Type A Lantibiotics: Nisin, Epidermin, and Gallidermin. Micromachines, 12(10), 1169. https://doi.org/10.3390/mi12101169








