3D Bioprinting of an In Vitro Model of a Biomimetic Urinary Bladder with a Contract-Release System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of the dECM Bioink
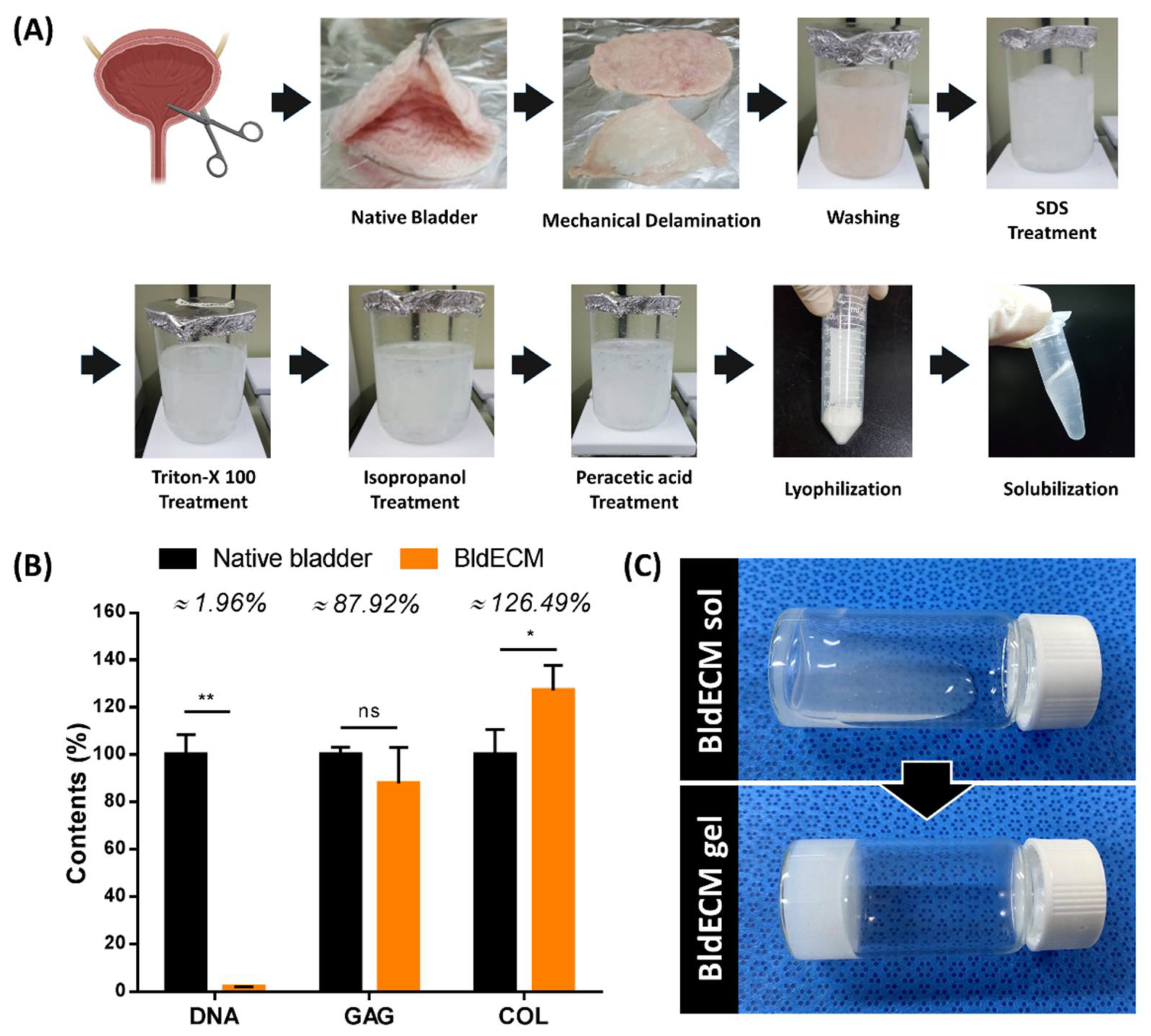
2.1.1. Decellularization Protocol of Bladder Tissue-Derived dECM (BldECM)
2.1.2. Biochemical Assay of BldECM
2.1.3. Preparation of BldECM Bioink
2.2. Cell Isolation and Culture
2.3. Fabrication of the Acrylic-Based Housing Platform
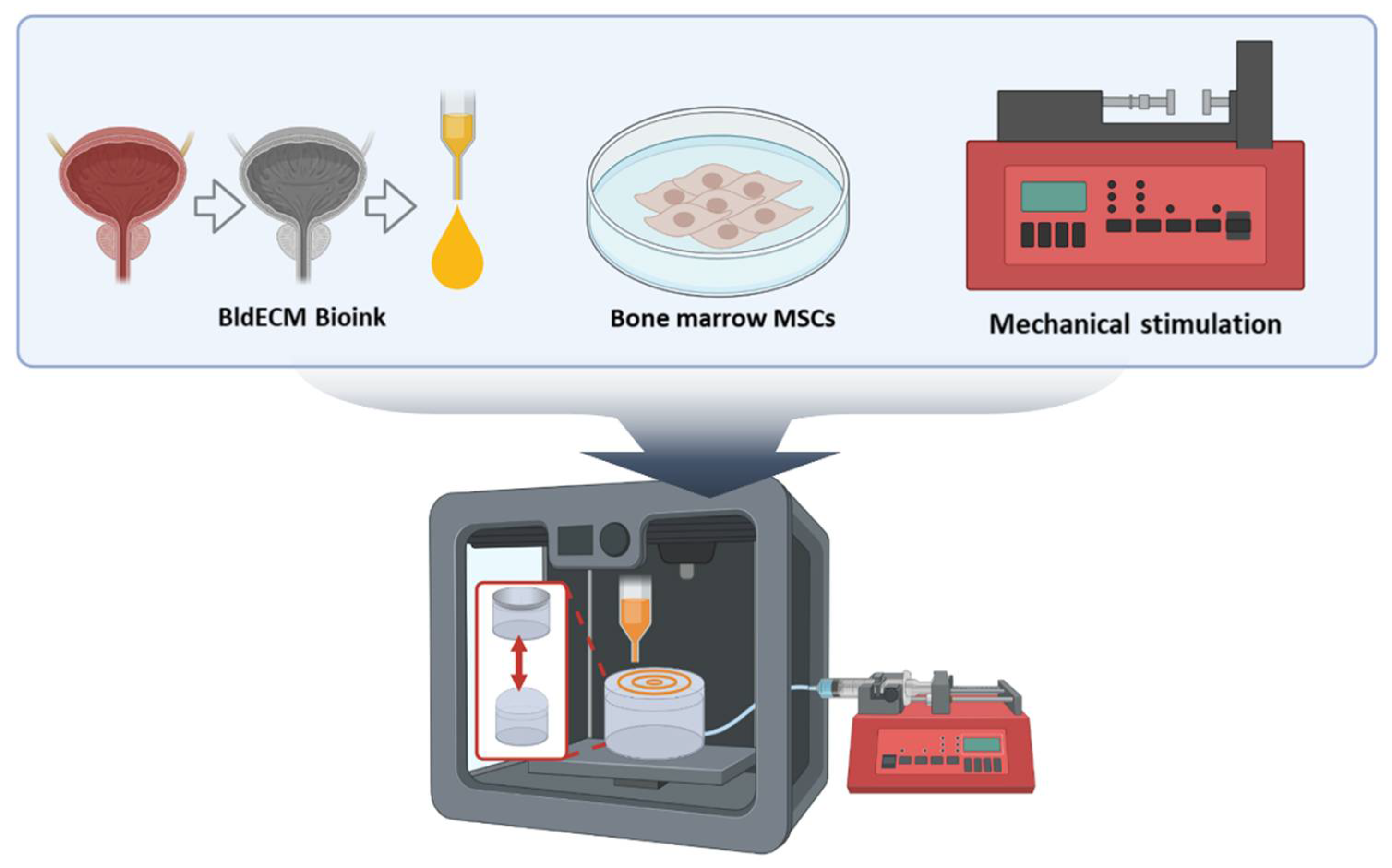
2.4. 3D Bioprinting Process of the In vitro Bladder Model
2.5. Establishing a Contract-Release System for Physiological Stimulation
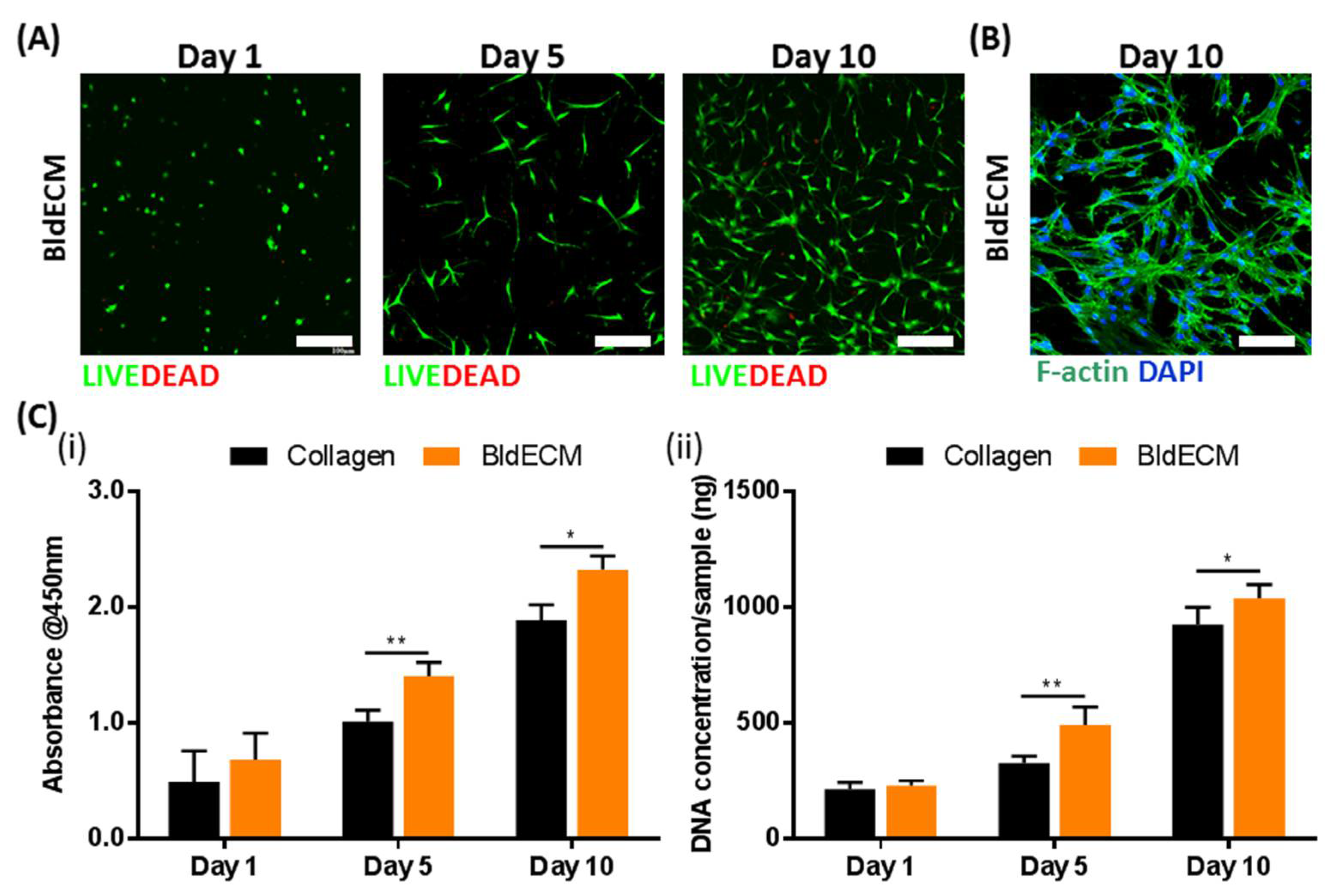
2.6. Live/Dead Assays
2.7. Cell Proliferation Rate
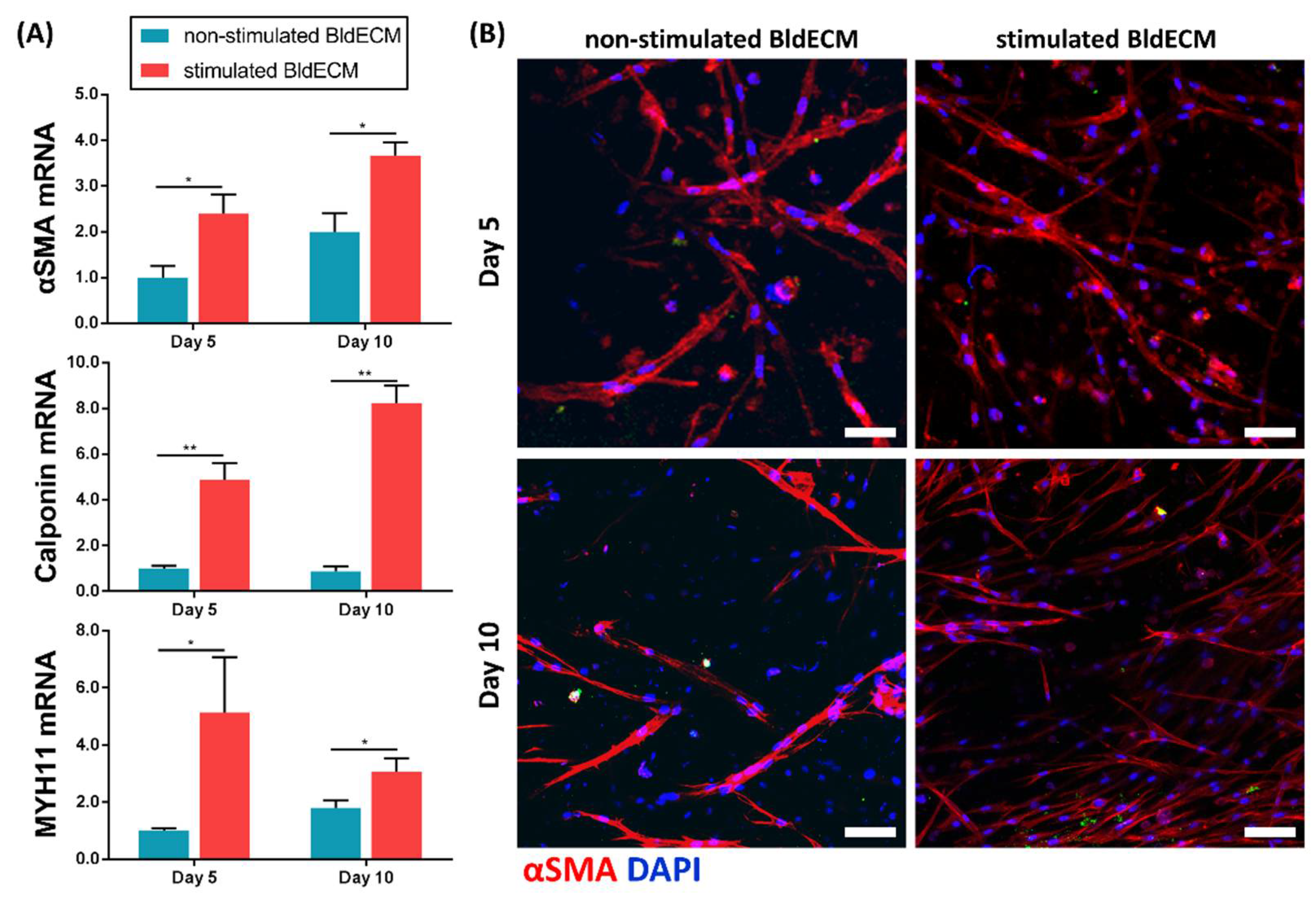
2.8. Real-Time Polymerase Chain Reactions
2.9. Immunofluorescence Staining
2.10. Statistical Analysis
3. Results and Discussion
3.1. Preparation of the Bladder-Derived dECM Bioink
3.2. Fabrication of a Bladder Mimicry Platform
3.3. Evaluation of the Biofunctionality of the Stem-Cell Laden Bladder dECM Bioink
3.4. Effect of Physiological Stimuli on MSC Myogenesis in the Engineered In Vitro Bladder Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hicks, R.M. The mammalian urinary bladderan accommodating organ. Biol. Rev. 1975, 50, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Baskin, L.S.; Howard, P.S.; Duckett, J.W.; Snyder, H.M.; Macarak, E.J. Bladder Smooth Muscle Cells in Culture: I. Identification and Characterization. J. Urol. 1993, 149, 190–197. [Google Scholar] [CrossRef]
- Baskin, L.; Howard, P.S.; Macarak, E. Effect of Physical Forces on Bladder Smooth Muscle and Urothelium. J. Urol. 1993, 150, 601–607. [Google Scholar] [CrossRef]
- Drake, M.J.; Williams, J.; Bijos, D.A. Voiding dysfunction due to detrusor underactivity: An overview. Nat. Rev. Urol. 2014, 11, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Hoag, N.; Gani, J. Underactive Bladder: Clinical Features, Urodynamic Parameters, and Treatment. Int. Neurourol. J. 2015, 19, 185–189. [Google Scholar] [CrossRef]
- Serrano-Aroca, Á.; Vera-Donoso, C.D.; Moreno-Manzano, V. Bioengineering Approaches for Bladder Regeneration. Int. J. Mol. Sci. 2018, 19, 1796. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-chip: Recent breakthroughs and future prospects. BioMed. Eng. Online 2020, 19, 9. [Google Scholar] [CrossRef] [Green Version]
- Huh, D.; Matthews Benjamin, D.; Mammoto, A.; Montoya-Zavala, M.; Hsin Hong, Y.; Ingber Donald, E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Yi, H.-G.; Kim, H.; Kwon, J.; Choi, Y.-J.; Jang, J.; Cho, D.-W. Application of 3D bioprinting in the prevention and the therapy for human diseases. Signal Transduct. Target. Ther. 2021, 6, 177. [Google Scholar] [CrossRef]
- Lee, S.; Chung, M.; Lee, S.-R.; Jeon, N.L. 3D brain angiogenesis model to reconstitute functional human blood–brain barrier in vitro. Biotechnol. Bioeng. 2020, 117, 748–762. [Google Scholar] [CrossRef] [PubMed]
- Lind, J.U.; Busbee, T.A.; Valentine, A.D.; Pasqualini, F.S.; Yuan, H.; Yadid, M.; Park, S.-J.; Kotikian, A.; Nesmith, A.P.; Campbell, P.H.; et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 2017, 16, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.Y.C.; Homan, K.A.; Robinson, S.S.; Kolesky, D.B.; Duarte, N.; Moisan, A.; Lewis, J.A. Renal reabsorption in 3D vascularized proximal tubule models. Proc. Natl. Acad. Sci. USA 2019, 116, 5399. [Google Scholar] [CrossRef] [Green Version]
- Wufuer, M.; Lee, G.; Hur, W.; Jeon, B.; Kim, B.J.; Choi, T.H.; Lee, S. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016, 6, 37471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffith, L.G.; Naughton, G. Tissue Engineering—Current Challenges and Expanding Opportunities. Science 2002, 295, 1009–1014. [Google Scholar] [CrossRef]
- Choi, Y.-J.; Park, H.; Ha, D.-H.; Yun, H.-S.; Yi, H.-G.; Lee, H. 3D Bioprinting of In Vitro Models Using Hydrogel-Based Bioinks. Polymers 2021, 13, 366. [Google Scholar] [CrossRef]
- Ha, D.-H.; Chae, S.; Lee, J.Y.; Kim, J.Y.; Yoon, J.; Sen, T.; Lee, S.-W.; Kim, H.J.; Cho, J.H.; Cho, D.-W. Therapeutic effect of decellularized extracellular matrix-based hydrogel for radiation esophagitis by 3D printed esophageal stent. Biomaterials 2021, 266, 120477. [Google Scholar] [CrossRef]
- Kim, B.S.; Das, S.; Jang, J.; Cho, D.-W. Decellularized Extracellular Matrix-based Bioinks for Engineering Tissue- and Organ-specific Microenvironments. Chem. Rev. 2020, 120, 10608–10661. [Google Scholar] [CrossRef]
- Pati, F.; Jang, J.; Ha, D.-H.; Won Kim, S.; Rhie, J.-W.; Shim, J.-H.; Kim, D.-H.; Cho, D.-W. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat. Commun. 2014, 5, 3935. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Wang, X.; Yan, Y.; Zheng, W.; Xiong, Z.; Lin, F.; Wu, R.; Zhang, R. Rapid Prototyping Three-Dimensional Cell/Gelatin/Fibrinogen Constructs for Medical Regeneration. J. Bioact. Compat. Polym. 2007, 22, 363–377. [Google Scholar] [CrossRef]
- Jia, J.; Richards, D.J.; Pollard, S.; Tan, Y.; Rodriguez, J.; Visconti, R.P.; Trusk, T.C.; Yost, M.J.; Yao, H.; Markwald, R.R.; et al. Engineering alginate as bioink for bioprinting. Acta Biomater. 2014, 10, 4323–4331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashammakhi, N.; Ahadian, S.; Xu, C.; Montazerian, H.; Ko, H.; Nasiri, R.; Barros, N.; Khademhosseini, A. Bioinks and bioprinting technologies to make heterogeneous and biomimetic tissue constructs. Mater. Today Bio 2019, 1, 100008. [Google Scholar] [CrossRef]
- Moreno-Manzano, V.; Zaytseva-Zotova, D.; López-Mocholí, E.; Briz-Redón, Á.; Løkensgard Strand, B.; Serrano-Aroca, Á. Injectable Gel Form of a Decellularized Bladder Induces Adipose-Derived Stem Cell Differentiation into Smooth Muscle Cells In Vitro. Int. J. Mol. Sci. 2020, 21, 8608. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Manzano, V.; Mellado-López, M.; Morera-Esteve, M.J.; Alastrue-Agudo, A.; Bisbal-Velasco, V.; Forteza-Vila, J.; Serrano-Aroca, Á.; Vera-Donoso, C.D. Human adipose-derived mesenchymal stem cells accelerate decellularized neobladder regeneration. Regen. Biomater. 2020, 7, 161–169. [Google Scholar] [CrossRef]
- Chae, S.; Lee, S.-S.; Choi, Y.-J.; Hong, D.H.; Gao, G.; Wang, J.H.; Cho, D.-W. 3D cell-printing of biocompatible and functional meniscus constructs using meniscus-derived bioink. Biomaterials 2021, 267, 120466. [Google Scholar] [CrossRef]
- Wazir, R.; Luo, D.-Y.; Dai, Y.; Yue, X.; Tian, Y.; Wang, K.-J. Expression and proliferation profiles of PKC, JNK and p38MAPK in physiologically stretched human bladder smooth muscle cells. Biochem. Biophys. Res. Commun. 2013, 438, 479–482. [Google Scholar] [CrossRef]
- Wazir, R.; Luo, D.-Y.; Tian, Y.; Yue, X.; Li, H.; Wang, K.-J. The purinergic component of human bladder smooth muscle cells’ proliferation and contraction under physiological stretch. Biochem. Biophys. Res. Commun. 2013, 437, 256–260. [Google Scholar] [CrossRef]
- Halper, J.; Kjaer, M. Basic Components of Connective Tissues and Extracellular Matrix: Elastin, Fibrillin, Fibulins, Fibrinogen, Fibronectin, Laminin, Tenascins and Thrombospondins. In Progress in Heritable Soft Connective Tissue Diseases; Halper, J., Ed.; Springer: Dordrecht, The Netherlands, 2014; pp. 31–47. [Google Scholar]
- Choudhury, D.; Tun, H.W.; Wang, T.; Naing, M.W. Organ-Derived Decellularized Extracellular Matrix: A Game Changer for Bioink Manufacturing? Trends Biotechnol. 2018, 36, 787–805. [Google Scholar] [CrossRef]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Shim, I.K.; Hwang, D.G.; Lee, Y.N.; Kim, M.; Kim, H.; Kim, S.-W.; Lee, S.; Kim, S.C.; Cho, D.-W.; et al. 3D cell printing of islet-laden pancreatic tissue-derived extracellular matrix bioink constructs for enhancing pancreatic functions. J. Mater. Chem. B 2019, 7, 1773–1781. [Google Scholar] [CrossRef]
- Saldin, L.T.; Cramer, M.C.; Velankar, S.S.; White, L.J.; Badylak, S.F. Extracellular matrix hydrogels from decellularized tissues: Structure and function. Acta Biomater. 2017, 49, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fry, C.H.; McCloskey, K.D. Spontaneous Activity and the Urinary Bladder. In Smooth Muscle Spontaneous Activity: Physiological and Pathological Modulation; Hashitani, H., Lang, R.J., Eds.; Springer: Singapore, 2019; pp. 121–147. [Google Scholar]
- Jang, T.-M.; Lee Joong, H.; Zhou, H.; Joo, J.; Lim Bong, H.; Cheng, H.; Kim Soo, H.; Kang, I.-S.; Lee, K.-S.; Park, E.; et al. Expandable and implantable bioelectronic complex for analyzing and regulating real-time activity of the urinary bladder. Sci. Adv. 2020, 6, eabc9675. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Yoon, J.J.; Park, E.K.; Kim, D.S.; Kim, S.-Y.; Cho, D.-W. Cell adhesion and proliferation evaluation of SFF-based biodegradable scaffolds fabricated using a multi-head deposition system. Biofabrication 2009, 1, 015002. [Google Scholar] [CrossRef]
- Sharma, K.; Dhar, N.; Thacker, V.V.; Simonet, T.M.; Signorino-Gelo, F.; Knott, G.W.; McKinney, J.D. Dynamic persistence of UPEC intracellular bacterial communities in a human bladder-chip model of urinary tract infection. eLife 2021, 10, e66481. [Google Scholar] [CrossRef] [PubMed]
- Chae, S.; Sun, Y.; Choi, Y.-J.; Ha, D.-H.; Jeon, I.; Cho, D.-W. 3D cell-printing of tendon-bone interface using tissue-derived extracellular matrix bioinks for chronic rotator cuff repair. Biofabrication 2021, 13, 035005. [Google Scholar] [CrossRef]
- Caddeo, S.; Boffito, M.; Sartori, S. Tissue Engineering Approaches in the Design of Healthy and Pathological In Vitro Tissue Models. Front. Bioeng. Biotechnol. 2017, 5, 40. [Google Scholar] [CrossRef] [Green Version]

| Primer | Sequence | |
|---|---|---|
| GAPDH | Forward | 5′ CCTGGAGAAACCTGCCAAGTAT 3′ |
| Reverse | 5′ CTCGGCCGCCTGCTT 3′ | |
| αSMA 1 | Forward | 5′ TGCCTGATGGGCAAGTGAT 3′ |
| Reverse | 5′ TCTCTGGGCAGCGGAAAC 3′ | |
| Calponin | Forward | 5′ GCCCGCCCACAACCA 3′ |
| Reverse | 5′ GTGGCCCTAGGCGGAATT 3′ | |
| MYH11 2 | Forward | 5′ ACAACCTGAGGGAGCGGTACT 3′ |
| Reverse | 5′ CACGCAGAAGAGGCCAGAGT 3′ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chae, S.; Kim, J.; Yi, H.-G.; Cho, D.-W. 3D Bioprinting of an In Vitro Model of a Biomimetic Urinary Bladder with a Contract-Release System. Micromachines 2022, 13, 277. https://doi.org/10.3390/mi13020277
Chae S, Kim J, Yi H-G, Cho D-W. 3D Bioprinting of an In Vitro Model of a Biomimetic Urinary Bladder with a Contract-Release System. Micromachines. 2022; 13(2):277. https://doi.org/10.3390/mi13020277
Chicago/Turabian StyleChae, Suhun, Jaewook Kim, Hee-Gyeong Yi, and Dong-Woo Cho. 2022. "3D Bioprinting of an In Vitro Model of a Biomimetic Urinary Bladder with a Contract-Release System" Micromachines 13, no. 2: 277. https://doi.org/10.3390/mi13020277







