Application of Microfluidic Systems for Breast Cancer Research
Abstract
:1. Introduction
1.1. Breast Cancer Physiology
1.2. Breast Cancer Types
2. Breast Cancer Metastasis
2.1. Invasion Modeling
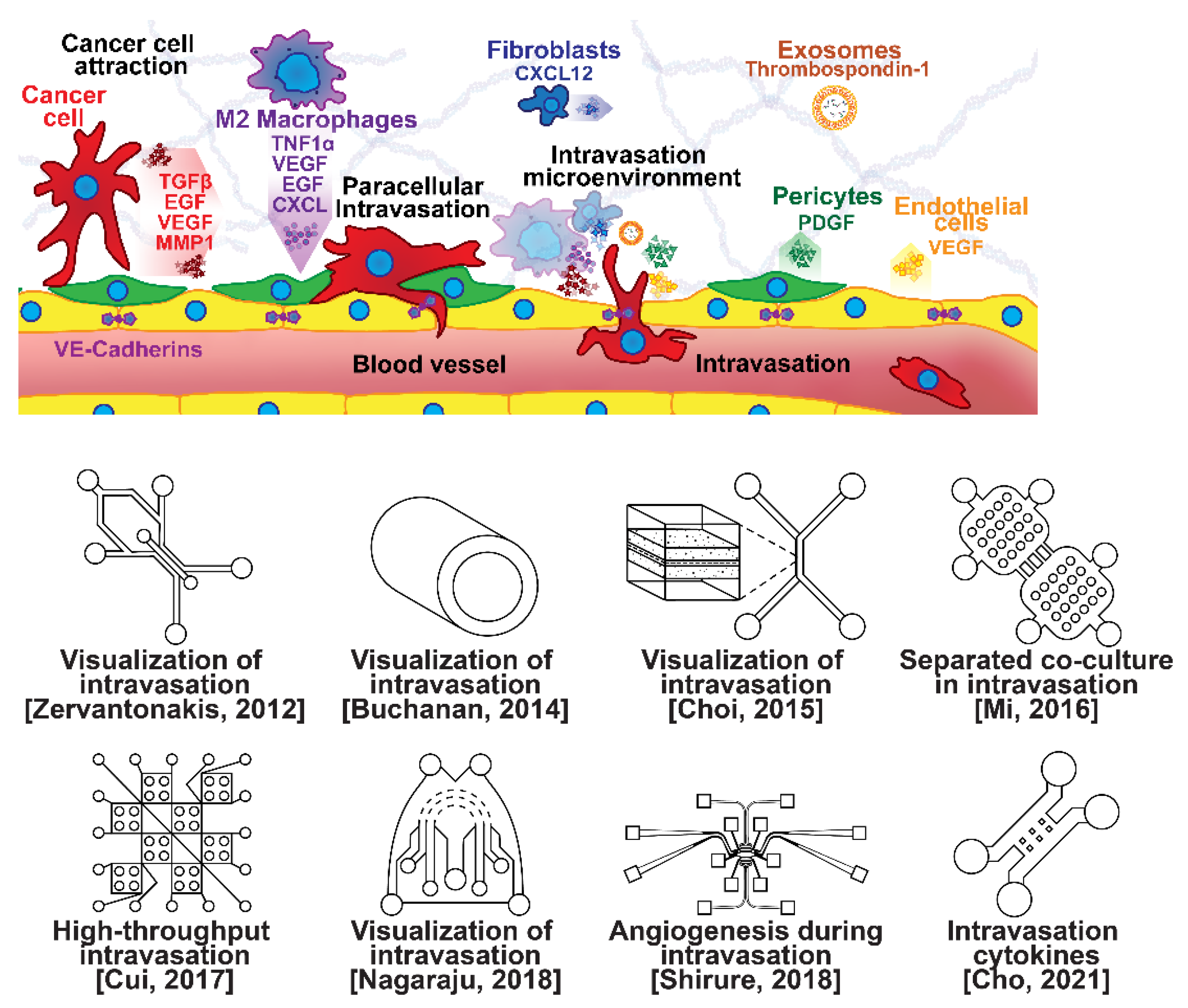
2.2. Intravasation Modeling
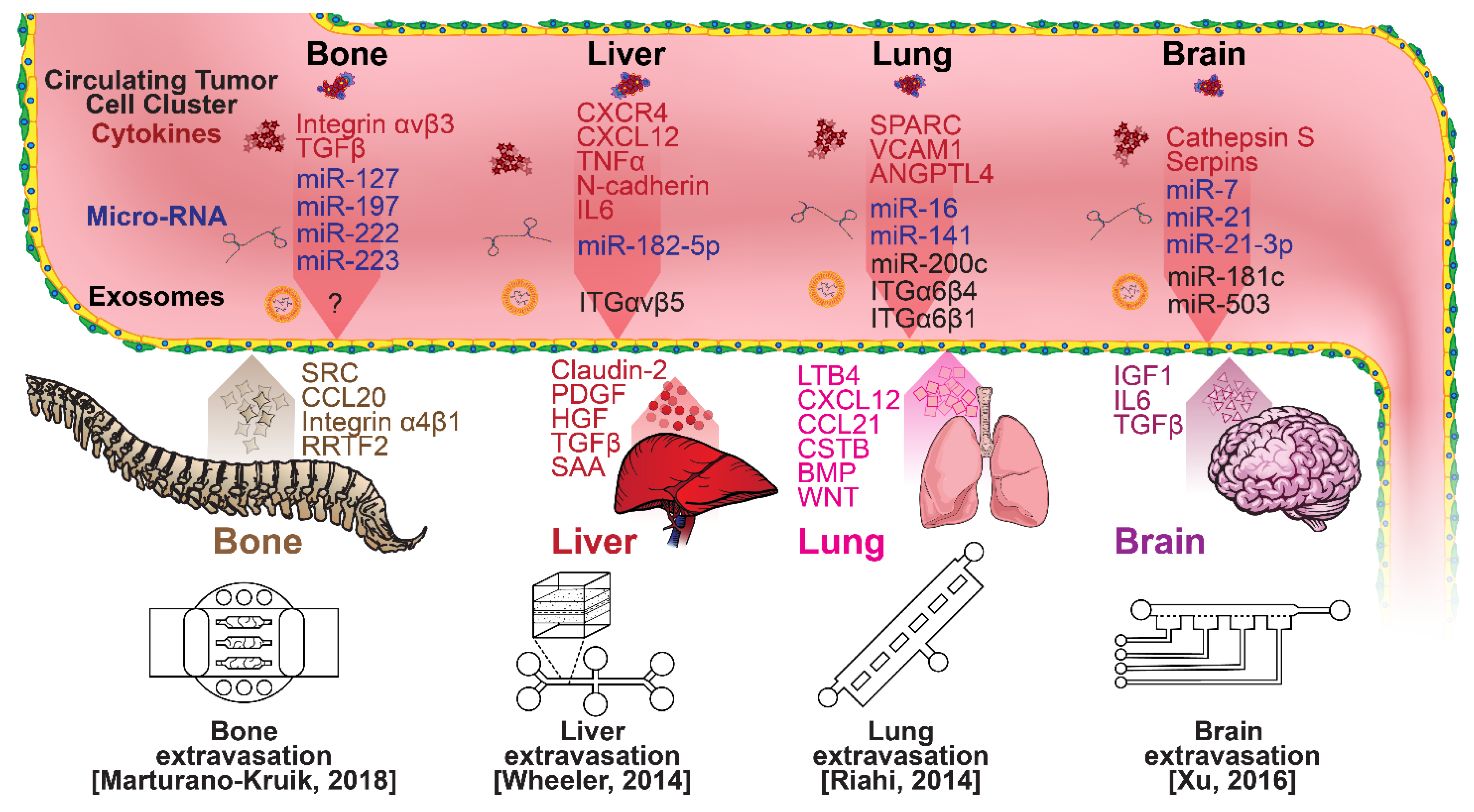
2.3. Extravasation Modeling
3. Detection Techniques of Breast Cancer
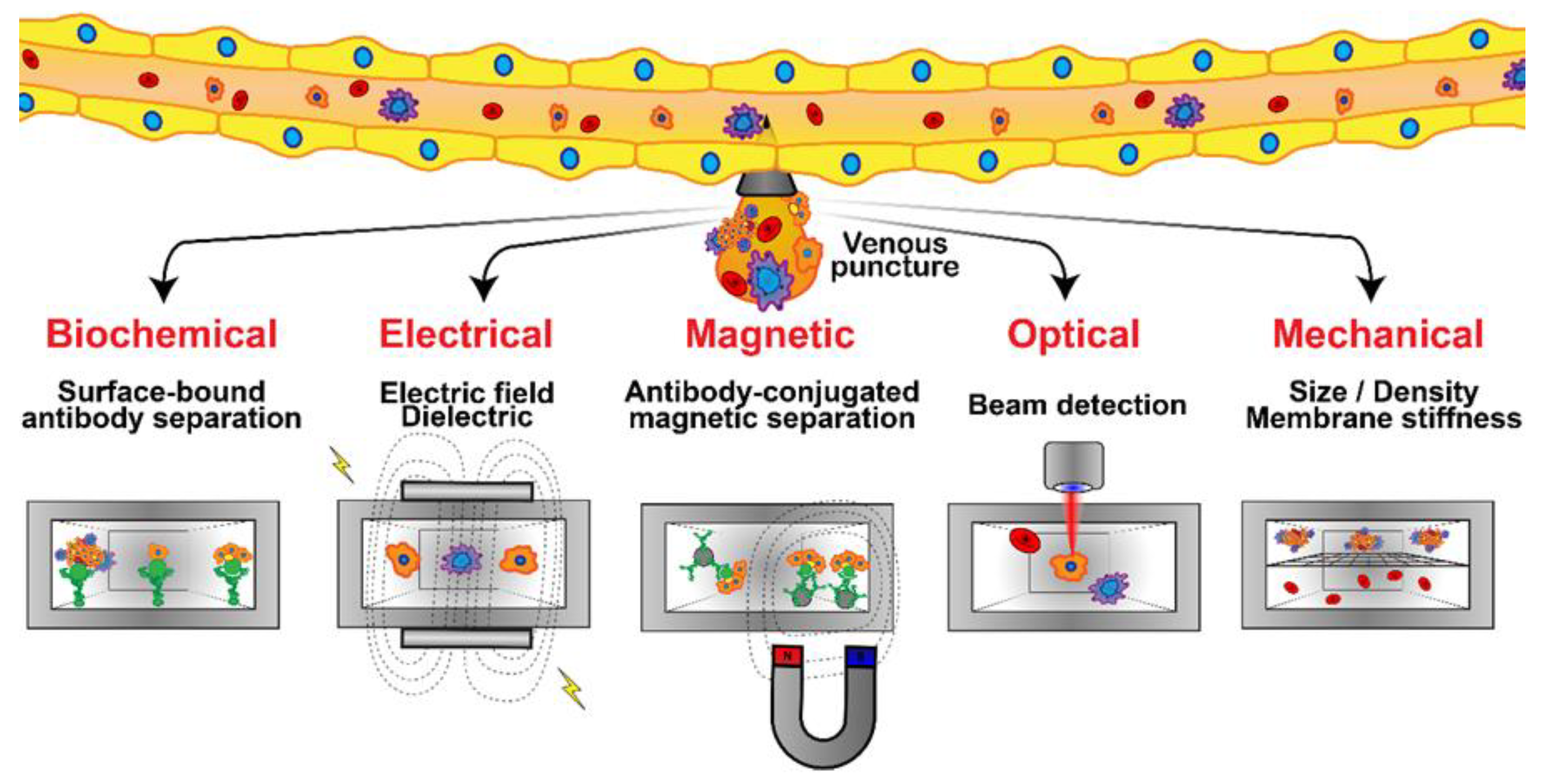
3.1. Detection of Breast Cancer CTCs
3.2. Detection of Breast Cancer Biomarkers
4. Breast Cancer Dormancy
4.1. Quiescence in Breast Cancer
4.2. Microfluidics for Quiescence Research
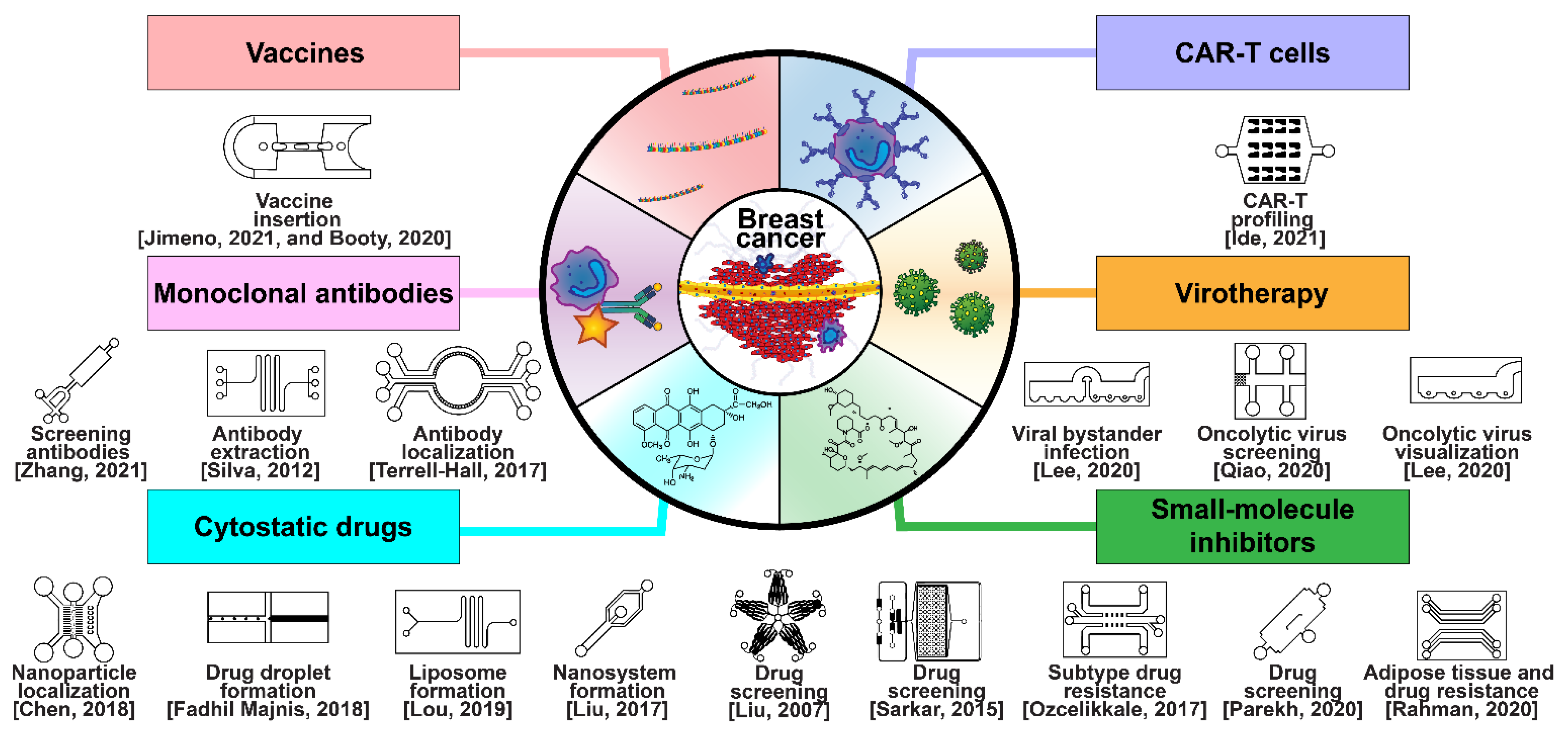
5. Breast Cancer Therapeutic Development
5.1. Drug Development and Delivery
5.2. Cancer Resistance to Treatment
6. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Thomas, R.S.; Black, M.B.; Li, L.; Healy, E.; Chu, T.-M.; Bao, W.; Andersen, M.E.; Wolfinger, R.D. A Comprehensive Statistical Analysis of Predicting In Vivo Hazard Using High-Throughput In Vitro Screening. Toxicol. Sci. 2012, 128, 398–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selimović, Š.; Dokmeci, M.R.; Khademhosseini, A. Organs-on-a-chip for drug discovery. Curr. Opin. Pharmacol. 2013, 13, 829–833. [Google Scholar] [CrossRef] [PubMed]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swartz, M.A.; Fleury, M.E. Interstitial Flow and Its Effects in Soft Tissues. Annu. Rev. Biomed. Eng. 2007, 9, 229–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holen, I.; Speirs, V.; Morrissey, B.; Blyth, K. In vivo models in breast cancer research: Progress, challenges and future directions. Dis. Model. Mech. 2017, 10, 359–371. [Google Scholar] [CrossRef] [Green Version]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Azamjah, N.; Soltan-Zadeh, Y.; Zayeri, F. Global Trend of Breast Cancer Mortality Rate: A 25-Year Study. Asian Pac. J. Cancer Prev. 2019, 20, 2015–2020. [Google Scholar] [CrossRef]
- Dong, M.; Cioffi, G.; Wang, J.; Waite, K.; Ostrom, Q.; Kruchko, C.; Lathia, J.; Rubin, J.; Berens, M.; Connor, J.; et al. Sex Differences in Cancer Incidence and Survival: A Pan-Cancer Analysis. Cancer Epidemiol. Biomark. Prev. 2020, 29, 1389–1397. [Google Scholar] [CrossRef]
- Franzen, N.; van Harten, W.H.; Retèl, V.P.; Loskill, P.; van den Eijnden-van Raaij, J.; IJzerman, M. Impact of organ-on-a-chip technology on pharmaceutical R&D costs. Drug Discov. Today 2019, 24, 1720–1724. [Google Scholar] [CrossRef]
- Chen, W.; Dong, J.; Haiech, J.; Kilhoffer, M.-C.; Zeniou, M. Cancer Stem Cell Quiescence and Plasticity as Major Challenges in Cancer Therapy. Stem Cells Int. 2016, 2016, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Strilic, B.; Offermanns, S. Intravascular Survival and Extravasation of Tumor Cells. Cancer Cell 2017, 32, 282–293. [Google Scholar] [CrossRef] [Green Version]
- Disibio, G.; French, S.W. Metastatic Patterns of Cancers: Results From a Large Autopsy Study. Arch. Pathol. Lab. Med. 2008, 132, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Soni, A.; Ren, Z.; Hameed, O.; Chanda, D.; Morgan, C.J.; Siegal, G.P.; Wei, S. Breast Cancer Subtypes Predispose the Site of Distant Metastases. Am. J. Clin. Pathol. 2015, 143, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, I.; Gupta, N.; Faizee, F.; Khare, V.M.; Tiwari, A.K.; Tang, Y. Biomimetic Microfluidic Platforms for the Assessment of Breast Cancer Metastasis. Front. Bioeng. Biotechnol. 2021, 9, 633671. [Google Scholar] [CrossRef] [PubMed]
- Krol, I.; Schwab, F.D.; Carbone, R.; Ritter, M.; Picocci, S.; De Marni, M.L.; Stepien, G.; Franchi, G.M.; Zanardi, A.; Rissoglio, M.D.; et al. Detection of clustered circulating tumour cells in early breast cancer. Br. J. Cancer 2021, 125, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Schirrmacher, V. From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment (Review). Int. J. Oncol. 2019, 13. [Google Scholar] [CrossRef]
- Saeki, K.; Chang, G.; Kanaya, N.; Wu, X.; Wang, J.; Bernal, L.; Ha, D.; Neuhausen, S.L.; Chen, S. Mammary cell gene expression atlas links epithelial cell remodeling events to breast carcinogenesis. Commun. Biol. 2021, 4, 660. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-H.; Wang, C.-H.; Hsu, K.-F.; Lee, G.-B. An Integrated Microfluidic Platform for Detecting BRCA1/BRCA2 Gene Mutation and Risk Assessment of Ovarian Cancer. In Proceedings of the 2021 21st International Conference on Solid-State Sensors, Actuators and Microsystems (Transducers), Orlando, FL, USA, 20–24 June 2021; pp. 1024–1027. [Google Scholar] [CrossRef]
- Pradhan, S.; Smith, A.M.; Garson, C.J.; Hassani, I.; Seeto, W.J.; Pant, K.; Arnold, R.D.; Prabhakarpandian, B.; Lipke, E.A. A Microvascularized Tumor-mimetic Platform for Assessing Anti-cancer Drug Efficacy. Sci. Rep. 2018, 8, 3171. [Google Scholar] [CrossRef]
- Ayuso, J.M.; Gillette, A.; Lugo-Cintrón, K.; Acevedo-Acevedo, S.; Gomez, I.; Morgan, M.; Heaster, T.; Wisinski, K.B.; Palecek, S.P.; Skala, M.C.; et al. Organotypic microfluidic breast cancer model reveals starvation-induced spatial-temporal metabolic adaptations. EBioMedicine 2018, 37, 144–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funamoto, K.; Zervantonakis, I.K.; Liu, Y.; Ochs, C.J.; Kim, C.; Kamm, R.D. A novel microfluidic platform for high-resolution imaging of a three-dimensional cell culture under a controlled hypoxic environment. Lab. Chip 2012, 12, 4855. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Soroush, F.; Sheffield, J.B.; Wang, B.; Prabhakarpandian, B.; Kiani, M.F. A Biomimetic Microfluidic Tumor Microenvironment Platform Mimicking the EPR Effect for Rapid Screening of Drug Delivery Systems. Sci. Rep. 2017, 7, 9359. [Google Scholar] [CrossRef] [PubMed]
- Nashimoto, Y.; Okada, R.; Hanada, S.; Arima, Y.; Nishiyama, K.; Miura, T.; Yokokawa, R. Vascularized cancer on a chip: The effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials 2020, 229, 119547. [Google Scholar] [CrossRef] [PubMed]
- Viale, G.; Regan, M.M.; Maiorano, E.; Mastropasqua, M.G.; Dell’Orto, P.; Rasmussen, B.B.; Raffoul, J.; Neven, P.; Orosz, Z.; Braye, S.; et al. Prognostic and Predictive Value of Centrally Reviewed Expression of Estrogen and Progesterone Receptors in a Randomized Trial Comparing Letrozole and Tamoxifen Adjuvant Therapy for Postmenopausal Early Breast Cancer: BIG 1-98. J. Clin. Oncol. 2007, 25, 3846–3852. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wu, J.; Ling, R.; Li, N. Quadruple negative breast cancer. Breast Cancer 2020, 27, 527–533. [Google Scholar] [CrossRef]
- Siddharth, S.; Sharma, D. Racial Disparity and Triple-Negative Breast Cancer in African-American Women: A Multifaceted Affair between Obesity, Biology, and Socioeconomic Determinants. Cancers 2018, 10, 514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwamoto, T.; Booser, D.; Valero, V.; Murray, J.L.; Koenig, K.; Esteva, F.J.; Ueno, N.T.; Zhang, J.; Shi, W.; Qi, Y.; et al. Estrogen Receptor (ER) mRNA and ER-Related Gene Expression in Breast Cancers That Are 1% to 10% ER-Positive by Immunohistochemistry. J. Clin. Oncol. 2012, 30, 729–734. [Google Scholar] [CrossRef]
- Ross, D.S.; Zehir, A.; Brogi, E.; Konno, F.; Krystel-Whittemore, M.; Edelweiss, M.; Berger, M.F.; Toy, W.; Chandarlapaty, S.; Razavi, P.; et al. Immunohistochemical analysis of estrogen receptor in breast cancer with ESR1 mutations detected by hybrid capture-based next-generation sequencing. Mod. Pathol. 2019, 32, 81–87. [Google Scholar] [CrossRef]
- Kumar, M.; Sahu, R.K.; Goyal, A.; Sharma, S.; Kaur, N.; Mehrotra, R.; Singh, U.R.; Hedau, S. BRCA1 Promoter Methylation and Expression-Associations with ER+, PR+ and HER2+ Subtypes of Breast Carcinoma. Asian Pac. J. Cancer Prev. 2017, 18, 3293. [Google Scholar] [CrossRef]
- Verdu, M.; Trias, I.; Roman, R.; Rodon, N.; Pubill, C.; Arraiza, N.; Martinez, B.; Garcia-Pelaez, B.; Serrano, T.; Puig, X. Cross-reactivity of EGFR Mutation-specific Immunohistochemistry Assay in HER2-positive Tumors. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 565–570. [Google Scholar] [CrossRef]
- Uliana, C.V.; Peverari, C.R.; Afonso, A.S.; Cominetti, M.R.; Faria, R.C. Fully disposable microfluidic electrochemical device for detection of estrogen receptor alpha breast cancer biomarker. Biosens. Bioelectron. 2018, 99, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, C.; Mao, X.; Brown-Clay, J.; Moreau, F.; Al Absi, A.; Wurzer, H.; Sousa, B.; Schmitt, F.; Berchem, G.; Janji, B.; et al. Hypoxia promotes breast cancer cell invasion through HIF-1α-mediated up-regulation of the invadopodial actin bundling protein CSRP2. Nat. Sci. Rep. 2018, 8, 10191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marín-Hernández, Á.; Gallardo-Pérez, J.C.; Hernández-Reséndiz, I.; Del Mazo-Monsalvo, I.; Robledo-Cadena, D.X.; Moreno-Sánchez, R.; Rodríguez-Enríquez, S. Hypoglycemia Enhances Epithelial-Mesenchymal Transition and Invasiveness, and Restrains the Warburg Phenotype, in Hypoxic HeLa Cell Cultures and Microspheroids: Hypoglycemia Restrains the Warburg Phenotype in Hypoxic Cancer Cells. J. Cell. Physiol. 2017, 232, 1346–1359. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, B.P. Epithelial-mesenchymal transition in breast cancer progression and metastasis. Chin. J. Cancer 2011, 30, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Padmanaban, V.; Krol, I.; Suhail, Y.; Szczerba, B.M.; Aceto, N.; Bader, J.S.; Ewald, A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019, 573, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-J.; Le Berre, M.; Lautenschlaeger, F.; Maiuri, P.; Callan-Jones, A.; Heuzé, M.; Takaki, T.; Voituriez, R.; Piel, M. Confinement and Low Adhesion Induce Fast Amoeboid Migration of Slow Mesenchymal Cells. Cell 2015, 160, 659–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Jiang, J.; Chen, B.; Wang, K.; Tang, Y.; Liang, X. Plasticity of cancer cell invasion: Patterns and mechanisms. Transl. Oncol. 2021, 14, 100899. [Google Scholar] [CrossRef]
- Estrella, V.; Chen, T.; Lloyd, M.; Wojtkowiak, J.; Cornnell, H.H.; Ibrahim-Hashim, A.; Bailey, K.; Balagurunathan, Y.; Rothberg, J.M.; Sloane, B.F.; et al. Acidity Generated by the Tumor Microenvironment Drives Local Invasion. Cancer Res. 2013, 73, 1524–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanley, C.J.; Henriet, E.; Sirka, O.K.; Thomas, G.J.; Ewald, A.J. Tumor-Resident Stromal Cells Promote Breast Cancer Invasion through Regulation of the Basal Phenotype. Mol. Cancer Res. 2020, 18, 1615–1622. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qiu, Z.; Li, F.; Wang, C. The relationship between MMP-2 and MMP-9 expression levels with breast cancer incidence and prognosis. Oncol. Lett. 2017, 14, 5865–5870. [Google Scholar] [CrossRef] [Green Version]
- Moon, H.; Ospina-Muñoz, N.; Noe-Kim, V.; Yang, Y.; Elzey, B.D.; Konieczny, S.F.; Han, B. Subtype-specific characterization of breast cancer invasion using a microfluidic tumor platform. PLoS ONE 2020, 15, e0234012. [Google Scholar] [CrossRef]
- Aw Yong, K.M.; Ulintz, P.J.; Caceres, S.; Cheng, X.; Bao, L.; Wu, Z.; Jiagge, E.M.; Merajver, S.D. Heterogeneity at the invasion front of triple negative breast cancer cells. Sci. Rep. 2020, 10, 5781. [Google Scholar] [CrossRef] [Green Version]
- Sung, K.E.; Yang, N.; Pehlke, C.; Keely, P.J.; Eliceiri, K.W.; Friedl, A.; Beebe, D.J. Transition to invasion in breast cancer: A microfluidic in vitro model enables examination of spatial and temporal effects. Integr Biol 2011, 3, 439–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, S.F.; Bishop, K.W.; Mintz, R.; Fang, L.; Achilefu, S. Calcium carbonate nanoparticles stimulate cancer cell reprogramming to suppress tumor growth and invasion in an organ-on-a-chip system. Sci. Rep. 2021, 11, 9246. [Google Scholar] [CrossRef]
- Toh, Y.-C.; Raja, A.; Yu, H.; van Noort, D. A 3D Microfluidic Model to Recapitulate Cancer Cell Migration and Invasion. Bioengineering 2018, 5, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yankaskas, C.L.; Thompson, K.N.; Paul, C.D.; Vitolo, M.I.; Mistriotis, P.; Mahendra, A.; Bajpai, V.K.; Shea, D.J.; Manto, K.M.; Chai, A.C.; et al. A microfluidic assay for the quantification of the metastatic propensity of breast cancer specimens. Nat. Biomed. Eng. 2019, 3, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Gioiella, F.; Urciuolo, F.; Imparato, G.; Brancato, V.; Netti, P.A. An Engineered Breast Cancer Model on a Chip to Replicate ECM-Activation In Vitro during Tumor Progression. Adv. Healthc. Mater. 2016, 5, 3074–3084. [Google Scholar] [CrossRef] [PubMed]
- Mosadegh, B.; Lockett, M.R.; Minn, K.T.; Simon, K.A.; Gilbert, K.; Hillier, S.; Newsome, D.; Li, H.; Hall, A.B.; Boucher, D.M.; et al. A paper-based invasion assay: Assessing chemotaxis of cancer cells in gradients of oxygen. Biomaterials 2015, 52, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Liu, Z.; Du, Z.; Yi, X.; Sun, W. Three-dimensional microfluidic tumor-macrophage system for breast cancer cell invasion. Biotechnol. Bioeng. 2019, 116, 1731–1741. [Google Scholar] [CrossRef]
- Lugo-Cintrón, K.M.; Gong, M.M.; Ayuso, J.M.; Tomko, L.A.; Beebe, D.J.; Virumbrales-Muñoz, M.; Ponik, S.M. Breast Fibroblasts and ECM Components Modulate Breast Cancer Cell Migration through the Secretion of MMPs in a 3D Microfluidic Co-Culture Model. Cancers 2020, 12, 1173. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Han, X.; Chen, R.; Zhang, K.; Li, Y.; Fruge, S.; Jang, J.h.; Ma, Y.; Qin, L. Microfluidic Mapping of Cancer Cell–Protein Binding Interaction. ACS Appl. Mater. Interfaces 2017, 9, 22143–22148. [Google Scholar] [CrossRef]
- Han, W.; Chen, S.; Yuan, W.; Fan, Q.; Tian, J.; Wang, X.; Chen, L.; Zhang, X.; Wei, W.; Liu, R.; et al. Oriented collagen fibers direct tumor cell intravasation. Proc. Natl. Acad. Sci. USA 2016, 113, 11208–11213. [Google Scholar] [CrossRef] [Green Version]
- Ginter, P.S.; Karagiannis, G.S.; Entenberg, D.; Lin, Y.; Condeelis, J.; Jones, J.; Oktay, M.H. Tumor Microenvironment of Metastasis (TMEM) Doorways Are Restricted to the Blood Vessel Endothelium in Both Primary Breast Cancers and Their Lymph Node Metastases. Cancers 2019, 11, 1507. [Google Scholar] [CrossRef] [Green Version]
- Vestweber, D.; Winderlich, M.; Cagna, G.; Nottebaum, A.F. Cell adhesion dynamics at endothelial junctions: VE-cadherin as a major player. Trends Cell Biol. 2009, 19, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, L.R.; Borriello, L.; Entenberg, D.; Condeelis, J.S.; Oktay, M.H.; Karagiannis, G.S. The emerging roles of macrophages in cancer metastasis and response to chemotherapy. J. Leukoc. Biol. 2019, 106, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Deng, Y.; Im, J.H.; Muschel, R.J.; Zou, Y.; Li, J.; Lang, R.A.; Pollard, J.W. A Distinct Macrophage Population Mediates Metastatic Breast Cancer Cell Extravasation, Establishment and Growth. PLoS ONE 2009, 4, e6562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linde, N.; Casanova-Acebes, M.; Sosa, M.S.; Mortha, A.; Rahman, A.; Farias, E.; Harper, K.; Tardio, E.; Reyes Torres, I.; Jones, J.; et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat. Commun. 2018, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Oshi, M.; Tokumaru, Y.; Asaoka, M.; Yan, L.; Satyananda, V.; Matsuyama, R.; Matsuhashi, N.; Futamura, M.; Ishikawa, T.; Yoshida, K.; et al. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci. Rep. 2020, 10, 16554. [Google Scholar] [CrossRef] [PubMed]
- Arwert, E.N.; Harney, A.S.; Entenberg, D.; Wang, Y.; Sahai, E.; Pollard, J.W.; Condeelis, J.S. A Unidirectional Transition from Migratory to Perivascular Macrophage Is Required for Tumor Cell Intravasation. Cell Rep. 2018, 23, 1239–1248. [Google Scholar] [CrossRef] [Green Version]
- Munir, M.T.; Kay, M.K.; Kang, M.H.; Rahman, M.M.; Al-Harrasi, A.; Choudhury, M.; Moustaid-Moussa, N.; Hussain, F.; Rahman, S.M. Tumor-Associated Macrophages as Multifaceted Regulators of Breast Tumor Growth. Int. J. Mol. Sci. 2021, 22, 6526. [Google Scholar] [CrossRef]
- Sainson, R.C.A.; Johnston, D.A.; Chu, H.C.; Holderfield, M.T.; Nakatsu, M.N.; Crampton, S.P.; Davis, J.; Conn, E.; Hughes, C.C.W. TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype. Blood 2008, 111, 4997–5007. [Google Scholar] [CrossRef]
- Yvette Drabsch & Peter ten Dijk TGF-β Signaling in Breast Cancer Cell Invasion and Bone Metastasis. J Mammary Gland Biol Neoplasia 2011, 16, 97–101. [CrossRef] [Green Version]
- Rodriguez-Vita, J.; Fischer, A. Notch signaling facilitates crossing of endothelial barriers by tumor cells. Mol. Cell. Oncol. 2017, 4, e1311828. [Google Scholar] [CrossRef] [Green Version]
- Lopes-Bastos, B.M.; Jiang, W.G.; Cai, J. Tumour–Endothelial Cell Communications: Important and Indispensable Mediators of Tumour Angiogenesis. Anticancer Res. 2016, 36, 1119–1126. [Google Scholar] [PubMed]
- Di Modica, M.; Regondi, V.; Sandri, M.; Iorio, M.V.; Zanetti, A.; Tagliabue, E.; Casalini, P.; Triulzi, T. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017, 384, 94–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanchan, R.K.; Siddiqui, J.A.; Mahapatra, S.; Batra, S.K.; Nasser, M.W. microRNAs Orchestrate Pathophysiology of Breast Cancer Brain Metastasis: Advances in Therapy. Mol. Cancer 2020, 19, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balzer, E.M.; Tong, Z.; Paul, C.D.; Hung, W.; Stroka, K.M.; Boggs, A.E.; Martin, S.S.; Konstantopoulos, K. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012, 26, 4045–4056. [Google Scholar] [CrossRef] [Green Version]
- Zervantonakis, I.K.; Hughes-Alford, S.K.; Charest, J.L.; Condeelis, J.S.; Gertler, F.B.; Kamm, R.D. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. USA 2012, 109, 13515–13520. [Google Scholar] [CrossRef] [Green Version]
- Buchanan, C.F.; Voigt, E.E.; Szot, C.S.; Freeman, J.W.; Vlachos, P.P.; Rylander, M.N. Three-Dimensional Microfluidic Collagen Hydrogels for Investigating Flow-Mediated Tumor-Endothelial Signaling and Vascular Organization. Tissue Eng. Part C Methods 2014, 20, 64–75. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Hyun, E.; Seo, J.; Blundell, C.; Kim, H.C.; Lee, E.; Lee, S.H.; Moon, A.; Moon, W.K.; Huh, D. A microengineered pathophysiological model of early-stage breast cancer. Lab. Chip 2015, 15, 3350–3357. [Google Scholar] [CrossRef]
- Mi, S.; Du, Z.; Xu, Y.; Wu, Z.; Qian, X.; Zhang, M.; Sun, W. Microfluidic co-culture system for cancer migratory analysis and anti-metastatic drugs screening. Sci. Rep. 2016, 6, 35544. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Guo, W.; Sun, Y.; Sun, B.; Hu, S.; Sun, D.; Lam, R.H.W. A microfluidic device for isolation and characterization of transendothelial migrating cancer cells. Biomicrofluidics 2017, 11, 014105. [Google Scholar] [CrossRef] [Green Version]
- Nagaraju, S.; Truong, D.; Mouneimne, G.; Nikkhah, M. Microfluidic Tumor–Vascular Model to Study Breast Cancer Cell Invasion and Intravasation. Adv. Healthc. Mater. 2018, 7, 1701257. [Google Scholar] [CrossRef] [PubMed]
- Shirure, V.S.; Bi, Y.; Curtis, M.B.; Lezia, A.; Goedegebuure, M.M.; Goedegebuure, S.P.; Aft, R.; Fields, R.C.; George, S.C. Tumor-on-a-chip platform to investigate progression and drug sensitivity in cell lines and patient-derived organoids. Lab. Chip 2018, 18, 3687–3702. [Google Scholar] [CrossRef]
- Cho, H.-Y.; Choi, J.-H.; Kim, K.-J.; Shin, M.; Choi, J.-W. Microfluidic System to Analyze the Effects of Interleukin 6 on Lymphatic Breast Cancer Metastasis. Front. Bioeng. Biotechnol. 2021, 8, 611802. [Google Scholar] [CrossRef]
- Leone, K.; Poggiana, C.; Zamarchi, R. The Interplay between Circulating Tumor Cells and the Immune System: From Immune Escape to Cancer Immunotherapy. Diagnostics 2018, 8, 59. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wang, J.; Kong, X.; Li, E.; Liu, Y.; Du, X.; Kang, Z.; Tang, Y.; Kuang, Y.; Yang, Z.; et al. CD47 Promotes Tumor Invasion and Metastasis in Non-small Cell Lung Cancer. Sci. Rep. 2016, 6, 29719. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, Q.; Liu, Q.; Wang, C.; Yao, R.; Wang, Y. CTC immune escape mediated by PD-L1. Med. Hypotheses 2016, 93, 138–139. [Google Scholar] [CrossRef] [PubMed]
- Twomey, J.; Zhang, B. Circulating Tumor Cells Develop Resistance to TRAIL-Induced Apoptosis Through Autophagic Removal of Death Receptor 5: Evidence from an In Vitro Model. Cancers 2019, 11, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, H.; Sung, J.Y.; Kim, S.-H.; Yun, U.-J.; Kim, H.; Jang, E.-J.; Yoo, H.-E.; Hong, E.K.; Goh, S.-H.; Moon, A.; et al. Fibronectin regulates anoikis resistance via cell aggregate formation. Cancer Lett. 2021, 508, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.J.; King, M.R. Fluid shear stress sensitizes cancer cells to receptor-mediated apoptosis via trimeric death receptors. New J. Phys. 2013, 15, 015008. [Google Scholar] [CrossRef] [Green Version]
- Ward, M.P.; Kane, E.L.; Norris, A.L.; Mohamed, B.M.; Kelly, T.; Bates, M.; Clarke, A.; Brady, N.; Martin, C.M.; Brooks, R.D.; et al. Platelets, immune cells and the coagulation cascade; friend or foe of the circulating tumour cell? Mol. Cancer 2021, 20, 59. [Google Scholar] [CrossRef]
- Liu, X.; Taftaf, R.; Kawaguchi, M.; Chang, Y.-F.; Chen, W.; Entenberg, D.; Zhang, Y.; Gerratana, L.; Huang, S.; Patel, D.B.; et al. Homophilic CD44 Interactions Mediate Tumor Cell Aggregation and Polyclonal Metastasis in Patient-Derived Breast Cancer Models. Cancer Discov. 2019, 9, 96–113. [Google Scholar] [CrossRef] [Green Version]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef]
- Hess, K.R.; Varadhachary, G.R.; Taylor, S.H.; Wei, W.; Raber, M.N.; Lenzi, R.; Abbruzzese, J.L. Metastatic patterns in adenocarcinoma. Cancer 2006, 106, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.L.; Abu Jalboush, S.; Lo, H.-W. Exosomal MicroRNAs and Organotropism in Breast Cancer Metastasis. Cancers 2020, 12, 1827. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, J.; Zhu, S.; Wu, J.; Chen, C.; Liu, Q.; Wei, W.; Zhang, Y.; Sun, S. Breast cancer subtypes predict the preferential site of distant metastases: A SEER based study. Oncotarget 2017, 8, 27990–27996. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Wang, C.; Xie, M.; Zhu, C.; Shu, Y.; Tang, J.; Guan, X. Heterogeneity of CTC contributes to the organotropism of breast cancer. Biomed. Pharmacother. 2021, 137, 111314. [Google Scholar] [CrossRef] [PubMed]
- Padua, D.; Zhang, X.H.-F.; Wang, Q.; Nadal, C.; Gerald, W.L.; Gomis, R.R.; Massagué, J. TGFβ Primes Breast Tumors for Lung Metastasis Seeding through Angiopoietin-like 4. Cell 2008, 133, 66–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.H.-F.; Wang, Q.; Gerald, W.; Hudis, C.A.; Norton, L.; Smid, M.; Foekens, J.A.; Massagué, J. Latent Bone Metastasis in Breast Cancer Tied to Src-Dependent Survival Signals. Cancer Cell 2009, 16, 67–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoneda, T.; Williams, P.J.; Hiraga, T.; Niewolna, M.; Nishimura, R. A Bone-Seeking Clone Exhibits Different Biological Properties from the MDA-MB-231 Parental Human Breast Cancer Cells and a Brain-Seeking Clone In Vivo and In Vitro. J. Bone Miner. Res. 2001, 16, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osmani, N.; Follain, G.; García León, M.J.; Lefebvre, O.; Busnelli, I.; Larnicol, A.; Harlepp, S.; Goetz, J.G. Metastatic Tumor Cells Exploit Their Adhesion Repertoire to Counteract Shear Forces during Intravascular Arrest. Cell Rep. 2019, 28, 2491–2500. [Google Scholar] [CrossRef] [PubMed]
- Yadavalli, S.; Jayaram, S.; Manda, S.S.; Madugundu, A.K.; Nayakanti, D.S.; Tan, T.Z.; Bhat, R.; Rangarajan, A.; Chatterjee, A.; Gowda, H.; et al. Data-Driven Discovery of Extravasation Pathway in Circulating Tumor Cells. Sci. Rep. 2017, 7, 43710. [Google Scholar] [CrossRef] [Green Version]
- Sang A Park and Young-Min Hyuncorresponding author Neutrophil Extravasation Cascade: What Can We Learn from Two-photon Intravital Imaging? Immune Netw. 2016, 16, 317–321. [CrossRef] [PubMed] [Green Version]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.H.-F.; Jin, X.; Malladi, S.; Zou, Y.; Wen, Y.H.; Brogi, E.; Smid, M.; Foekens, J.A.; Massagué, J. Selection of Bone Metastasis Seeds by Mesenchymal Signals in the Primary Tumor Stroma. Cell 2013, 154, 1060–1073. [Google Scholar] [CrossRef] [Green Version]
- Hosaka, K.; Yang, Y.; Seki, T.; Fischer, C.; Dubey, O.; Fredlund, E.; Hartman, J.; Religa, P.; Morikawa, H.; Ishii, Y.; et al. Pericyte–fibroblast transition promotes tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, E5618–E5627. [Google Scholar] [CrossRef] [Green Version]
- Lugassy, C.; Kleinman, H.; Barnhill, R. Pericyte mimicry: An embryogenesis-derived program of extravascular tumor cell migration. In Tumor Vascularization; Elsevier: Amsterdam, The Netherlands, 2020; pp. 49–88. ISBN 978-0-12-819494-2. [Google Scholar]
- Hurtado, P.; Martínez-Pena, I.; Piñeiro, R. Dangerous Liaisons: Circulating Tumor Cells (CTCs) and Cancer-Associated Fibroblasts (CAFs). Cancers 2020, 12, 2861. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, W.; Lu, D.; Wu, Z.; Duan, H.; Luo, Y.; Feng, J.; Yang, D.; Fu, L.; Yan, X. CD146, an epithelial-mesenchymal transition inducer, is associated with triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 1127–1132. [Google Scholar] [CrossRef] [Green Version]
- Mostert, B.; Kraan, J.; Bolt-de Vries, J.; van der Spoel, P.; Sieuwerts, A.M.; Schutte, M.; Timmermans, A.M.; Foekens, R.; Martens, J.W.M.; Gratama, J.-W.; et al. Detection of circulating tumor cells in breast cancer may improve through enrichment with anti-CD146. Breast Cancer Res. Treat. 2011, 127, 33–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imbert, A.-M.; Garulli, C.; Choquet, E.; Koubi, M.; Aurrand-Lions, M.; Chabannon, C. CD146 Expression in Human Breast Cancer Cell Lines Induces Phenotypic and Functional Changes Observed in Epithelial to Mesenchymal Transition. PLoS ONE 2012, 7, e43752. [Google Scholar] [CrossRef]
- Mayo, V.; Bowles, A.; Wubker, L.; Ortiz, I.; Cordoves, A.; Cote, R.; Correa, D.; Agarwal, A. Human-derived osteoblast-like cells and pericyte-like cells induce distinct metastatic phenotypes in primary breast cancer cells. Exp. Biol. Med. 2021, 246, 971–985. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; LaFortune, T.A.; Krishnamurthy, S.; Esteva, F.J.; Cristofanilli, M.; Liu, P.; Lucci, A.; Singh, B.; Hung, M.-C.; Hortobagyi, G.N.; et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Reverses Mesenchymal to Epithelial Phenotype and Inhibits Metastasis in Inflammatory Breast Cancer. Clin. Cancer Res. 2009, 15, 6639–6648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.; Chen, A.; He, W.; Li, Z.; Zhang, G.; Liu, Z.; Liu, G.; Liu, X.; He, S.; Xiao, G.; et al. BMP-2 induces EMT and breast cancer stemness through Rb and CD44. Cell Death Discov. 2017, 3, 17039. [Google Scholar] [CrossRef]
- Armbrecht, L.; Rutschmann, O.; Szczerba, B.M.; Nikoloff, J.; Aceto, N.; Dittrich, P.S. Quantification of Protein Secretion from Circulating Tumor Cells in Microfluidic Chambers. Adv. Sci. 2020, 7, 1903237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Park, S.; Hyun, K.A.; Jung, H.-I. Microfluidic recapitulation of circulating tumor cell–neutrophil clusters via double spiral channel-induced deterministic encapsulation. Lab. Chip 2021, 21, 3483–3497. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.E.; Clark, A.M.; Taylor, D.P.; Young, C.L.; Pillai, V.C.; Stolz, D.B.; Venkataramanan, R.; Lauffenburger, D.; Griffith, L.; Wells, A. Spontaneous dormancy of metastatic breast cancer cells in an all human liver microphysiologic system. Br. J. Cancer 2014, 111, 2342–2350. [Google Scholar] [CrossRef] [PubMed]
- Riahi, R.; Yang, Y.L.; Kim, H.; Jiang, L.; Wong, P.K.; Zohar, Y. A microfluidic model for organ-specific extravasation of circulating tumor cells. Biomicrofluidics 2014, 8, 024103. [Google Scholar] [CrossRef] [Green Version]
- Aleman, J.; Skardal, A. A multi-site metastasis-on-a-chip microphysiological system for assessing metastatic preference of cancer cells: ALEMAN AND SKARDAL. Biotechnol. Bioeng. 2019, 116, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Miermont, A.; Lim, C.T.; Kamm, R.D. A 3D microvascular network model to study the impact of hypoxia on the extravasation potential of breast cell lines. Sci. Rep. 2018, 8, 17949. [Google Scholar] [CrossRef] [PubMed]
- Marturano-Kruik, A.; Nava, M.M.; Yeager, K.; Chramiec, A.; Hao, L.; Robinson, S.; Guo, E.; Raimondi, M.T.; Vunjak-Novakovic, G. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl. Acad. Sci. USA 2018, 115, 1256–1261. [Google Scholar] [CrossRef] [Green Version]
- Jeon, J.S.; Bersini, S.; Gilardi, M.; Dubini, G.; Charest, J.L.; Moretti, M.; Kamm, R.D. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. USA 2015, 112, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Bersini, S.; Jeon, J.S.; Dubini, G.; Arrigoni, C.; Chung, S.; Charest, J.L.; Moretti, M.; Kamm, R.D. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 2014, 35, 2454–2461. [Google Scholar] [CrossRef]
- Mei, X.; Middleton, K.; Shim, D.; Wan, Q.; Xu, L.; Ma, Y.-H.V.; Devadas, D.; Walji, N.; Wang, L.; Young, E.W.K.; et al. Microfluidic platform for studying osteocyte mechanoregulation of breast cancer bone metastasis. Integr. Biol. 2019, 11, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, Z.; Yu, Y.; Sizdahkhani, S.; Ho, W.S.; Yin, F.; Wang, L.; Zhu, G.; Zhang, M.; Jiang, L.; et al. A dynamic in vivo-like organotypic blood-brain barrier model to probe metastatic brain tumors. Sci. Rep. 2016, 6, 36670. [Google Scholar] [CrossRef] [Green Version]
- Wang, L. Early Diagnosis of Breast Cancer. Sensors 2017, 17, 1572. [Google Scholar] [CrossRef]
- Houssami, N.; Ciatto, S.; Martinelli, F.; Bonardi, R.; Duffy, S.W. Early detection of second breast cancers improves prognosis in breast cancer survivors. Ann. Oncol. 2009, 20, 1505–1510. [Google Scholar] [CrossRef] [PubMed]
- Jaglan, P.; Dass, R.; Duhan, M. Breast Cancer Detection Techniques: Issues and Challenges. J. Inst. Eng. India Ser. B 2019, 100, 379–386. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Alimirzaie, S.; Bagherzadeh, M.; Akbari, M.R. Liquid biopsy in breast cancer: A comprehensive review. Clin. Genet. 2019, 95, 643–660. [Google Scholar] [CrossRef]
- Akgönüllü, S.; Bakhshpour, M.; Pişkin, A.K.; Denizli, A. Microfluidic Systems for Cancer Diagnosis and Applications. Micromachines 2021, 12, 1349. [Google Scholar] [CrossRef]
- Guo, B.; Oliver, T.G. Partners in Crime: Neutrophil–CTC Collusion in Metastasis. Trends Immunol. 2019, 40, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; Zhou, J.; Kenny, L.; Papautsky, I.; Punyadeera, C. Capture of Circulating Tumour Cell Clusters Using Straight Microfluidic Chips. Cancers 2019, 11, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duoma, S.; Van Laar, T.; Zevenhoven, J.; Meuwissen, R.; Van Garderen, E.; Peeper, D.S. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TRKB. Nature 2004, 430, 1034–1039. [Google Scholar] [CrossRef]
- Sachdev, D.; Zhang, X.; Matise, I.; Gaillard-Kelly, M.; Yee, D. The type I insulin-like growth factor receptor regulates cancer metastasis independently of primary tumor growth by promoting invasion and survival. Oncogene 2010, 29, 251–262. [Google Scholar] [CrossRef] [Green Version]
- Ghadially, H.; Brown, L.; Lloyd, C.; Lewis, L.; Lewis, A.; Dillon, J.; Sainson, R.; Jovanovic, J.; Tigue, N.J.; Bannister, D.; et al. MHC class I chain-related protein A and B (MICA and MICB) are predominantly expressed intracellularly in tumour and normal tissue. Br. J. Cancer 2017, 116, 1208–1217. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Zhang, X.H.-F.; Massagué, J. Macrophage Binding to Receptor VCAM-1 Transmits Survival Signals in Breast Cancer Cells that Invade the Lungs. Cancer Cell 2011, 20, 538–549. [Google Scholar] [CrossRef] [Green Version]
- Bernson, E.; Christenson, K.; Pesce, S.; Pasanen, M.; Marcenaro, E.; Sivori, S.; Thorén, F.B. Downregulation of HLA Class I Renders Inflammatory Neutrophils More Susceptible to NK Cell-Induced Apoptosis. Front. Immunol. 2019, 10, 2444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmiedel, D.; Mandelboim, O. NKG2D Ligands–Critical Targets for Cancer Immune Escape and Therapy. Front. Immunol. 2018, 9, 2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mammadova-Bach, E.; Mangin, P.; Lanza, F.; Gachet, C. Platelet integrin Alpha6beta1 controls lung metastasis through direct binding to cell-derived ADAM9. JCI Insight 2015, 1, e88245. [Google Scholar] [CrossRef] [Green Version]
- Takagi, S.; Sato, S.; Oh-hara, T.; Takami, M.; Koike, S.; Mishima, Y.; Hatake, K.; Fujita, N. Platelets promote tumor growth and metastasis via direct interaction between Aggrus/podoplanin and CLEC-2. PLoS ONE 2013, 8, e73609. [Google Scholar] [CrossRef] [PubMed]
- Gil-Bernabe, A.M.; Ferjancic, S.; Tlalka, M.; Zhao, L.; Allen, P.D.; Im, J.H.; Watson, K.; Hill, S.A.; Amirkhosravi, A.; Francis, J.L. Recruitment of monocytes/macrophages by tissue factor-mediated coagulation is essential for metastatic cell survival and premetastatic niche establishment in mice. Blood 2012, 119, 3164–3175. [Google Scholar] [CrossRef] [Green Version]
- Furlow, P.W.; Xhang, S.; Soong, T.D.; Halberg, N.; Goodarzi, H.; Mangrum, C.; Wum, Y.G.; Elemento, O.; Tavazoie, S.F. Mechanosensitive Pannexin-1 channels mediate microvascular metastatic cell survival. Nat. Cell Biol. 2015, 17, 943–952. [Google Scholar] [CrossRef] [Green Version]
- Vora, D.H.H.; Patel, N.A.; Rajvik, K.N.; Mehta, S.V.; Brahmbhatt, B.V.; Shah, M.J.; Shukla, S.N.; Shah, P.M. Cytokeratin and Vimentin Expression in Breast Cancer. Int. J. Biol. Markers 2009, 24, 9. [Google Scholar] [CrossRef]
- Gradilone, A.; Raimondi, C.; Nicolazzo, C.; Petracca, A.; Gandini, O.; Vincenzi, B.; Naso, G.; Aglianò, A.M.; Cortesi, E.; Gazzaniga, P. Circulating tumour cells lacking cytokeratin in breast cancer: The importance of being mesenchymal. J. Cell. Mol. Med. 2011, 15, 1066–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi, N.; Oloomi, M.; Orafa, Z. Circulating Tumor Cells Detection in Patients with Early Breast Cancer Using MACS Immunomagnetic Flow Cytometry. Avicenna J. Med. Biotechnol. 2020, 12, 9. [Google Scholar]
- Tarhan, M.O.; Gonel, A.; Kucukzeybek, Y.; Erten, C.; Cuhadar, S.; Yigit, S.C.; Atay, A.; Somali, I.; Dirican, A.; Demir, L.; et al. Prognostic Significance of Circulating Tumor Cells and Serum CA15-3 Levels in Metastatic Breast Cancer, Single Center Experience, Preliminary Results. Asian Pac. J. Cancer Prev. 2013, 14, 1725–1729. [Google Scholar] [CrossRef] [Green Version]
- Charbonneau, H.; Tonks, N.K.; Walsh, K.A.; Fischer, E.H. The leukocyte common antigen (CD45): A putative receptor-linked protein tyrosine phosphatase. Proc. Natl. Acad. Sci. USA 1988, 85, 7182–7186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peeters, D.J.E.; Van den Eynden, G.G.; van Dam, P.-J.; Prové, A.; Benoy, I.H.; van Dam, P.A.; Vermeulen, P.B.; Pauwels, P.; Peeters, M.; Van Laere, S.J.; et al. Circulating tumour cells in the central and the peripheral venous compartment in patients with metastatic breast cancer. Br. J. Cancer 2011, 104, 1472–1477. [Google Scholar] [CrossRef]
- Mendelaar, P.A.J.; Kraan, J.; Van, M.; Zeune, L.L.; Terstappen, L.W.M.M.; Oomen-de Hoop, E.; Martens, J.W.M.; Sleijfer, S. Defining the dimensions of circulating tumor cells in a large series of breast, prostate, colon, and bladder cancer patients. Mol. Oncol. 2021, 15, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wong, K.H.K.; Khankhel, A.H.; Zeinali, M.; Reategui, E.; Phillips, M.J.; Luo, X.; Aceto, N.; Fachin, F.; Hoang, A.N.; et al. Microfluidic isolation of platelet-covered circulating tumor cells. Lab. Chip 2017, 17, 3498–3503. [Google Scholar] [CrossRef] [PubMed]
- Au, S.H.; Edd, J.; Stoddard, A.E.; Wong, K.H.K.; Fachin, F.; Maheswaran, S.; Haber, D.A.; Stott, S.L.; Kapur, R.; Toner, M. Microfluidic isolation of circulating tumor cell clusters by size and asymmetry. Sci. Rep. 2017, 7, 2433. [Google Scholar] [CrossRef] [PubMed]
- Gertler, R.; Rosenberg, R.; Fuehrer, K.; Dahm, M.; Nekarda, H.; Siewert, J.R. Detection of Circulating Tumor Cells in Blood Using an Optimized Density Gradient Centrifugation. In Molecular Staging of Cancer; Allgayer, H., Heiss, M.M., Schildberg, F.W., Eds.; Recent Results in Cancer Research; Springer: Berlin/Heidelberg, Germany, 2003; Volume 162, pp. 149–155. ISBN 978-3-642-63945-6. [Google Scholar]
- Deliorman, M.; Janahi, F.K.; Sukumar, P.; Glia, A.; Alnemari, R.; Fadl, S.; Chen, W.; Qasaimeh, M.A. AFM-compatible microfluidic platform for affinity-based capture and nanomechanical characterization of circulating tumor cells. Microsyst. Nanoeng. 2020, 6, 20. [Google Scholar] [CrossRef] [Green Version]
- Alexandrova, A.; Antonova, N.; Marina, S.; Shamray, E.A.; Cherkashina, O.V. Evaluation of the elastic properties and topography of leukocytes’ surface in patients with type 2 diabetes mellitus using atomic force microscope. Ser. Biomech. 2017, 31, 16–24. [Google Scholar]
- Zhou, Z.L.; Hui, T.H.; Tang, B.; Ngan, A.H.W. Accurate measurement of stiffness of leukemia cells and leukocytes using an optical trap by a rate-jump method. RSC Adv. 2014, 4, 8453. [Google Scholar] [CrossRef] [Green Version]
- Han, S.-I.; Joo, Y.-D.; Han, K.-H. An electrorotation technique for measuring the dielectric properties of cells with simultaneous use of negative quadrupolar dielectrophoresis and electrorotation. The Analyst 2013, 138, 1529. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Broglio, K.R.; Guarneri, V.; Jackson, S.; Fritsche, H.A.; Islam, R.; Dawood, S.; Reuben, J.M.; Kau, S.-W.; Lara, J.M.; et al. Circulating Tumor Cells in Metastatic Breast Cancer: Biologic Staging Beyond Tumor Burden. Clin. Breast Cancer 2007, 7, 34–42. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Stopeck, A.; Reuben, J.M. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef] [Green Version]
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrínyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, H.; Murray, M.; Turner, J.N.; Caggana, M. Isolation of tumor cells using size and deformation. J. Chromatogr. A 2009, 1216, 8289–8295. [Google Scholar] [CrossRef] [PubMed]
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 2015, 12, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Marrella, A.; Fedi, A.; Varani, G.; Vaccari, I.; Fato, M.; Firpo, G.; Guida, P.; Aceto, N.; Scaglione, S. High blood flow shear stress values are associated with circulating tumor cells cluster disaggregation in a multi-channel microfluidic device. PLoS ONE 2021, 16, e0245536. [Google Scholar] [CrossRef]
- Regmi, S.; Fu, A.; Luo, K.Q. High Shear Stresses under Exercise Condition Destroy Circulating Tumor Cells in a Microfluidic System. Sci. Rep. 2017, 7, 39975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, H.; Zhao, C.; Song, K.; Liu, D.; Ma, W.; Yu, X.; Su, H.; Zhang, Z.; Zohar, Y.; Lee, Y.-K. A nonlinear two-degree-of-freedom mass–damper–spring model to predict the isolation of circulating tumor cells in microfluidic-elasto-filtration devices. Microfluid. Nanofluidics 2019, 23, 72. [Google Scholar] [CrossRef]
- Gascoyne, P.; Shim, S. Isolation of Circulating Tumor Cells by Dielectrophoresis. Cancers 2014, 6, 545–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Oliveira, R.A.G.; Nicoliche, C.Y.N.; Pasqualeti, A.M.; Shimizu, F.M.; Ribeiro, I.R.; Melendez, M.E.; Carvalho, A.L.; Gobbi, A.L.; Faria, R.C.; Lima, R.S. Low-Cost and Rapid-Production Microfluidic Electrochemical Double-Layer Capacitors for Fast and Sensitive Breast Cancer Diagnosis. Anal. Chem. 2018, 90, 12377–12384. [Google Scholar] [CrossRef]
- Plouffe, B.D.; Murthy, S.K.; Lewis, L.H. Fundamentals and application of magnetic particles in cell isolation and enrichment: A review. Rep. Prog. Phys. 2015, 78, 016601. [Google Scholar] [CrossRef] [PubMed]
- Wyatt Shields IV, C.; Reyes, C.D.; López, G.P. Microfluidic cell sorting: A review of the advances in the separation of cells from debulking to rare cell isolation. Lab. Chip 2015, 15, 1230–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atajanov, A.; Zhbanov, A.; Yang, S. Sorting and manipulation of biological cells and the prospects for using optical forces. Micro Nano Syst. Lett. 2018, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Zhu, D.; Chen, M.; Chen, K.; Liu, H.; Liu, W.; Yang, Y. Precise and non-invasive circulating tumor cell isolation based on optical force using homologous erythrocyte binding. Lab. Chip 2019, 19, 2549–2556. [Google Scholar] [CrossRef]
- Zheng, X.; Jiang, L.; Schroeder, J.; Stopeck, A.; Zohar, Y. Isolation of viable cancer cells in antibody-functionalized microfluidic devices. Biomicrofluidics 2014, 8, 024119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Cheung, L.S.-L.; Schroeder, J.A.; Jiang, L.; Zohar, Y. A high-performance microsystem for isolating circulating tumor cells. Lab. Chip 2011, 11, 3269. [Google Scholar] [CrossRef] [PubMed]
- Yap, Y.-S.; Leong, M.C.; Chua, Y.W.; Loh, K.W.J.; Lee, G.E.; Lim, E.H.; Dent, R.; Ng, R.C.H.; Lim, J.H.-C.; Singh, G.; et al. Detection and prognostic relevance of circulating tumour cells (CTCs) in Asian breast cancers using a label-free microfluidic platform. PLoS ONE 2019, 14, e0221305. [Google Scholar] [CrossRef]
- Murlidhar, V.; Rivera-Báez, L.; Nagrath, S. Affinity Versus Label-Free Isolation of Circulating Tumor Cells: Who Wins? Small 2016, 12, 4450–4463. [Google Scholar] [CrossRef]
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur. J. Cancer 2021, 144, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Malla, R.R.; Pandrangi, S.; Kumari, S.; Gavara, M.M.; Badana, A.K. Exosomal tetraspanins as regulators of cancer progression and metastasis and novel diagnostic markers. Asia Pac. J. Clin. Oncol. 2018, 14, 383–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomiyama, T.; Yang, G.-X.; Zhao, M.; Zhang, W.; Tanaka, H.; Wang, J.; Leung, P.S.; Okazaki, K.; He, X.-S.; Lu, Q.; et al. The modulation of co-stimulatory molecules by circulating exosomes in primary biliary cirrhosis. Cell. Mol. Immunol. 2017, 14, 276–284. [Google Scholar] [CrossRef] [Green Version]
- Lynch, S.; Santos, S.G.; Campbell, E.C.; Nimmo, A.M.S.; Botting, C.; Prescott, A.; Antoniou, A.N.; Powis, S.J. Novel MHC Class I Structures on Exosomes. J. Immunol. 2009, 183, 1884–1891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jella, K.K.; Yu, L.; Yue, Q.; Friedman, D.; Duke, B.J.; Alli, A.A. Exosomal GAPDH from Proximal Tubule Cells Regulate ENaC Activity. PLoS ONE 2016, 11, e0165763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Shi, K.; Yang, S.; Liu, J.; Zhou, Q.; Wang, G.; Song, J.; Li, Z.; Zhang, Z.; Yuan, W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer 2018, 17, 147. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Dang, W.; Zhang, S.; Yue, W.; Yang, L.; Zhai, X.; Yan, Q.; Lu, J. The role of exosomal noncoding RNAs in cancer. Mol. Cancer 2019, 18, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, T.; Kamada, H.; Kanasaki, S.; Maeda, Y.; Nagano, K.; Abe, Y.; Inoue, M.; Yoshioka, Y.; Tsutsumi, Y.; Katayama, S.; et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 2013, 68, 5. [Google Scholar]
- Chairoungdua, A.; Smith, D.; Pochard, P.; Hull, M.; Caplan, M. Exosome release of B-catenin: A novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 2010, 190, 1079–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krawczyk, M.A.; Pospieszynska, A.; Styczewska, M.; Bien, E.; Sawicki, S.; Marino Gammazza, A.; Fucarino, A.; Gorska-Ponikowska, M. Extracellular Chaperones as Novel Biomarkers of Overall Cancer Progression and Efficacy of Anticancer Therapy. Appl. Sci. 2020, 10, 6009. [Google Scholar] [CrossRef]
- Olejarz, W.; Dominiak, A. Tumor-Derived Exosomes in Immunosuppression and Immunotherapy. J. Immunol. Res. 2020, 2020, 6272498. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Bai, X.; Ni, J.; Zhang, H.; Duan, W.; Graham, P.; Li, Y. Exosomes and breast cancer drug resistance. Cell Death Dis. 2020, 11, 987. [Google Scholar] [CrossRef]
- Huang, J.; Ding, Z.; Luo, Q.; Xu, W. Cancer cell-derived exosomes promote cell proliferation and inhibit cell apoptosis of both normal lung fibroblasts and non-small cell lung cancer cell through delivering alpha-smooth muscle actin. Am. J. Transl. Res. 2019, 11, 14. [Google Scholar]
- Tang, Q.; Cheng, J.; Cao, X.; Surowy, H.; Burwinkel, B. Blood-based DNA methylation as biomarker for breast cancer: A systematic review. Clin. Epigenetics 2016, 8, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Tang, W.; Yang, F. Cancer Liquid Biopsy Using Integrated Microfluidic Exosome Analysis Platforms. Biotechnol. J. 2020, 15, 1900225. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, R.; Naghibosadat, M.; Rauf, S.; Korbie, D.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Detecting Exosomes Specifically: A Multiplexed Device Based on Alternating Current Electrohydrodynamic Induced Nanoshearing. Anal. Chem. 2014, 86, 11125–11132. [Google Scholar] [CrossRef] [Green Version]
- Sina, A.A.I.; Vaidyanathan, R.; Dey, S.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Real time and label free profiling of clinically relevant exosomes. Sci. Rep. 2016, 6, 30460. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.; Tian, H.; Li, X.; Jin, D.; Li, X.; Kong, J.; Yang, C.; Yang, X.; Lu, Y.; Luo, Y.; et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS ONE 2017, 12, e0175050. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Chi, J.; Zheng, L.; Ma, B.; Li, Z.; Wang, S.; Zhao, C.; Liu, H. Efficient isolation and sensitive quantification of extracellular vesicles based on an integrated ExoID-Chip using photonic crystals. Lab. Chip 2019, 19, 2897–2904. [Google Scholar] [CrossRef]
- Gao, Y.; Qiang, L.; Chu, Y.; Han, Y.; Zhang, Y.; Han, L. Microfluidic chip for multiple detection of miRNA biomarkers in breast cancer based on three-segment hybridization. AIP Adv. 2020, 10, 045022. [Google Scholar] [CrossRef]
- Terzi, M.Y.; Izmirli, M.; Gogebakan, B. The cell fate: Senescence or quiescence. Mol. Biol. Rep. 2016, 43, 1213–1220. [Google Scholar] [CrossRef]
- Coller, H.A.; Sang, L.; Roberts, J.M. A New Description of Cellular Quiescence. PLoS Biol. 2006, 4, e83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, A.; Laurenti, E.; Oser, G.; van der Wath, R.C.; Blanco-Bose, W.; Jaworski, M.; Offner, S.; Dunant, C.F.; Eshkind, L.; Bockamp, E.; et al. Hematopoietic Stem Cells Reversibly Switch from Dormancy to Self-Renewal during Homeostasis and Repair. Cell 2008, 135, 1118–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Clevers, H. Coexistence of Quiescent and Active Adult Stem Cells in Mammals. Science 2010, 327, 542–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Fujimaki, K.; Mitchell, G.C.; Kwon, J.S.; Della Croce, K.; Langsdorf, C.; Zhang, H.H.; Yao, G. Exit from quiescence displays a memory of cell growth and division. Nat. Commun. 2017, 8, 321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, J.S.; Everetts, N.J.; Wang, X.; Wang, W.; Della Croce, K.; Xing, J.; Yao, G. Controlling Depth of Cellular Quiescence by an Rb-E2F Network Switch. Cell Rep. 2017, 20, 3223–3235. [Google Scholar] [CrossRef] [Green Version]
- Llorens-Bobadilla, E.; Zhao, S.; Baser, A.; Saiz-Castro, G.; Zwadlo, K.; Martin-Villalba, A. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell 2015, 17, 329–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodgers, J.T.; King, K.Y.; Brett, J.O.; Cromie, M.J.; Charville, G.W.; Maguire, K.K.; Brunson, C.; Mastey, N.; Liu, L.; Tsai, C.-R.; et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to GAlert. Nature 2014, 510, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Orford, K.W.; Scadden, D.T. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat. Rev. Genet. 2008, 9, 115–128. [Google Scholar] [CrossRef]
- Sampieri, K.; Fodde, R. Cancer stem cells and metastasis. Semin. Cancer Biol. 2012, 22, 187–193. [Google Scholar] [CrossRef]
- Zhou, J.; Wulfkuhle, J.; Zhang, H.; Gu, P.; Yang, Y.; Deng, J.; Margolick, J.B.; Liotta, L.A.; Petricoin, E.; Zhang, Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl. Acad. Sci. USA 2007, 104, 16158–16163. [Google Scholar] [CrossRef] [Green Version]
- Lim, P.K.; Bliss, S.A.; Patel, S.A.; Taborga, M.; Dave, M.A.; Gregory, L.A.; Greco, S.J.; Bryan, M.; Patel, P.S.; Rameshwar, P. Gap Junction–Mediated Import of MicroRNA from Bone Marrow Stromal Cells Can Elicit Cell Cycle Quiescence in Breast Cancer Cells. Cancer Res. 2011, 71, 1550–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Kennedy, M.; Payne, S.; Kennedy, K.; Seewaldt, V.L.; Pizzo, S.V.; Bachelder, R.E. Model of Tumor Dormancy/Recurrence after Short-Term Chemotherapy. PLoS ONE 2014, 9, e98021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef]
- Bliss, S.A.; Sinha, G.; Sandiford, O.A.; Williams, L.M.; Engelberth, D.J.; Guiro, K.; Isenalumhe, L.L.; Greco, S.J.; Ayer, S.; Bryan, M.; et al. Mesenchymal Stem Cell–Derived Exosomes Stimulate Cycling Quiescence and Early Breast Cancer Dormancy in Bone Marrow. Cancer Res. 2016, 76, 5832–5844. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Reed-Newman, T.; Anant, S.; Ramasamy, T.S. Regulatory Role of Quiescence in the Biological Function of Cancer Stem Cells. Stem Cell Rev. Rep. 2020, 16, 1185–1207. [Google Scholar] [CrossRef]
- Faley, S.L.; Copland, M.; Wlodkowic, D.; Kolch, W.; Seale, K.T.; Wikswo, J.P.; Cooper, J.M. Microfluidic single cell arrays to interrogate signalling dynamics of individual, patient-derived hematopoietic stem cells. Lab. Chip 2009, 9, 2659. [Google Scholar] [CrossRef] [Green Version]
- Lecault, V.; VanInsberghe, M.; Sekulovic, S.; Knapp, D.J.H.F.; Wohrer, S.; Bowden, W.; Viel, F.; McLaughlin, T.; Jarandehei, A.; Miller, M.; et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat. Methods 2011, 8, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Kai, K.; Choi, D.S.; Iwamoto, T.; Nguyen, Y.H.; Wong, H.; Landis, M.D.; Ueno, N.T.; Chang, J.; Qin, L. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proc. Natl. Acad. Sci. USA 2012, 109, 18707–18712. [Google Scholar] [CrossRef] [Green Version]
- Ma, N.; Kamalakshakurup, G.; Aghaamoo, M.; Lee, A.P.; Digman, M.A. Label-Free Metabolic Classification of Single Cells in Droplets Using the Phasor Approach to Fluorescence Lifetime Imaging Microscopy. Cytometry A 2019, 95, 93–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argüello-Miranda, O.; Marchand, A.J.; Kennedy, T.; Russo, M.A.X.; Noh, J. Cell cycle–independent integration of stress signals by Xbp1 promotes Non-G1/G0 quiescence entry. J. Cell Biol. 2022, 221, e202103171. [Google Scholar] [CrossRef]
- Kang, D.-K.; Lu, J.; Zhang, W.; Chang, E.; Eckert, M.A.; Ali, M.M.; Zhao, W.; Li, X. Microfluidic devices for stem cell analysis. In Microfluidic Devices for Biomedical Applications; Elsevier: Amsterdam, The Netherlands, 2021; pp. 437–487. ISBN 978-0-12-819971-8. [Google Scholar]
- Liu, B.; Wang, X.; Jiang, L.; Xu, J.; Zohar, Y.; Yao, G. Extracellular Fluid Flow Induces Shallow Quiescence through Physical and Biochemical Cues. biorXiv 2021. [Google Scholar] [CrossRef]
- Fares, J.E.; El Tomb, P.; Khalil, L.E.; Atwani, R.W.; Moukadem, H.A.; Awada, A.; El Saghir, N.S. Metronomic chemotherapy for patients with metastatic breast cancer: Review of effectiveness and potential use during pandemics. Cancer Treat. Rev. 2020, 89, 102066. [Google Scholar] [CrossRef] [PubMed]
- Foldi, J.; Silber, A.; Reisenbichler, E.; Singh, K.; Fischbach, N.; Persico, J.; Adelson, K.; Katoch, A.; Horowitz, N.; Lannin, D.; et al. Neoadjuvant durvalumab plus weekly nab-paclitaxel and dose-dense doxorubicin/cyclophosphamide in triple-negative breast cancer. Npj Breast Cancer 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- McClendon, A.K.; Dean, J.L.; Rivadeneira, D.B.; Yu, J.E.; Reed, C.A.; Gao, E.; Farber, J.L.; Force, T.; Koch, W.J.; Knudsen, E.S. CDK4/6 inhibition antagonizes the cytotoxic response to anthracycline therapy. Cell Cycle 2012, 11, 2747–2755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrelli, F.; Ghidini, A.; Pedersini, R.; Cabiddu, M.; Borgonovo, K.; Parati, M.C.; Ghilardi, M.; Amoroso, V.; Berruti, A.; Barni, S. Comparative efficacy of palbociclib, ribociclib and abemaciclib for ER+ metastatic breast cancer: An adjusted indirect analysis of randomized controlled trials. Breast Cancer Res. Treat. 2019, 174, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Nunes, M.R.; Stearns, V. CDK4/6 Inhibitors: Game Changers in the Management of Hormone Receptor–Positive Advanced Breast Cancer? Oncology 2018, 15, 216–222. [Google Scholar]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Huang, C.-S.; Mano, M.S.; Loibl, S.; Mamounas, E.P.; Untch, M.; Wolmark, N.; Rastogi, P.; Schneeweiss, A.; Redondo, A.; et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N. Engl. J. Med. 2019, 380, 617–628. [Google Scholar] [CrossRef]
- Eyvazi, S.; Farajnia, S.; Dastmalchi, S.; Kanipour, F.; Zarredar, H.; Bandehpour, M. Antibody Based EpCAM Targeted Therapy of Cancer, Review and Update. Curr. Cancer Drug Targets 2018, 18, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Pondé, N.; Aftimos, P.; Piccart, M. Antibody-Drug Conjugates in Breast Cancer: A Comprehensive Review. Curr. Treat. Options Oncol. 2019, 20, 37. [Google Scholar] [CrossRef]
- Philipson, B.I.; O’Connor, R.S.; May, M.J.; June, C.H.; Albelda, S.M.; Milone, M.C. 4-1BB costimulation promotes CAR T cell survival through noncanonical NF-κB signaling. Sci. Signal. 2020, 13, eaay8248. [Google Scholar] [CrossRef]
- Guedan, S.; Posey, A.D.; Shaw, C.; Wing, A.; Da, T.; Patel, P.R.; McGettigan, S.E.; Casado-Medrano, V.; Kawalekar, O.U.; Uribe-Herranz, M.; et al. Enhancing CAR T cell persistence through ICOS and 4-1BB costimulation. JCI Insight 2018, 3, e96976. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, F.; Cao, J.; Wang, X.; Cheng, H.; Qi, K.; Wang, G.; Xu, K.; Zheng, J.; Fu, Y.-X.; et al. A chimeric antigen receptor with antigen-independent OX40 signaling mediates potent antitumor activity. Sci. Transl. Med. 2021, 13, eaba7308. [Google Scholar] [CrossRef] [PubMed]
- Melzer, M.; Lopez-Martinez, A.; Altomonte, J. Oncolytic Vesicular Stomatitis Virus as a Viro-Immunotherapy: Defeating Cancer with a “Hammer” and “Anvil. ” Biomedicines 2017, 5, 8. [Google Scholar] [CrossRef]
- Pol, J.G.; Lévesque, S.; Workenhe, S.T.; Gujar, S.; Le Boeuf, F.; Clements, D.R.; Fahrner, J.-E.; Fend, L.; Bell, J.C.; Mossman, K.L.; et al. Trial Watch: Oncolytic viro-immunotherapy of hematologic and solid tumors. OncoImmunology 2018, 7, e1503032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamani, P.; Navashenaq, J.G.; Nikpoor, A.R.; Hatamipour, M.; Oskuee, R.K.; Badiee, A.; Jaafari, M.R. MPL nano-liposomal vaccine containing P5 HER2/neu-derived peptide pulsed PADRE as an effective vaccine in a mice TUBO model of breast cancer. J. Controlled Release 2019, 303, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Soon, R.H.; Lim, C.T.; Khoo, B.L.; Han, J. Microfluidic modelling of the tumor microenvironment for anti-cancer drug development. Lab. Chip 2019, 19, 369–386. [Google Scholar] [CrossRef] [PubMed]
- Venugopal Menon, N.; Lim, S.B.; Lim, C.T. Microfluidics for personalized drug screening of cancer. Curr. Opin. Pharmacol. 2019, 48, 155–161. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, X.; Zou, J.; Jia, C.; Hu, Y.; Du, H.; Wang, H. Evaluation of photodynamic therapy efficiency using an in vitro three-dimensional microfluidic breast cancer tissue model. Lab. Chip 2015, 15, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, D.; Wang, Y.; Lin, S.; Jiang, Y. A novel 3D breast-cancer-on-chip platform for therapeutic evaluation of drug delivery systems. Anal. Chim. Acta 2018, 1036, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Ozcelikkale, A.; Moon, H.; Linnes, M.; Han, B. In vitro microfluidic models of tumor microenvironment to screen transport of drugs and nanoparticles: In vitro microfluidic models of tumor microenvironment. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1460. [Google Scholar] [CrossRef]
- Tomeh, M.A.; Zhao, X. Recent Advances in Microfluidics for the Preparation of Drug and Gene Delivery Systems. Mol. Pharm. 2020, 17, 4421–4434. [Google Scholar] [CrossRef]
- Sharma, I.; Thakur, M.; Singh, S.; Tripathi, A. Microfluidic Devices as a Tool for Drug Delivery and Diagnosis: A Review. Int. J. Appl. Pharm. 2021, 13, 95–102. [Google Scholar] [CrossRef]
- Fadhil Majnis, M.; Francis, H.; Zilati Ku Shaari, K. Droplet formation in microchannels at low values of the capillary and the reynolds numbers. Mater. Today Proc. 2018, 5, 21765–21771. [Google Scholar] [CrossRef]
- He, J.; Wei, Z.; Wang, L.; Tomova, Z.; Babu, T.; Wang, C.; Han, X.; Fourkas, J.T.; Nie, Z. Hydrodynamically Driven Self-Assembly of Giant Vesicles of Metal Nanoparticles for Remote-Controlled Release. Angew. Chem. Int. Ed. 2013, 52, 2463–2468. [Google Scholar] [CrossRef]
- Lou, G.; Anderluzzi, G.; Woods, S.; Roberts, C.W.; Perrie, Y. A novel microfluidic-based approach to formulate size-tuneable large unilamellar cationic liposomes: Formulation, cellular uptake and biodistribution investigations. Eur. J. Pharm. Biopharm. 2019, 143, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhu, Z.; Wang, X.; Gonçalves, D.; Zhang, B.; Hierlemann, A.; Hunziker, P. Microfluidics-based single-step preparation of injection-ready polymeric nanosystems for medical imaging and drug delivery. Nanoscale 2015, 7, 16983–16993. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Bernuz, C.R.; Fan, J.; Li, W.; Correia, A.; Hirvonen, J.; Santos, H.A. A Nano-in-Nano Vector: Merging the Best of Polymeric Nanoparticles and Drug Nanocrystals. Adv. Funct. Mater. 2017, 27, 1604508. [Google Scholar] [CrossRef]
- Li, W.; Liu, D.; Zhang, H.; Correia, A.; Mäkilä, E.; Salonen, J.; Hirvonen, J.; Santos, H.A. Microfluidic assembly of a nano-in-micro dual drug delivery platform composed of halloysite nanotubes and a pH-responsive polymer for colon cancer therapy. Acta Biomater. 2017, 48, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Hajba, L.; Guttman, A. Continuous-flow-based microfluidic systems for therapeutic monoclonal antibody production and organ-on-a-chip drug testing. J. Flow Chem. 2017, 7, 118–123. [Google Scholar] [CrossRef]
- Zhang, W.; Li, Q.; Jia, F.; Hu, Z.; Wei, Z. A Microfluidic Chip for Screening and Sequencing of Monoclonal Antibody at a Single-Cell Level. Anal. Chem. 2021, 93, 10099–10105. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.F.C.; Azevedo, A.M.; Fernandes, P.; Chu, V.; Conde, J.P.; Aires-Barros, M.R. Design of a microfluidic platform for monoclonal antibody extraction using an aqueous two-phase system. J. Chromatogr. A 2012, 1249, 1–7. [Google Scholar] [CrossRef]
- Bourguignon, N.; Attallah, C.; Karp, P.; Booth, R.; Peñaherrera, A.; Payés, C.; Oggero, M.; Pérez, M.S.; Helguera, G.; Lerner, B. Production of monoclonal antibodies in microfluidic devices. Integr. Biol. 2018, 10, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Wright, B.D.; Whittenberg, J.; Desai, A.; DiFelice, C.; Kenis, P.J.A.; Lapi, S.E.; Reichert, D.E. Microfluidic Preparation of a 89 Zr-Labeled Trastuzumab Single-Patient Dose. J. Nucl. Med. 2016, 57, 747–752. [Google Scholar] [CrossRef] [Green Version]
- Terrell-Hall, T.B.; Nounou, M.I.; El-Amrawy, F.; Griffith, J.I.G.; Lockman, P.R. Trastuzumab distribution in an in-vivo and in-vitro model of brain metastases of breast cancer. Oncotarget 2017, 8, 83734–83744. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, R.; Shen, B.; Li, N.; Zhou, H.; Wang, W.; Zhao, Y.; Huang, M.; Fang, P.; Wang, S.; et al. High-throughput functional screening for next-generation cancer immunotherapy using droplet-based microfluidics. Sci. Adv. 2021, 7, eabe3839. [Google Scholar] [CrossRef]
- Elsemary, M.T.; Maritz, M.F.; Smith, L.E.; Thierry, B. Microfluidic purification of T lymphocytes separated from blood for chimeric antigen receptor T-cell manufacturing. Cytotherapy 2019, 21, e4. [Google Scholar] [CrossRef]
- Fajrial, A.K.; He, Q.Q.; Wirusanti, N.I.; Slansky, J.E.; Ding, X. A review of emerging physical transfection methods for CRISPR/Cas9-mediated gene editing. Theranostics 2020, 10, 5532–5549. [Google Scholar] [CrossRef]
- Ide, H.; Espulgar, W.V.; Saito, M.; Aoshi, T.; Koyama, S.; Takamatsu, H.; Tamiya, E. Profiling T cell interaction and activation through microfluidics-assisted serial encounter with APCs. Sens. Actuators B Chem. 2021, 330, 129306. [Google Scholar] [CrossRef]
- Jimeno, A.; Baranda, J.; Mita, M.; Gordon, M.; Taylor, M.; Iams, W.; Janku, F.; Matulonis, U.; Bernstein, H.; Loughhead, S.; et al. Initial results of a first-in-human, dose escalation study of a cell-based vaccine in HLA A*02+ patients (pts) with recurrent, locally advanced or metastatic HPV16+ solid tumors: SQZ-PBMC-HPV-101. J. Clin. Oncol. 2021, 39, 2536. [Google Scholar] [CrossRef]
- Booty, M.; Stockmann, A.; Pryor, O.; Myint, M.; Trumpfheller, C.; Nicolini, V.; Klein, C.; Codarri, L.; Umana, P.; Sharei, A.; et al. 141 PBMC-based cancer vaccines generated with microfluidics squeezing demonstrate synergistic and durable tumor reduction in combination with PD1 checkpoint and FAP targeted IL-2 variants. J. Immunother. Cancer 2020, 8, A155. [Google Scholar] [CrossRef]
- Chao, B.H.; Sung, K.E.; Yin, J.; Beebe, D.J. Abstract C236: The inhibitory effect of tumor microenvironment on oncolytic virotherapy in lung cancer. Mol. Cancer Ther. 2011, 10, C236. [Google Scholar] [CrossRef]
- Lee, S.W.; Lee, K.J.; Jeong, S.Y.; Joo, C.H.; Lee, H.; Jeong, G.S. Evaluation of Bystander Infection of Oncolytic Virus using a Medium Flow Integrated 3D In Vitro Microphysiological System. Adv. Biosyst. 2020, 4, 1900143. [Google Scholar] [CrossRef]
- Qiao, H.; Chen, X.; Wang, Q.; Zhang, J.; Huang, D.; Chen, E.; Qian, H.; Zhong, Y.; Tang, Q.; Chen, W. Tumor localization of oncolytic adenovirus assisted by pH-degradable microgels with JQ1-mediated boosting replication and PD-L1 suppression for enhanced cancer therapy. Biomater. Sci. 2020, 8, 2472–2480. [Google Scholar] [CrossRef]
- Paterson, K.; Zanivan, S.; Glasspool, R.; Coffelt, S.B.; Zagnoni, M. Microfluidic technologies for immunotherapy studies on solid tumours. Lab. Chip 2021, 21, 2306–2329. [Google Scholar] [CrossRef]
- Lee, K.J.; Lee, S.W.; Woo, H.-N.; Cho, H.M.; Yu, D.B.; Jeong, S.Y.; Joo, C.H.; Jeong, G.S.; Lee, H. Real-time monitoring of oncolytic VSV properties in a novel in vitro microphysiological system containing 3D multicellular tumor spheroids. PLoS ONE 2020, 15, e0235356. [Google Scholar] [CrossRef]
- Fan, Y.; Li, M.; Ma, K.; Hu, Y.; Jing, J.; Shi, Y.; Li, E.; Dong, D. Dual-target MDM2/MDMX inhibitor increases the sensitization of doxorubicin and inhibits migration and invasion abilities of triple-negative breast cancer cells through activation of TAB1/TAK1/p38 MAPK pathway. Cancer Biol. Ther. 2019, 20, 617–632. [Google Scholar] [CrossRef] [PubMed]
- Lovitt, C.J.; Shelper, T.B.; Avery, V.M. Doxorubicin resistance in breast cancer cells is mediated by extracellular matrix proteins. BMC Cancer 2018, 18, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, L.; Jia, N.; Gao, Y.; Hu, H.; Zhao, X.; Chen, D.; Qiao, M. Multi-Modulation of Doxorubicin Resistance in Breast Cancer Cells by Poly(l-histidine)-Based Multifunctional Micelles. Pharmaceutics 2019, 11, 385. [Google Scholar] [CrossRef] [Green Version]
- Marinello, P.C.; Panis, C.; Silva, T.N.X.; Binato, R.; Abdelhay, E.; Rodrigues, J.A.; Mencalha, A.L.; Lopes, N.M.D.; Luiz, R.C.; Cecchini, R.; et al. Metformin prevention of doxorubicin resistance in MCF-7 and MDA-MB-231 involves oxidative stress generation and modulation of cell adaptation genes. Sci. Rep. 2019, 9, 5864. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cai, H.; Chen, W.; Guan, Q.; He, J.; Guo, Z.; Li, J. A Qualitative Transcriptional Signature for Predicting Extreme Resistance of ER-Negative Breast Cancer to Paclitaxel, Doxorubicin, and Cyclophosphamide Neoadjuvant Chemotherapy. Front. Mol. Biosci. 2020, 7, 34. [Google Scholar] [CrossRef]
- Merry, C.R.; McMahon, S.; Forrest, M.E.; Bartels, C.F.; Saiakhova, A.; Bartel, C.A.; Scacheri, P.C.; Thompson, C.L.; Jackson, M.W.; Harris, L.N.; et al. Transcriptome-wide identification of mRNAs and lincRNAs associated with trastuzumab-resistance in HER2-positive breast cancer. Oncotarget 2016, 7, 53230–53244. [Google Scholar] [CrossRef] [PubMed]
- Yazdanifar, M.; Zhou, R.; Grover, P.; Williams, C.; Bose, M.; Moore, L.; Wu, S.; Maher, J.; Dreau, D.; Mukherjee, P. Overcoming Immunological Resistance Enhances the Efficacy of A Novel Anti-tMUC1-CAR T Cell Treatment against Pancreatic Ductal Adenocarcinoma. Cells 2019, 8, 1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaurasiya, S.; Fong, Y. Viroimmunotherapy for breast cancer: Promises, problems and future directions. Cancer Gene Ther. 2021, 28, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.N.; Fry, T.J. Mechanisms of resistance to CAR T cell therapy. Nat. Rev. Clin. Oncol. 2019, 16, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.D.; Elsen, S.; Krysko, D.V.; Vandenabeele, P.; de Witte, P.; Agostinis, P. Resistance to anticancer vaccination effect is controlled by a cancer cell-autonomous phenotype that disrupts immunogenic phagocytic removal. Oncotarget 2015, 6, 26841–26860. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Wang, L.; Zhong, R.; Li, B.; Ye, N.; Liu, X.; Lin, B. Parallel microfluidic networks for studying cellular response to chemical modulation. J. Biotechnol. 2007, 131, 286–292. [Google Scholar] [CrossRef]
- Sarkar, S.; Cohen, N.; Sabhachandani, P.; Konry, T. Phenotypic drug profiling in droplet microfluidics for better targeting of drug-resistant tumors. Lab. Chip 2015, 15, 4441–4450. [Google Scholar] [CrossRef] [Green Version]
- Ozcelikkale, A.; Shin, K.; Noe-Kim, V.; Elzey, B.D.; Dong, Z.; Zhang, J.-T.; Kim, K.; Kwon, I.C.; Park, K.; Han, B. Differential response to doxorubicin in breast cancer subtypes simulated by a microfluidic tumor model. J. Controlled Release 2017, 266, 129–139. [Google Scholar] [CrossRef]
- Qi, R.; Zhu, G.; Wang, Y.; Wu, S.; Li, S.; Zhang, D.; Bu, Y.; Bhave, G.; Han, R.; Liu, X. Microfluidic device for the analysis of MDR cancerous cell-derived exosomes’ response to nanotherapy. Biomed. Microdevices 2019, 21, 35. [Google Scholar] [CrossRef]
- Parekh, K.; Noghabi, H.S.; Lopez, J.A.; Li, P.C.H. Microfluidic chip enables single-cell measurement for multidrug resistance in triple-negative breast cancer cells. Cancer Drug Resist 2020, 3, 113–127. [Google Scholar] [CrossRef] [Green Version]
- Parekh, K.; Sharifi, H.; Khamenehfar, A.; Beischlag, T.V.; Payer, R.T.M.; Li, P.C.H. The microfluidic capture of single breast cancer cells for multi-drug resistance assays. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 628, pp. 113–127. ISBN 978-0-12-817090-8. [Google Scholar]
- Rahman, S.M.; Campbell, J.M.; Coates, R.N.; Render, K.M.; Byrne, C.E.; Martin, E.C.; Melvin, A.T. Evaluation of intercellular communication between breast cancer cells and adipose-derived stem cells via passive diffusion in a two-layer microfluidic device. Lab. Chip 2020, 20, 2009–2019. [Google Scholar] [CrossRef] [PubMed]



| Reference | Cell Used | Culture Type | Field of Investigation | Device Properties | Findings |
|---|---|---|---|---|---|
| Cheng [19] | Acellular | 2D | Cancer formation | Extract cell-free DNA from plasma to detect BRCA1 and BRCA2 mutations. | Successful detection of BRCA1/2 mutations with a minimum detectable number of copies of 20,000. |
| Uses four distinct primers in parallel to provide point of care risk assessment. | |||||
| Dimensions not described, no ECM used, utilizes pressure differential pumping. | |||||
| Pradhan [20] | MCF7 and MDA-MB-231 cancer, hBTEC endothelial, BJ5ta fibroblast | 3D | Cancer formation | Two distinct chips with normo- and pathophysiologic vascular layout, respectively, used to assess anti-cancer drug delivery and tumor reaction to treatment. | Cells found to elongate and align along flow, dependent on the cell line. |
| Model of cancer–stromal–endothelial interactions within a pillar-filled tumor region adjacent to vessels. | MCF7 found to have significant resistance to anti-cancer drug paclitaxel in low-perfusion chip design. | ||||
| 100 μm channel width, PEG–fibrinogen hydrogel matrix, high perfusion layout experiences 40–50 s−1 shear rate, low perfusion chip experiences 10–20 s−1 shear rate. | Both MCF7 and MDA-MB-231 found to have significant resistance to anti-cancer drug doxorubicin in low-perfusion chip design. | ||||
| Ayuso [21] | MCF10A cancer, HMF fibroblasts | 3D | Cancer formation | Ductal carcinoma in situ in a central vessel surrounded by stroma-filled matrix between two empty channels for perfusion, metabolism, mobility, and gene expression investigation. | Hypoxia-activated Tirapazamine selectively destroys DCIS cells. |
| Breast cancer cells, fibroblasts, and hydrogel model of luminal mammary duct. | Hypoxia-activated glycolysis transcriptome upregulation. | ||||
| Dimensions not described, collagen hydrogel, static conditions with daily media change. | |||||
| Funamoto [22] | MDA-MB-231 cancer | 3D | Cancer formation | Five-channel chip with central tumor model, surrounded by two media perfusion channels, which are, in turn, surrounded by two gas perfusion channels. | Hypoxia found to significantly increase cancer mobility. |
| Breast cancer cells in 3D culture with controlled oxygen perfusion to investigate hypoxia effects. | |||||
| 1.3 mm tumor channel width, 0.5 mm media and gas channel widths, collagen I hydrogel, 30 μL/h flow. | |||||
| Tang [23] | MDA-MB-231 and MCF-7 cancer, primary human-breast-tumor-associated endothelial cells | 3D | Cancer formation | Microfluidic chip modeled on 2D projection of tumor vasculature to investigate enhanced permeability and retention (EPR) found in tumors. | TNF-α found to significantly increase the permeability of endothelial cells. |
| Breast cancer and endothelial cell biomimetic tumor microenvironment. | Tumor cell co-culture significantly increases the permeability of endothelial cells. | ||||
| 100 μm channel width, fibronectin ECM, 60 uL/h or 0–90 s−1 shear rate. | Liposome extravasation through endothelial cells found to significantly increase during tumor co-culture. | ||||
| Nashimoto [24] | MCF-7 and GFP MDA-MB-231 cancer, RFP HUVECs, normal human lung fibroblasts, SW620 with luciferase, Hepg2 | Spheroids | Cancer formation | 2 or 3 cell type co-culture spheroids of various cancers in 96-well plate transferred into microfluidic chip for angiogenesis. | Fibroblast co-culture induced angiogenic sprouts. |
| 1 mm width culture channel, fibrin-collagen matrix, 30 μL/h. | Flow reduces anti-cancer drug paclitaxel efficacy and leads to less necrotic tumors. | ||||
| Uliana [32] | Acellular | 2D | Cancer formation | Disposable microfluidic electrochemical array device to detect estrogen receptor alpha (ERα). | USD 0.20 cost of manufacture. |
| Highly decorated magnetic particles and protein–DNA interaction detected using electrodes to quantify cancer signals. | Ultralow detection limit of 10.0 fg mL−1 for the determination of ERα in calf serum. | ||||
| 3 mm wide and 200 mm long, no ECM, 6000 μL/h. | Good recoveries for detection of the biomarker in MCF-7 cell lysate. | ||||
| Moon [44] | MCF-7, MDA-MB-231, and SUM-159PT cancer | 3D | Cancer formation | Hydrogel tube seeded with breast cancers to quantify breast cancer subtype motility. | SUM-159PT found to be most invasive cell line. |
| 500 μm diameter, collagen I hydrogel, static conditions. | CD24 expression was elevated in 3D compared with 2D cultures. | ||||
| Yong [45] | SUM149, HCC1937, MDA-MB-231, and BT549 cancer | 3D | Cancer formation | A hydrogel channel with two perfused vessels, one seeded with breast cancer cells to quantify the directional migration of cancer cell lines. | MDA-MB-231 found to be most invasive. |
| 200 μm diameter, collagen hydrogel, static conditions. | 305 genes identified as altered during invasion. | ||||
| Wang [251] | SK-BR-3 cancer, HEK293FT cells, human T cells, Jurkat cells | Suspended cells | Cancer formation | A pair of chips, one that generates droplets containing cells and antibodies or lentivirus, another that electrically sorts droplets based on fluorescence. | CD40 antibody developed. |
| Continuous flow droplet-based lentivirus transduction and antibody screening. | Active anti-Her2 × anti-CD3 BiTE antibodies developed using antibody library. | ||||
| 40 μm deep channels, no ECM used, 600–1800 μL/h. | |||||
| Sung [46] | MCF-DCIS.com cancer cells, GFP Human mammary fibroblasts | 3D | Invasion | Mammary endothelial and fibroblast cells co-cultured in Y-shaped compartmentalized model. | Co-culture promotes invasion, diminishing as distance between cell types increases |
| Second harmonic collagen imaging provides an index of transition to invasion. | Soluble factors shown to begin migration, while cell–cell contact between cancer and stromal cells completed the transition to invasion. | ||||
| 1.5 mm width, Matrigel and collagen I matrix, static culture. | |||||
| Lam [47] | RFP MDA-MB-231, skin fibroblasts | 3D | Invasion | Two parallel 3D models with breast cancer and fibroblast co-cultures to study the role of acidity in the transition to invasion. | Calcium bicarbonate buffer nanoparticles inhibit acid-induced invasion. |
| Channels between 200–1000 μm wide, fibrin hydrogel, 36 μL/h | Buffering selectively inhibited the growth of the MDA-MB-231. | ||||
| Toh [48] | MX-1, MCF7 cancer | 3D | Invasion | Tumor cells encased in collagen exposed to flowing chemoattractant signals on either side. | Cells found to exhibit collective, amoeboid, and mesenchymal-like motility. |
| Real-time monitoring of cell extravasation | Distinct populations of collagen-penetrating invasive cancer cells and collagen-avoidant migratory cancer cells identified. | ||||
| 1 cm long, 600 μm wide, and 100 μm tall, positively charged collagen and a negatively charged HEMA-MMA-MAA terpolymer matrix, 30 μL/h. | |||||
| Yankaskas [49] | ZR75-1, MDA-MB-468, MDA-MB-436, Hs578t, BT-549, and MDA-MB-231 cancer cells, MCF-10A, T47, and human mammary epithelial cells, and HCC1428 | 3D | Invasion | Continuous flow of breast cancer cells near parallel apertures through which chemoattractants diffuse to induce migration. | Identified motility- and survival-related genes. |
| Quantifies extravasation potential, abundance, and proliferative index of breast cancer cell subtypes. | MDA-MB-231 found to be most migratory. | ||||
| Width of 20 µm and a height of 10 µm, collagen matrix, flow not specified. | Self-sorted migratory cells found to be significantly more proliferative and preferentially localize to bone in comparison with non-sorted cells. | ||||
| Gioiella [50] | MCF7 cancer cells, normal and cancer activated fibroblasts | 3D | Invasion | Breast cancer tumor model using epithelial and stromal cell co-culture. | Hyaluronic acid and fibronectin overexpression are associated with invasion |
| Macromolecule and collagen production quantified. | Fibroblasts significantly lose diffusivity when activated by nearby tumor cells. | ||||
| 370 μm long, 780 μm wide, 300 μm deep, other channel is 1200 μm long, 1370 μm wide, 300 μm deep, crosslinked type A porcine-derived gelatin matrix, 180 µL/h. | |||||
| Mosadegh [51] | A549 | 3D | Invasion | Stacked paper seeded with hydrogel and cancer cells, used to examine hypoxia-induced migration. | Oxygen acts as a chemoattractant for cancer cells. |
| No channels, Matrigel matrix, no perfusion. | Distinct rates of oxygen chemotaxis between cell lines. | ||||
| Lugo-Cintron [53] | MDA-MB-231 cancer cells, normal and cancer activated fibroblasts | 3D | Invasion | Large channel containing fibroblasts in microenvironment with a central channel seeded with breast cancer cells. | Fibronectin-rich matrix associated with increased migration. |
| 3D breast cancer and fibroblast tumor model to visualize invasion. | MMP secretion associated with increased migration. | ||||
| 400 μm wide central channel, rat-tail collagen type I, fibronectin, and fibrin matrix, static culture. | |||||
| Buchanan [72] | MDA-MB-231 cancer cells, telomerase immortalized microvascular endothelial (TIME) cells | 3D | Intravasation | Microchannel embedded within a collagen hydrogel as a vessel model with shear quantified using microparticle image velocimetry. | Tumor cells significantly increase the expression of proangiogenic genes in response to co-culture with endothelial cells under low flow conditions. |
| Microfluidic tumor vascular model for co-culture of tumor and endothelial cells under varying flow shear stress conditions | Endothelial cells develop a confluent endothelium on the microchannel lumen that maintains integrity under physiological flow shear stresses. | ||||
| 850 μm diameter, Collagen 1 matrix, 180 μL/h. | |||||
| Choi [73] | Human primary mammary fibroblasts, HMT-3522 cells | Spheroids, 3D | Intravasation | Intravasation model using a two-channel stroma and endothelial co-culture device later seeded with human mammary ductal epithelial cells and mammary fibroblasts co-culture breast tumor spheroids. | Paclitaxel anti-cancer drug shown to inhibit progression through cytotoxicity. |
| Visualization of attachment and intravasation. | |||||
| 1 mm wide, 3 mm long, and 200 μm tall, collagen hydrogel, 60 μL/h. | |||||
| Nagaraju [76] | MDA-MB-231 and MCF7 cancer cells, HUVECs | 3D | Intravasation | Chip with distinct tumor, stroma, and vascular channels to model invasion and intravasation into media-filled vascular channel. | Endothelial cells significantly increase the migration of tumor cells. |
| 200 μm wide channel, collagen matrix, flow rate not specified. | Tumor signaling significantly reduces vessel diameter and increases endothelial permeability. | ||||
| Absence of endothelial cells significantly alters the secretion of ANG-2 and angiogenin. | |||||
| Zervantonakis [71] | MDA231 cancer cells, HT1080 fibroblasts | 3D | Intravasation | Three-channel tumor model with an endothelial channel and a tumor channel separated by a cell-free 3D ECM channel to model intravasation towards endothelial cells. | Macrophage-secreted tumor necrosis factor alpha significantly increases intravasation rate. |
| 500 μm wide, 20 mm in length, and 120 μm in height, collagen matrix, flow not specified. | Endothelium provides a barrier to intravasation, regulated by tumor microenvironment factors. | ||||
| Cui [75] | MDA-MB-231 cancer cells, primary human vascular endothelial cells | 3D | Intravasation | 32 independent cell collection microchambers with endothelial layer for characterization of trans-endothelial migration. | Migratory cancer cells significantly alter Palladin expression, F-actin orientation, and cell aspect ratio. |
| 4 mm by 4 mm chip, poly-D-lysine and fibronectin matrix, 1200 µL/h. | Different cancer lines show significantly distinct sensitivity to shear stress impact on trans-endothelial migration | ||||
| Shirure [77] | MDA-MB-231 and MCF-7 cancer cells, Endothelial colony forming cell-derived endothelial cells, normal human lung fibroblasts, colorectal cancer cell line Caco-2 | 3D and Spheroid | Intravasation | Two tumor model chambers separated by one fibroblast and endothelial cell vascular model chamber to model arterial capillary intravasation. | VEGF and TGFβ significantly elevated during intravasation and migration. |
| 355 μm wide, fibrin matrix, 10 mm H2O pressure head. | Tumor cells expressing mesenchymal-like transcriptome invade into vascular chamber significantly more efficiently. | ||||
| Jiang [146] | Lung, breast, and melanoma cancer blood samples | Suspended cells | Circulating signals and cells | Chip using deterministic lateral displacement to enrich platelet-CTC aggregates in conjugation with platelet antibodies for isolation. | 60% reliable isolation of breast cancer CTC clusters. |
| 24 parallel channels that are 150 μm in depth, no ECM, 1000 μL/h. | |||||
| Au [147] | Primary breast cancer CTCs and CTC clusters | Suspended cells | Circulating signals and cells | Two-stage continuous isolation of CTC clusters using asymmetry induced rotation. | 2–100+ cells recovered from whole blood. |
| 99% recovery of large clusters, cell viability over 87%. | |||||
| Deliorman [149] | PC3 human prostate cancer cell line | Suspended cells | Circulating signals and cells | Surface-bound antibodies for EpCAM, PSA, and PSMA to isolate CTCs for AFM measurements. | CTCs from metastatic cancer have decreased elasticity and increased deformability compared with tumor cells. |
| 900 μm wide, 85 μm deep, and 48 mm long, no ECM, 1200 μL/h. | Fewer multiple adhesion events in CTCs compared with tumor cells. | ||||
| Sarioglu [158] | MDA-MB-231 | Suspended cells | Circulating signals and cells | Detection of CTCs independent of tumor-specific markers using bifurcating traps under low-shear conditions. | 30–40% successful detection of CTC clusters. |
| 4096 parallel 60 μm wide traps, no ECM, 2.5 mL/h. | RNA sequencing identifies macrophages within CTC clusters are tissue-derived from the primary site. | ||||
| de Oliveira [163] | Acellular | Suspended cells | Circulating signals and cells | CTC detection using double-layer capillary capacitors to quantify CA 15-3. | 5µL sample volume and 92.0 μU mL−1 detection limit. |
| Antibody-anchored magnetic bead capture of CTCs for analysis. | USD 0.97 cost of manufacture. | ||||
| 800 μm outer diameter, 545 μm inner diameter, 200 μm electrode gap, no ECM, no flow. | |||||
| Marrella [159] | MDA-MB-231 | Suspended cells | Circulating signals and cells | Multi-channel device simultaneously analyze correlation between shear stress and CTC cluster behavior. | Higher values of wall shear stress significantly correlated with decreased CTC viability to metastasize. |
| 1 mm wide and 120 mm long, no ECM, 0–20 dyne/cm2. | High shear stress significantly disaggregates CTC clusters within 6 h. | ||||
| Regmi [160] | MDA-MB-231 and UACC-893 cancer cells, lung cancer A549, ovarian cancer 2008, leukemia K562 cells | Suspended cells | Circulating signals and cells | Microfluidic circulatory system to produce relevant shear stress levels on CTCs and CTC clusters to investigate disaggregation under shear. | 60 dynes/cm2 during exercise leads to necrosis or apoptosis in 90% of CTCs after 4 h of circulation, significantly more than 15 dynes/cm2. |
| 1mm diameter tube, no ECM, 15–60 dyne/cm2. | High shear significantly reduces metastatic potential and drug resistance of breast cancer cells. | ||||
| Uliana [32] | MCF-7 cells, a human breast cancer cell line | Suspended cells | Circulating signals and cells | Disposable microfluidic electrochemical arrays using electrode-bound DNA and antibody-conjugated magnetic beads to detect estrogen receptor alpha in plasma. | 10.0 fg mL−1 detection limit. |
| 1.5 mm diameter electrode, no ECM, 6000 μL/h. | 94.7–108% recovery of estrogen receptor alpha. | ||||
| USD 0.20 cost of manufacture. | |||||
| Vaidyanathan [187] | BT-474 and MDA-MB-231 cancer cells, PC3 prostate cancer cells | Suspended cells | Circulating signals and cells | Multiplexed electrohydrodynamic detection of exosome targets in a chip. | Isolation of exosomal samples from HER2 and prostate-specific antigen. |
| 400 μm wide, 300 μm tall, 25 mm length, no ECM, 420 μL/h. | 8300 exosomes/µL detection sensitivity. | ||||
| Gao [191] | Acellular | Suspended cells | Circulating signals and cells | Multiple detections of miRNAs from multiple samples using three-segment hybridization detection in a microfluidic chip. | Successful detection of four breast cancer biomarker miRNAs. |
| 50 independent channels on a 25 by 75 mm2 chip, no ECM, flow not described. | Detection limit of 1 pM in 30 min. | ||||
| Armbrecht [110] | MCF-7, SK-BR-3, and BR16 cancer cells, LM2 variant cell line | Suspended cells | Organotropism and extravasation | Integrated capture, isolation, and membrane analysis of CTCs in a chip. | 95% isolation efficiency, granulocyte growth-stimulating factor detection efficiency at 1.5 ng/mL−1. |
| 30 independent 75 μm width chambers, no ECM, 12 μL/h. | |||||
| Park [111] | MCF-7 and MDA-MB-231 cancer cells, HL-60 promyelocytic leukemia cell line | Suspended cells | Organotropism and extravasation | Inertial-force-assisted droplet generation using spirals to generate CTC clusters with known cell type ratios. | E-cadherin, VCAM-1, and mRNA expression characterized between cluster compositions. |
| 140 μm width and five-loop structure, no ECM, 1200 μL/h. | |||||
| Riahi [113] | MDA-MB-231 cancer cells, HUVECs, human mammary epithelial cells, MCF7 | Suspended cells, 3D | Organotropism and extravasation | Organ-specific extravasation of CTCs through an endothelial layer using localized chemokine gradients in a chip. | CXCL12 found to significantly increase extravasation. |
| 150 μm by 500 μm by 3 cm, Matrigel matrix, 50 μL/h. | |||||
| Aleman Sarkal [114] | HUVECs, HCT-116 colorectal cancer, HEPG2, A549 | Suspended cells, 3D | Organotropism and extravasation | Organ-specific extravasation of CTCs modeled using bioengineered 3D organoids of five different tissues. | HCT115 CRC cells preferentially extravasate into liver and lung cells, as seen in vivo. |
| 200 μm width, HA/gelatin matrix, 600 μL/h. | |||||
| Song [115] | MDA-MB-231, MCF-10A, and MCF-7 cancer cells, HUVECs, normal human lung fibroblasts | Suspended cells, 3D | Organotropism and extravasation | Microvascular network model to quantify the extravasation of breast cell lines in distinct oxygen conditions. | HIF-1a confirmed through knockout and siRNA to significantly increase the transmigration capacity in breast cell lines and regulate apoptotic-related cellular processes. |
| 1 mm wide and 150 µm deep, fibrin matrix, static conditions. | In hypoxia, HIF-1a levels increased alongside changes in morphology and an increase in cancer viability and metastatic potential. | ||||
| Marturano-Kruik [116] | GFP MDA-MB-231, Firefly MDA-MB-231, HUVECs, human bone marrow mesenchymal stem cells | 3D | Organotropism and extravasation | Perfused bone perivascular niche on a chip to measure progression and drug resistance during metastasis. | Bone-marrow-derived mesenchymal stem cells shown to transition towards perivascular cell lineages and support capillary formation. |
| Dimensions not shown, decellularized bone ECM matrix, 15 μL/h. | Interstitial flow within bone perivascular niche persists with low proliferation and high drug resistance. | ||||
| Jeon [117] | MDA-MB-231 cancer, GFP HUVECs, human bone marrow mesenchymal stem cells | 3D | Organotropism and extravasation. | Microfluidic bone, vascular, and myoblast model to analyze breast cancer organotropism. | Extravasation rate and permeability found to be significantly distinct between bone, myoblast, and unconditioned matrix models. |
| 1.3 mm wide and 200 μm deep, fibrin ECM, 120 μm/h. | A3 adenosine receptor disruption resulted in significantly higher extravasation rates to myoblast-containing matrix | ||||
| Bersini [118] | GFP MDA-MB-231 cancer, RFP HUVECs, human bone marrow mesenchymal stem cells | 3D | Organotropism and extravasation | Osteo-differentiated bone marrow-derived mesenchymal stem cells and endothelial cell bone microenvironment model of organotropism of breast cancer CTCs in a chip. | CTCs extravasated into the bone model 77.5% of the time at a distance of 50.8 µm, compared with 37.6% of the time at a distance of 31.8 µm into collagen control. |
| Eight parallel 225 µm by 150 µm gel regions, collagen hydrogel ECM, static conditions. | Bone secreted signals CXCR2 and CXCL5 found to influence extravasation rate and travel distance. | ||||
| Mei [119] | MDA-MB-231 cancer cells, HUVECs, MLO-Y4 osteocytes, RAW264.7 osteoclasts | 3D | Organotropism and extravasation | Breast cancer, endothelial cell, and osteocyte-like cell model of bone extravasation using oscillatory shear in a chip. | 3.71-fold increase in calcium response in 82.3% of osteocytes compared to continuous flow. |
| 1 mm by 200 μm static channel in addition to a 500 μm by 500 μm vascular model. Channel, collagen, and Matrigel matrix, 1.5 Pa wall shear stress. | Mechanical stimulation reduced extravasation distance 32.4% and frequency by 53.5%. | ||||
| Xu [120] | MDA-MB-231 and M624 cells cancer, primary rat brain microvascular endothelial cells (BMECs), primary rat cerebral astrocytes | 3D | Circulating signals and cells | Blood–brain barrier model in a chip. | Cancer cell and astrocyte interactions increase brain tumor migration between brain and vascular compartments. |
| 200 μm by 400 μm, collagen matrix, 0.1 dyne/cm2 and 60 μL/h. | |||||
| Liu [270] | MCF-7 and MCF-7adm cancer | 3D | Treatment | Five parallel gradient-generating networks on a chip using dam and weir structures for cell positioning and seeding to investigate anti-cancer drugs. | GSH levels of two breast cancer cell lines were reduced during arsenic trioxide treatment, resulting in increased chemotherapy sensitivity, and vice versa was found with N-acetyl cystine. |
| 2 mm by 1 mm by 30 μm, no ECM, 20 μL/h. | |||||
| Parekh [275] | MDA-MB-231 | Suspended particles | Treatment | Single breast cancer cell selection, drug loading, and fluorescence measurement on a chip. | Cyclosporine A found to greatly increase the cellular uptake of anti-cancer drugs by reducing drug efflux. |
| 30 mm by 30 mm chip, no ECM, no flow. | |||||
| Sarkar [271] | MCF-7 | Suspended particles | Treatment | Droplet docking microfluidic microarray to encapsulate single cells to investigate anti-cancer drug influx, efflux, and cytotoxicity. | Confirmation of previous findings and further classification of drug resistance between breast cancer cell lines. |
| No dimensions given, no ECM, no flow. | Observed increased drug resistance during the homotypic fusion of cell-sensitive and resistant cell types within droplets. | ||||
| Zhang [244] | 2A4 hybridoma cells | Suspended particles | Treatment | Integrating chip for screening heavy and light chain combinations in antibodies using single-cell trapping, qPCR, and fluorescence to quantify specificity. | Anti-CD45 monoclonal antibodies identified using hybridoma cells. |
| 50 μm width, no ECM, 180 μL/h. | |||||
| Cheng [233] | T47D and BT549 cancer, primary HUVECs | Spheroids | Treatment | Isolated tumor spheroids trapped next to endothelial cell vascular tissue model to analyze nanoparticle drug delivery systems. | The nanoparticle drug Ds-PEG-FA/DOX found to penetrate spheroids of BT549 cancer but not T47D. |
| 300 μm long, 200 μm wide, 100 μm deep, basement membrane extract matrix, flow rate not specified. | |||||
| Qi [273] | MDA-MB-231 and MDA-MB-231/MDR | Subcellular and 2D | Treatment | Antibody-laden pillars for the specific capture of exosomes for isolation. | CD63-laden exosomes isolated at 70% efficiency. |
| 4 mm channel length, no ECM, 600 μL/h. | Drug content of isolated exosomes found to be significantly different between cell lines and between treatments. | ||||
| Qiao [257] | A549 Human lung adeno-carcinoma, HEK293T human kidney cell line | Suspended particles | Treatment | Microfluidic encapsulation of oncolytic adenovirus and the BET bromodomain inhibitor JQ1 within PVA microgels for injection into tumors. | Found to extend persistence and accumulation of oncolytic adenovirus within tumors. |
| Device dimensions not specified, no ECM, flow rate not specified. | PVA microgels inhibited PD-L1 expression to overcome immune suppression. | ||||
| Ide [252] | BW5147 (H2k), JCRB9002, and BW5147 | Suspended cells | Treatment | Lymph node and T cell–APC interaction model for cell collection and analysis. | Calcium ion flux fluorescent dye in T cells used as metric of activation during serial contact with APCs. |
| 20 μm well diameter, no ECM, 4200 μL/h. | T cell activation during contact with OVA 257–264 peptide presenting quantified APCs. | ||||
| Lou [239] | Bone marrow cells | Suspended particles | Treatment | Tunable liposome formation in a microfluidic channel. | Cationic liposomes of 50–750 nm composed of combinations of tailored phospholipid ratios. |
| Hydrodynamic focusing cartridges of 10 and 65 µm width, no ECM, 900 mL/h. | Macrophage liposome uptake is modulated by area and volume, and biodistribution in mice showed that <50 nm particles increase clearance rate. | ||||
| Liu [240] | HeLa | 3D | Treatment | Continuous flow co-precipitation of polymers to produce injectable “nanosystems”. | Nanosystem loaded with photosensitive zinc successfully localized to cancer cells to enable photodynamic therapy. |
| 200 μm wide and 45 μm deep, no ECM, 7200 μL/h. | |||||
| Lee [259] | MRC5 fibroblasts, human lung cancer A549 cells | Spheroid | Treatment | Cancer, fibroblast, and endothelial spheroid model for oncolytic virus infection on a chip. | Cancer cells transfected substantially more than bystander cells, which is not observed in 2D cell culture |
| 500 μm spheroid chambers, collagen I matrix, 0.3 dyne/cm2. | HUVEC IFN-B secretion is delayed in 2D compared to the microfluidic model. | ||||
| Terrell [248] | MDA-MB-231-HER2+ cancer, CTX-TNA2 rat astrocytes | 3D | Treatment | Blood–brain barrier model in a chip to investigate monoclonal antibody localization. | Tailored antibody trastuzumab found to localize 3% to healthy brain and 5% to tumor model brain. |
| 200 μm width, fibronectin matrix, 60 μL/h. | Rate of uptake quantified as 2.7 × 103 to healthy brain and 1.28 × 104 to cancerous brain. | ||||
| He [238] | Acellular | Suspended particles | Treatment | Hydrodynamic flow for self-assembly of amphiphilic nanoparticles capable of forming a “micelle”. | 500 nm to 2 μm giant vesicles formed. |
| Channel size not described, no ECM, 5400 μL/h. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frankman, Z.D.; Jiang, L.; Schroeder, J.A.; Zohar, Y. Application of Microfluidic Systems for Breast Cancer Research. Micromachines 2022, 13, 152. https://doi.org/10.3390/mi13020152
Frankman ZD, Jiang L, Schroeder JA, Zohar Y. Application of Microfluidic Systems for Breast Cancer Research. Micromachines. 2022; 13(2):152. https://doi.org/10.3390/mi13020152
Chicago/Turabian StyleFrankman, Zachary D., Linan Jiang, Joyce A. Schroeder, and Yitshak Zohar. 2022. "Application of Microfluidic Systems for Breast Cancer Research" Micromachines 13, no. 2: 152. https://doi.org/10.3390/mi13020152
APA StyleFrankman, Z. D., Jiang, L., Schroeder, J. A., & Zohar, Y. (2022). Application of Microfluidic Systems for Breast Cancer Research. Micromachines, 13(2), 152. https://doi.org/10.3390/mi13020152






