The Effect of a Rotating Cone on Horseradish Peroxidase Aggregation on Mica Revealed by Atomic Force Microscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Enzyme
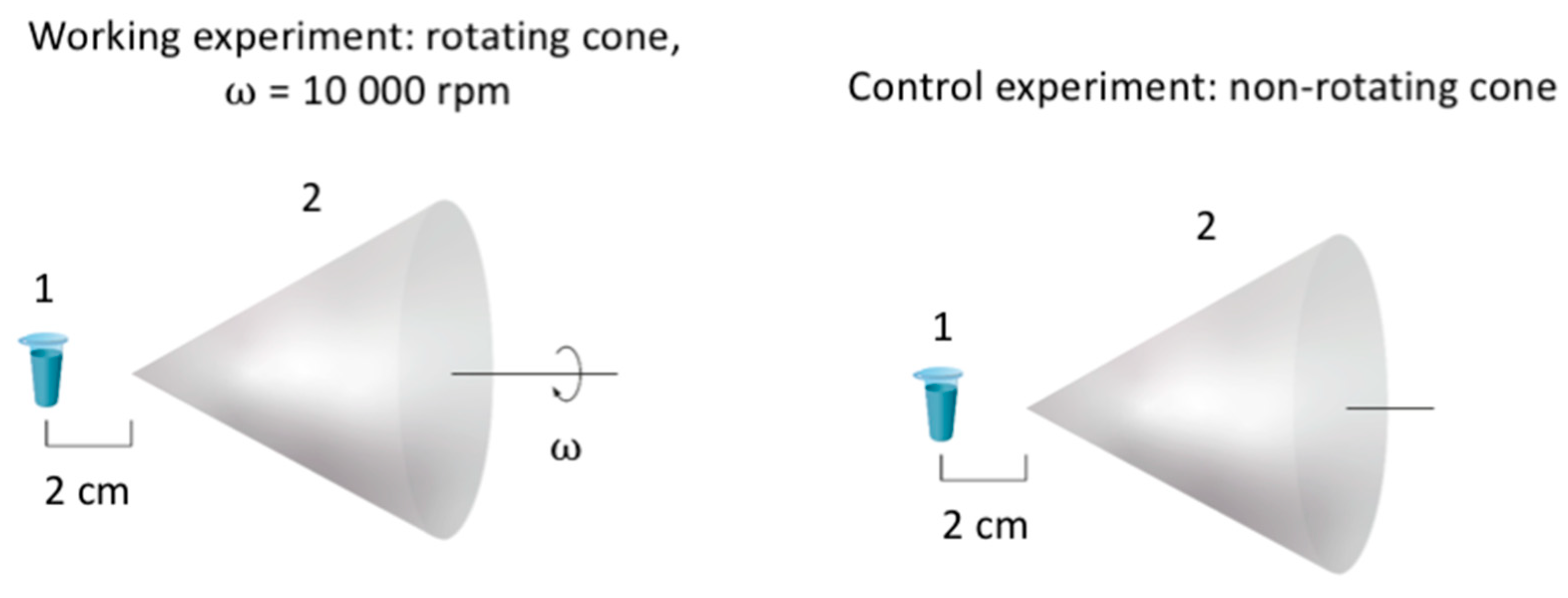
2.2. Experimental Setup
2.3. Atomic Force Microscopy
2.4. Spectrophotometry
3. Results
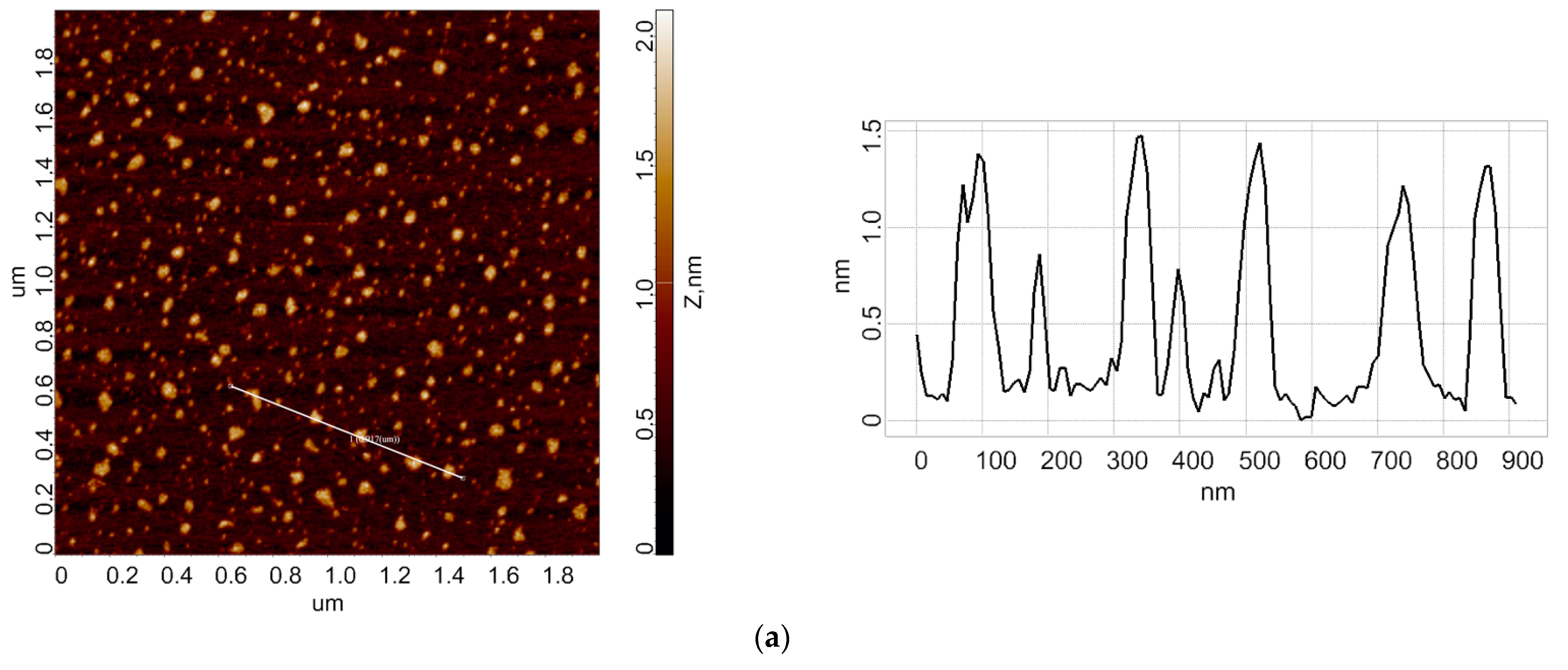
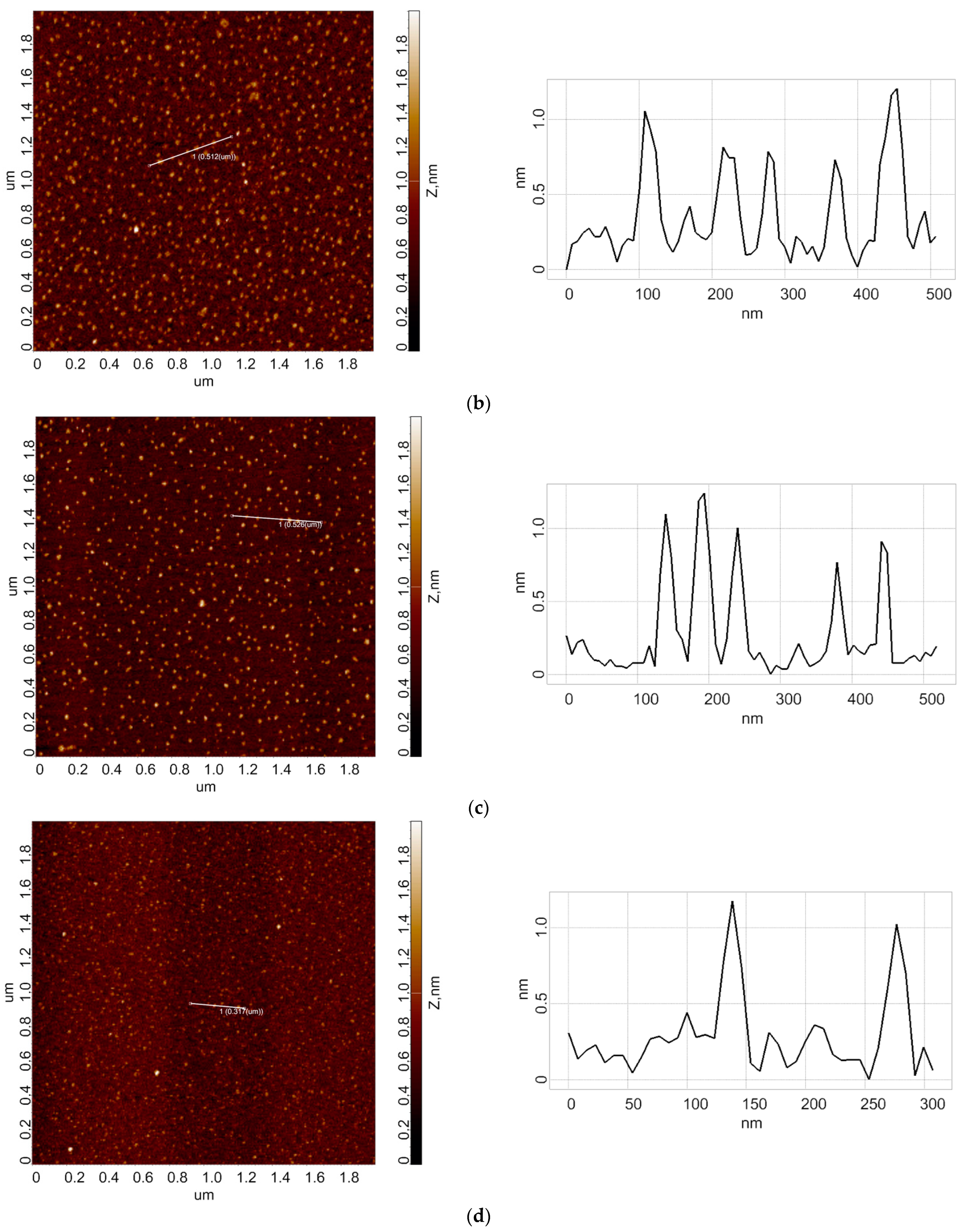
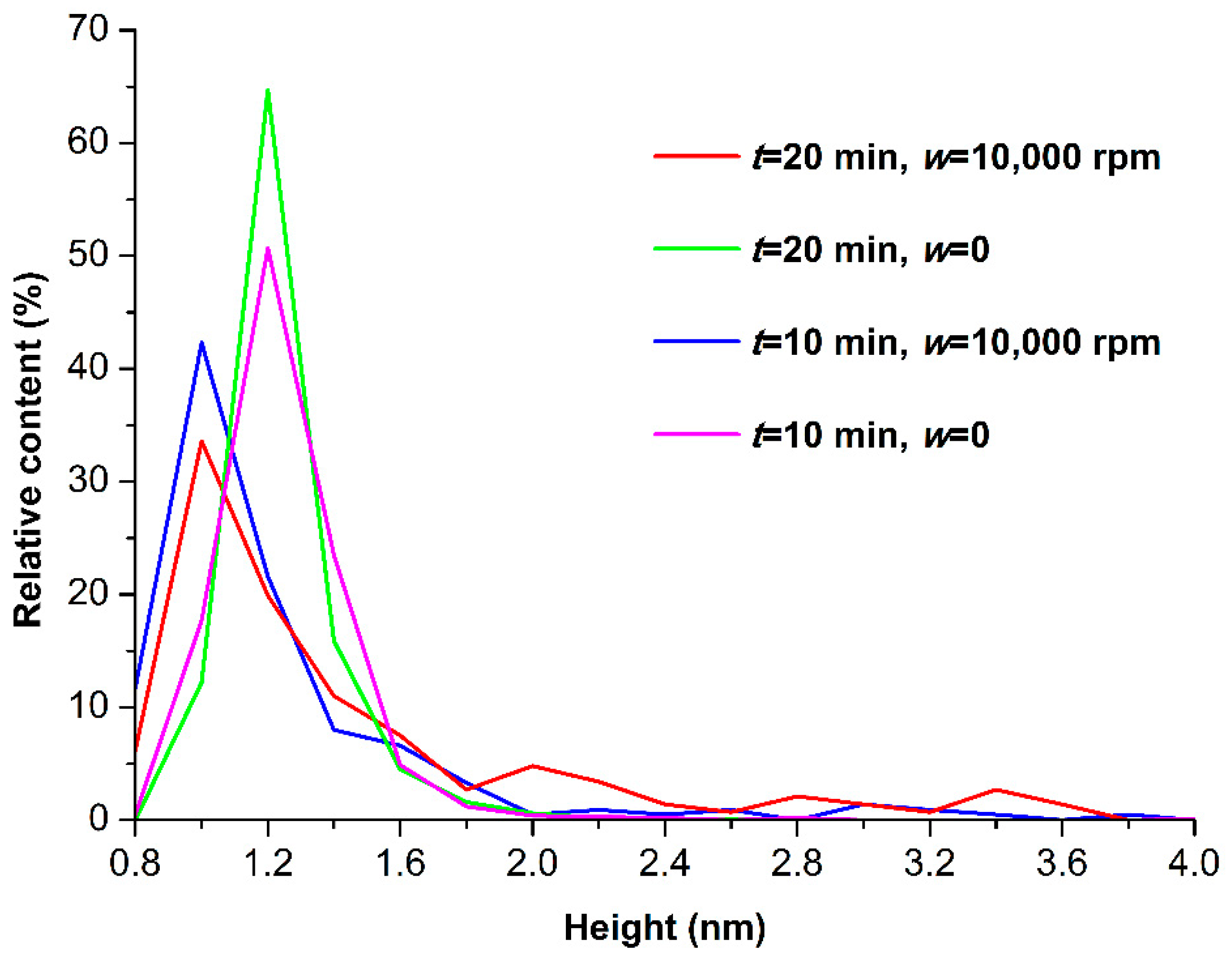
3.1. Atomic Force Microscopy
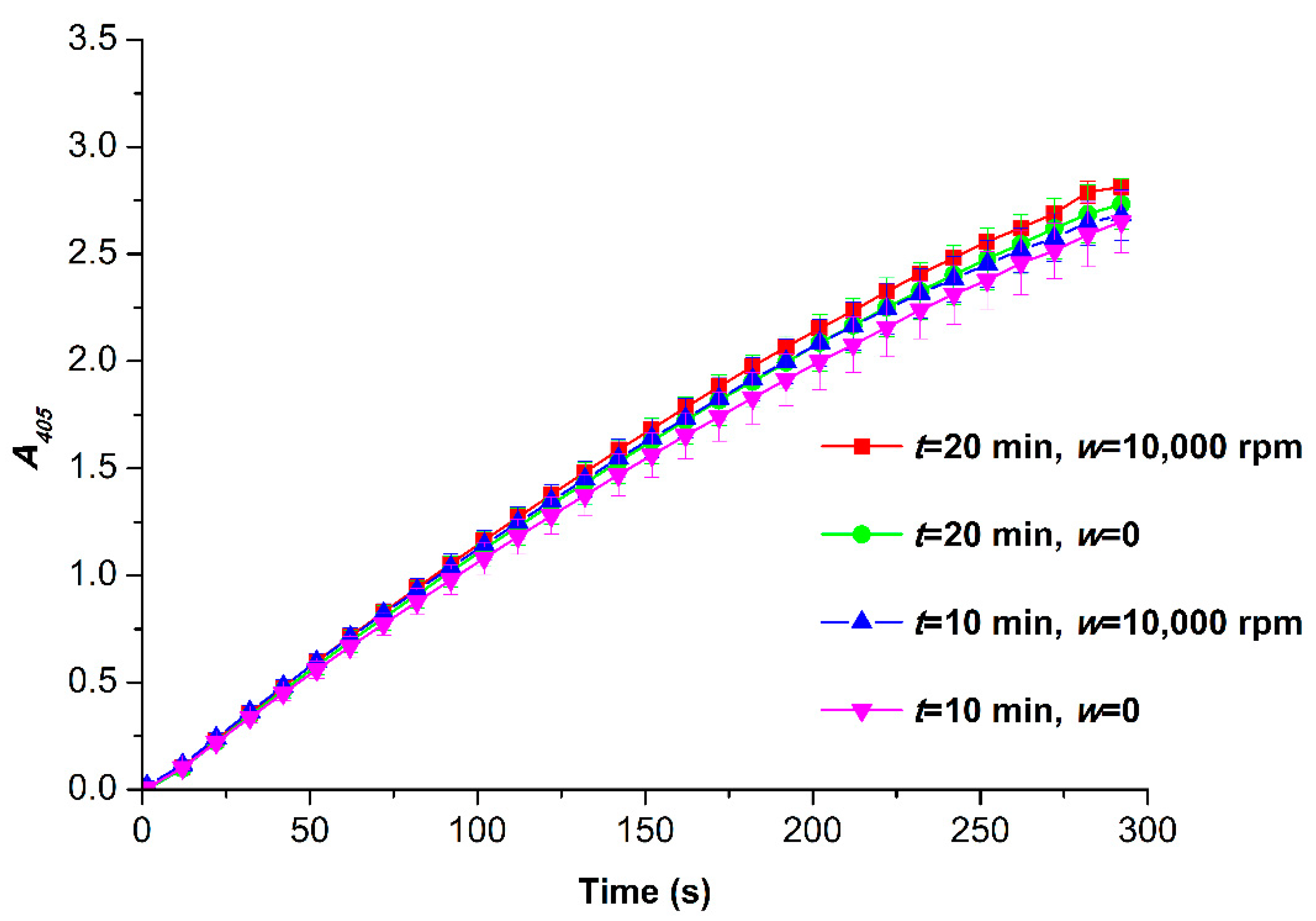
3.2. Spectrophotometry
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choi, D.; Lee, H.; Im, D.J.; Kang, I.S.; Lim, G.; Kim, D.S.; Kang, K.H. Spontaneous electrical charging of droplets by conventional pipetting. Sci. Rep. 2013, 3, 2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, H.; Zhang, Y.; Guo, L.; Wang, P.; He, X.; Dai, G.; Zheng, H.; Chen, C.; Wang, A.C.; Xu, C.; et al. Quantifying the triboelectric series. Nat. Commun. 2019, 10, 1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowell, J. The role of material transfer in contact electrification. J. Phys. D Appl. Phys. 1977, 10, L233. [Google Scholar] [CrossRef]
- Diaz, A.F.; Felix-Navarro, R.M. A semi-quantitative tribo-electric series for polymeric materials: The influence of chemical structure and properties. J. Electrost. 2004, 62, 277–290. [Google Scholar] [CrossRef]
- Matsusaka, S.; Masuda, H. Electrostatics of particles. Adv. Powder Technol. 2003, 14, 143–166. [Google Scholar] [CrossRef]
- Armitage, J.L.; Ghanbarzadeh, A.; Bryant, M.G.; Neville, A. Investigating the Influence of Friction and Material Wear on Triboelectric Charge Transfer in Metal–Polymer Contacts. Tribol. Lett. 2022, 70, 46. [Google Scholar] [CrossRef]
- Elfström, N.; Juhasz, R.; Sychugov, I.; Engfeldt, T.; Karlström, A.E.; Linnros, J. Surface charge sensitivity of silicon nanowires: Size dependence. Nano Lett. 2007, 7, 2608–2612. [Google Scholar] [CrossRef] [Green Version]
- Malsagova, K.A.; Pleshakova, T.O.; Kozlov, A.F.; Galiullin, R.A.; Popov, V.P.; Tikhonenko, F.V.; Glukhov, A.V.; Ziborov, V.S.; Shumov, I.D.; Petrov, O.F.; et al. Detection of Influenza Virus Using a SOI-Nanoribbon Chip, Based on an N-Type Field-Effect Transistor. Biosensors 2021, 11, 119. [Google Scholar] [CrossRef]
- Zheng, G.; Patolsky, F.; Cui, Y.; Wang, W.U.; Lieber, C.M. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat. Biotechnol. 2005, 23, 1294–1301. [Google Scholar] [CrossRef]
- IAsys Cuvette System User’s Guide, 1st ed.; Fisons plc.: Loughborough, UK, 1993.
- Ziborov, V.S.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Ivanova, I.A.; Valueva, A.A.; Tatur, V.Y.; Negodailov, A.N.; Lukyanitsa, A.A.; Ivanov, Y.D. Investigation of the Influence of Liquid Motion in a Flow-Based System on an Enzyme Aggregation State with an Atomic Force Microscopy Sensor: The Effect of Glycerol Flow. Appl. Sci. 2020, 10, 4825. [Google Scholar] [CrossRef]
- Ispas, C.R.; Crivat, G.; Andreescu, S. Review: Recent Developments in Enzyme-Based Biosensors for Biomedical Analysis. Anal. Lett. 2012, 45, 168–186. [Google Scholar] [CrossRef]
- Metzler, D.E. Biochemistry. The Chemical Reactions of Living Cells, 1st ed.; Academic Press: Cambridge, UK, 1977. [Google Scholar]
- Rogozhin, V.V.; Kutuzova, G.D.; Ugarova, N.N. Inhibition of horseradish peroxidase by N-ethylamide of o-sulfobenzoylacetic acid. Russian J. Bioorg. Chem. 2000, 26, 138–141. [Google Scholar] [CrossRef]
- Gavrilenko, T.I.; Ryzhkova, N.A.; Parkhomenko, A.N. Myeloperoxidase and its role in development of ischemic heart disease. Ukr. J. Cardiol. 2014, 4, 119–126. [Google Scholar]
- Yao, Y.; Zhang, B.; Pang, H.; Wang, Y.; Fu, H.; Chen, X.; Wang, Y. The effect of radio frequency heating on the inactivation and structure of horseradish peroxidase. Food Chem. 2023, 398, 133875. [Google Scholar] [CrossRef]
- Lopes, L.C.; Barreto, M.T.; Gonçalves, K.M.; Alvarez, H.M.; Heredia, M.F.; De Souza, R.O.M.; Cordeiro, Y.; Dariva, C.; Fricks, A.T. Stability and structural changes of horseradish peroxidase: Microwave versus conventional heating treatment. Enzym. Microb. Technol. 2015, 69, 10–18. [Google Scholar] [CrossRef]
- Marko-Varga, G.; Johansson, K.; Gorton, L. Enzyme-based biosensor as a selective detection unit in column liquid chromatography. J. Chromatogr. A 1994, 660, 153–167. [Google Scholar] [CrossRef]
- Hernández-Cancel, G.; Suazo-Dávila, D.; Medina-Guzmán, J.; Rosado-González, M.; Díaz-Vázquez, L.M.; Griebenow, K. Chemically glycosylation improves the stability of an amperometric horseradish peroxidase biosensor. Anal. Chim. Acta 2015, 854, 129–139. [Google Scholar] [CrossRef] [Green Version]
- Duan, N.D.; Li, C.; Song, M.; Wang, Z.; Zhu, C.; Wu, S. Signal amplification of SiO2 nanoparticle loaded horseradish peroxidase for colorimetric detection of lead ions in water. Stereochim. Acta A Mol. Biomol. Spectrosc. 2022, 265, 120342. [Google Scholar] [CrossRef]
- Nayak, S.; Kale, P.; Balasubramanian, P. Inhibition assays of horseradish peroxidase by hexavalent chromium and other heavy metals. Int. J. Environ. Anal. Chem. 2020, 1–13. [Google Scholar] [CrossRef]
- Rahemi, V.; Trashin, S.; Hafideddine, Z.; van Doorslaer, S.; Meynen, V.; Gorton, L.; de Wael, K. Amperometric Flow-Injection Analysis of Phenols Induced by Reactive Oxygen Species Generated under Daylight Irradiation of Titania Impregnated with Horseradish Peroxidase. Anal. Chem. 2020, 92, 3643–3649. [Google Scholar] [CrossRef]
- Ignatenko, O.V.; Sjölander, A.; Hushpulian, D.M.; Kazakov, S.V.; Ouporov, I.V.; Chubar, T.A.; Poloznikov, A.A.; Ruzgas, T.; Tishkov, V.I.; Gorton, L.; et al. Electrochemistry of chemically trapped dimeric and monomeric recombinant horseradish peroxidase. Adv. Biosens. Bioelectron. 2013, 2, 25–34. [Google Scholar]
- Welinder, K.G. Amino acid sequence studies of horseradish peroxidase. amino and carboxyl termini, cyanogen bromide and tryptic fragments, the complete sequence, and some structural characteristics of horseradish peroxidase. Cent. Eur. J. Biochem. 1979, 96, 483–502. [Google Scholar] [CrossRef]
- Tams, J.W.; Welinder, K.G. Mild chemical deglycosylation of horseradish peroxidase yields a fully active, homogeneous enzyme. Anal. Biochem. 1995, 228, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Shannon, L.M.; Kay, E.; Lew, J.Y. Peroxidase isozymes from horseradish roots I. Isolation and physical properties. J. Biol. Chem. 1966, 241, 2166–2172. [Google Scholar] [CrossRef]
- Veitch, N.C.; Smith, A.T. Horseradish peroxidase. Adv. Inorg. Chem. 2000, 51, 107–162. [Google Scholar] [CrossRef]
- Davies, P.F.; Rennke, H.G.; Cotran, R.S. Influence of molecular charge upon the endocytosis and intracellular fate of peroxidase activity in cultured arterial endothelium. J. Cell Sci. 1981, 49, 69–86. [Google Scholar] [CrossRef]
- Gajhede, M.; Schuller, D.J.; Henriksen, A.; Smith, A.T.; Poulos, T.L. Crystal structure of horseradish peroxidase C at 2.15 Å resolution. Nat. Struct. Mol. Biol. 1997, 4, 1032–1038. [Google Scholar] [CrossRef]
- Pleshakova, T.O.; Bukharina, N.S.; Archakov, A.I.; Ivanov, Y.D. Atomic Force Microscopy for Protein Detection and Their Physicochemical Characterization. Int. J. Mol. Sci. 2018, 19, 1142. [Google Scholar] [CrossRef] [Green Version]
- Dufrêne, Y.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Müller, D.G. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295–307. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Frantsuzov, P.A.; Zöllner, A.; Medvedeva, N.V.; Archakov, A.I.; Reinle, W.; Bernhardt, R. Atomic Force Microscopy Study of Protein–Protein Interactions in the Cytochrome CYP11A1 (P450scc)-Containing Steroid Hydroxylase System. Nanoscale Res. Lett. 2011, 6, 54. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, Y.D.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Ivanova, I.A.; Valueva, A.A.; Tatur, V.Y.; Smelov, M.V.; Ivanova, N.D.; Ziborov, V.S. AFM imaging of protein aggregation in studying the impact of knotted electromagnetic field on a peroxidase. Sci. Rep. 2020, 10, 9022. [Google Scholar] [CrossRef] [PubMed]
- Braga, P.C.; Ricci, D. Atomic Force Microscopy. Biology: Biomedical Methods and Applications. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2003; Volume 242, p. 382. [Google Scholar]
- Laskowski, D.; Strzelecki, J.; Pawlak, K.; Dahm, H.; Balter, A. Effect of ampicillin on adhesive properties of bacteria examined by atomic force microscopy. Micron 2018, 112, 84–90. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Tatur, V.Y.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Valueva, A.A.; Ivanova, I.A.; Ershova, M.O.; Ivanova, N.D.; Repnikov, V.V.; et al. Effect of Spherical Elements of Biosensors and Bioreactors on the Physicochemical Properties of a Peroxidase Protein. Polymers 2021, 13, 1601. [Google Scholar] [CrossRef] [PubMed]
- Kiselyova, O.I.; Yaminsky, I.; Ivanov, Y.D.; Kanaeva, I.P.; Kuznetsov, V.Y.; Archakov, A.I. AFM study of membrane proteins, cytochrome P450 2B4, and NADPH–Cytochrome P450 reductase and their complex formation. Arch. Biochem. Biophys. 1999, 371, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pleshakova, T.O.; Kaysheva, A.L.; Shumov, I.D.; Ziborov, V.S.; Bayzyanova, J.M.; Konev, V.A.; Uchaikin, V.F.; Archakov, A.I.; Ivanov, Y.D. Detection of hepatitis C virus core protein in serum using aptamer-functionalized AFM chips. Micromachines 2019, 10, 129. [Google Scholar] [CrossRef] [Green Version]
- Sanders, S.A.; Bray, R.C.; Smith, A.T. pH-dependent properties of a mutant horseradish peroxidase isoenzyme C in which Arg38 has been replaced with lysine. Eur. J. Biochem. 1994, 224, 1029–1037. [Google Scholar] [CrossRef]
- Enzymatic Assay of Peroxidase (EC 1.11.1.7) 2,20-Azino-Bis(3-Ethylbenzthiazoline-6-Sulfonic Acid) as a Substrate Sigma Prod. No. P-6782. Available online: https://www.sigmaaldrich.com/RU/en/technical-documents/protocol/protein-biology/enzymeactivity-assays/enzymatic-assay-of-peroxidase-abts-as-substrate (accessed on 18 February 2022).
- Ziborov, V.S.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Valueva, A.A.; Ivanova, I.A.; Ershova, M.O.; Larionov, D.I.; Evdokimov, A.N.; Tatur, V.Y.; et al. The Impact of Fast-Rise-Time Electromagnetic Field and Pressure on the Aggregation of Peroxidase upon Its Adsorption onto Mica. Appl. Sci. 2021, 11, 11677. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Tatur, V.Y.; Shumov, I.D.; Kozlov, A.F.; Valueva, A.A.; Ivanova, I.A.; Ershova, M.O.; Ivanova, N.D.; Stepanov, I.N.; Lukyanitsa, A.A.; et al. The Effect of a Dodecahedron-Shaped Structure on the Properties of an Enzyme. J. Funct. Biomater. 2022, 13, 166. [Google Scholar] [CrossRef]
- Caliga, R.; Maniu, C.L.; Mihăşan, M. ELF-EMF exposure decreases the peroxidase catalytic efficiency in vitro. Open Life Sci. 2016, 11, 71–77. [Google Scholar] [CrossRef]
- Pershin, S.M. The Physical Basis of the Weak Fields Interaction with Bio Objects Is the Quantum Differences of H2O Spin-isomers. Online Biophysical Blog. 2012. Available online: http://www.biophys.ru/archive/congress2012/proc-p27.htm (accessed on 18 February 2022).
- Pershin, S.M. Quantum differences of ortho and para spin isomers of H2O as a physical basis of anomalous properties of water. Nanostructures Math. Phys. Model. 2012, 7, 103–120. [Google Scholar]
- Bunkin, A.F.; Nurmatov, A.A.; Pershin, S.M.; Vigasin, A.A. Four-photon coherent spectroscopy of orientational motion of H2O molecules in liquid water. J. Raman Spectrosc. 2005, 36, 145–147. [Google Scholar] [CrossRef]
- Bunkin, A.F.; Nurmatov, A.A.; Pershin, S.M. Coherent four-photon spectroscopy of low-frequency molecular librations in a liquid. Physics-Uspekhi 2006, 49, 855–861. [Google Scholar] [CrossRef]
- Pershin, S.M. Two-liquid water. Phys. Wave Phenom. 2005, 13, 192–208. [Google Scholar]
- Artmann, G.M.; Kelemen, C.; Porst, D.; Büldt, G.; Chien, S. Temperature Transitions of Protein Properties in Human Red Blood Cells. Biophys. J. 1998, 75, 3179–3183. [Google Scholar] [CrossRef]
- Pershin, S.M. Conversion of ortho-para H2O isomers in water and a jump in erythrocyte fluidity through a microcapillary at a temperature of 36.6 ± 0.3 °C. Phys. Wave Phenom. 2009, 17, 241–250. [Google Scholar] [CrossRef]
- Pershin, S.M. A New Conception of the Action of EMF on Water/Aqueous Solutions, Taking into Account the Quantum Differences of the Ortho/Para of Spin Isomers of H2O. Online Biophysical Blog. Available online: http://www.biophys.ru/archive/sarov2013/proc-p17.pdf (accessed on 3 October 2022).
- Pershin, S.M.; Bunkin, A.F.; Golo, V.L. H2O monomers in channels of icelike water structures. J. Exp. Theor. Phys. 2012, 115, 1008–1011. [Google Scholar] [CrossRef]
- Lednev, V.V.; Belova, N.A.; Rozhdestvenskaya, Z.E.; Tiras, K.P. Bioeffects of weak variable magnetic fields and biological precursors of earthquakes. Geophys. Process. Biosph. 2003, 2, 3–11. [Google Scholar]
- Lednev, V.V.; Belova, N.A.; Ermakov, A.M.; Akimov, E.B.; Tonevitsky, A.G. Modulation of cardiac rhythm in the humans exposed to extremely weak alternating magnetic fields. Biophysics 2008, 53, 648–654. [Google Scholar] [CrossRef]
- Morelli, A.; Ravera, S.; Panfoli, I.; Pepe, I.M. Effects of extremely low frequency electromagnetic fields on membrane-associated enzymes. Arch. Biochem. Biophys. 2005, 441, 191–198. [Google Scholar] [CrossRef]
- Thumm, S.; Löschinger, M.; Glock, S.; Hämmerle, H.; Rodemann, H.P. Induction of cAMP-dependent protein kinase A activity in human skin fibroblasts and rat osteoblasts by extremely low-frequency electromagnetic fields. Radiat. Environ. Biophys. 1999, 38, 195–199. [Google Scholar] [CrossRef]
- Andrade, J.D.; Hlady, V.; Wei, A.P. Adsorption of complex proteins at interfaces. Pure Appl. Chem. 1992, 64, 1777–1781. [Google Scholar] [CrossRef] [Green Version]
- Younes-Metzler, O.; Ben, R.N.; Giorgi, J.B. The adsorption of antifreeze glycoprotein fraction 8 on dry and wet mica. Colloids Surf. B Biointerfaces 2011, 82, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.W.; Cho, C.H. Role of Hydration Water in Protein Unfolding. Biophys. J. 1999, 77, 3311–3318. [Google Scholar] [CrossRef] [Green Version]
- Morón, M.C. Protein hydration shell formation: Dynamics of water in biological systems exhibiting nanoscopic cavities. J. Mol. Liquids 2021, 337, 116584. [Google Scholar] [CrossRef]
- Lind, P.A.; Daniel, R.M.; Monk, C.; Dunn, R.V. Esterase catalysis of substrate vapour: Enzyme activity occurs at very low hydration. Biochim. Biophys. Acta 2004, 1702, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gorenstein, D.G. Structure and dynamics of cytochrome c in nonaqueous solvents by 2D NH-exchange NMR spectroscopy. J. Am. Chem. Soc. 1993, 115, 6843–6850. [Google Scholar] [CrossRef]
- Ivanova, I.A.; Ershova, M.O.; Shumov, I.D.; Valueva, A.A.; Ivanov, Y.D.; Pleshakova, T.O. Atomic Force Microscopy Study of the Temperature and Storage Duration Dependencies of Horseradish Peroxidase Oligomeric State. Biomedicines 2022, 10, 2645. [Google Scholar] [CrossRef]
- Samygina, V.R.; Sokolov, A.V.; Bourenkov, G.; Petoukhov, M.V.; Pulina, M.O.; Zakharova, E.T.; Vasilyev, V.B.; Bartunik, H.; Svergun, D.I. Ceruloplasmin: Macromolecular assemblies with iron-containing acute phase proteins. PLoS ONE 2013, 8, e67145. [Google Scholar] [CrossRef] [Green Version]
- Sopova, J.V.; Koshel, E.I.; Belashova, T.A.; Zadorsky, S.P.; Sergeeva, A.V.; Siniukova, V.A.; Shenfeld, A.A.; Velizhanina, M.E.; Volkov, K.V.; Nizhnikov, A.A.; et al. RNA-binding protein FXR1 is presented in rat brain in amyloid form. Sci. Rep. 2019, 9, 18983. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanov, Y.D.; Tatur, V.Y.; Shumov, I.D.; Kozlov, A.F.; Valueva, A.A.; Ivanova, I.A.; Ershova, M.O.; Ivanova, N.D.; Stepanov, I.N.; Lukyanitsa, A.A.; et al. The Effect of a Rotating Cone on Horseradish Peroxidase Aggregation on Mica Revealed by Atomic Force Microscopy. Micromachines 2022, 13, 1947. https://doi.org/10.3390/mi13111947
Ivanov YD, Tatur VY, Shumov ID, Kozlov AF, Valueva AA, Ivanova IA, Ershova MO, Ivanova ND, Stepanov IN, Lukyanitsa AA, et al. The Effect of a Rotating Cone on Horseradish Peroxidase Aggregation on Mica Revealed by Atomic Force Microscopy. Micromachines. 2022; 13(11):1947. https://doi.org/10.3390/mi13111947
Chicago/Turabian StyleIvanov, Yuri D., Vadim Y. Tatur, Ivan D. Shumov, Andrey F. Kozlov, Anastasia A. Valueva, Irina A. Ivanova, Maria O. Ershova, Nina D. Ivanova, Igor N. Stepanov, Andrei A. Lukyanitsa, and et al. 2022. "The Effect of a Rotating Cone on Horseradish Peroxidase Aggregation on Mica Revealed by Atomic Force Microscopy" Micromachines 13, no. 11: 1947. https://doi.org/10.3390/mi13111947




