Novel Quick Cell Patterning Using Light-Responsive Gas-Generating Polymer and Fluorescence Microscope
Abstract
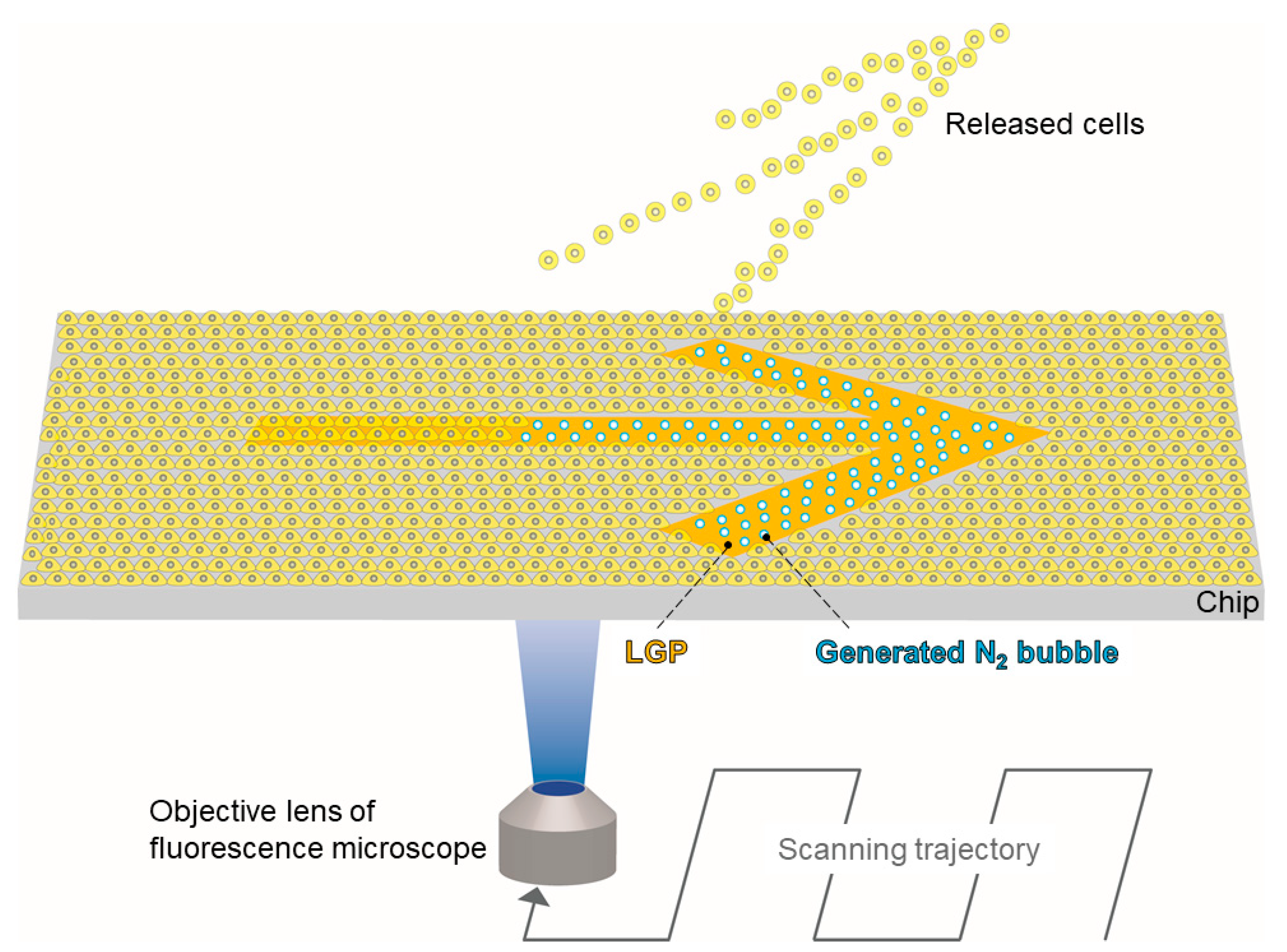
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Fabrication Method for LGP-Implanted Chip
2.3. Cell Releasing from the LGP-Implanted Chip Coated with Fibronectin, Collagen, and Poly-D-Lysine
3. Results and Discussion
3.1. Characterization of LGP-Implanted Chip
3.2. Cell Patterning by LGP-Implanted Chip Coated by Fibronectin, Collagen, and Poly-D-Lysine
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Théry, M. Micropatterning as a Tool to Decipher Cell Morphogenesis and Functions. J. Cell Sci. 2010, 123, 4201–4213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, M.; Racine, V.; Piel, M.; Pepin, A.; Dimitrov, A.; Chen, Y.; Sibarita, J.-B.; Bornens, M. Anisotropy of Cell Adhesive Microenvironment Governs Cell Internal Organization and Orientation of Polarity. Proc. Natl. Acad. Sci. USA 2006, 103, 19771–19776. [Google Scholar] [CrossRef] [Green Version]
- Théry, M.; Racine, V.; Pépin, A.; Piel, M.; Chen, Y.; Sibarita, J.-B.; Bornens, M. The Extracellular Matrix Guides the Orientation of the Cell Division Axis. Nat. Cell Biol. 2005, 7, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Sazan, H.; Kenigsberg, A.; Schori, H.; Piperno, S.; Shpaisman, H.; Shefi, O. Large-Scale Acoustic-Driven Neuronal Patterning and Directed Outgrowth. Sci. Rep. 2020, 10, 4932. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Ikeda, K.; Yamaguchi, M.; Kudoh, S.N.; Yokoyama, K.; Satoh, R.; Ito, D.; Nagayama, M.; Uchida, T.; Gohara, K. Neuronal Cell Patterning on a Multi-Electrode Array for a Network Analysis Platform. Biomaterials 2013, 34, 5210–5217. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Kato-Negishi, M.; Takeuchi, S. Assembly and Connection of Micropatterned Single Neurons for Neuronal Network Formation. Micromachines 2018, 9, 235. [Google Scholar] [CrossRef] [Green Version]
- Tenje, M.; Cantoni, F.; Porras Hernández, A.M.; Searle, S.S.; Johansson, S.; Barbe, L.; Antfolk, M.; Pohlit, H. A Practical Guide to Microfabrication and Patterning of Hydrogels for Biomimetic Cell Culture Scaffolds. Organs-A-Chip 2020, 2, 100003. [Google Scholar] [CrossRef]
- Tian, C.; Tu, Q.; Liu, W.; Wang, J. Recent Advances in Microfluidic Technologies for Organ-on-a-Chip. TrAC Trends Anal. Chem. 2019, 117, 146–156. [Google Scholar] [CrossRef]
- Esch, M.B.; Ueno, H.; Applegate, D.R.; Shuler, M.L. Modular, Pumpless Body-on-a-Chip Platform for the Co-Culture of GI Tract Epithelium and 3D Primary Liver Tissue. Lab Chip 2016, 16, 2719–2729. [Google Scholar] [CrossRef]
- Chen, L.; Yang, Y.; Ueno, H.; Esch, M.B. Body-in-a-Cube: A Microphysiological System for Multi-Tissue Co-Culture with near-Physiological Amounts of Blood Surrogate. Microphysiol. Syst. 2020, 4, 1. [Google Scholar] [CrossRef]
- Natarajan, A.; Stancescu, M.; Dhir, V.; Armstrong, C.; Sommerhage, F.; Hickman, J.J.; Molnar, P. Patterned Cardiomyocytes on Microelectrode Arrays as a Functional, High Information Content Drug Screening Platform. Biomaterials 2011, 32, 4267–4274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, N.; Sekine, R.; Funano, S.; Sato, A.; Carretero, N.T.; Ebisuya, M.; Tanaka, Y. Vacuum Microcasting of 2-Methacryloyloxyethyl Phosphorylcholine Polymer for Stable Cell Patterning. BioTechniques 2020, 69, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Yamashita, T.; Sato, A.; Vogel, V.; Tanaka, Y. Simple Agarose Micro-Confinement Array and Machine-Learning-Based Classification for Analyzing the Patterned Differentiation of Mesenchymal Stem Cells. PLoS ONE 2017, 12, e0173647. [Google Scholar] [CrossRef] [PubMed]
- Hacohen, A.; Jessel, H.R.; Richter-Levin, A.; Shefi, O. Patterning of Particles and Live Cells at Single Cell Resolution. Micromachines 2020, 11, 505. [Google Scholar] [CrossRef]
- Shigeto, H.; Yamada, E.; Kitamatsu, M.; Ohtsuki, T.; Iizuka, A.; Akiyama, Y.; Yamamura, S. Analysis of Single Nucleotide-Mutated Single-Cancer Cells Using the Combined Technologies of Single-Cell Microarray Chips and Peptide Nucleic Acid-DNA Probes. Micromachines 2020, 11, 628. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Kishi, H.; Tokimitsu, Y.; Kondo, S.; Honda, R.; Rao, S.R.; Omori, M.; Tamiya, E.; Muraguchi, A. Single-Cell Microarray for Analyzing Cellular Response. Anal. Chem. 2005, 77, 8050–8056. [Google Scholar] [CrossRef]
- Yamamura, S.; Yamada, E.; Kimura, F.; Miyajima, K.; Shigeto, H. Separation and Analysis of Adherent and Non-Adherent Cancer Cells Using a Single-Cell Microarray Chip. Sensors 2017, 17, 2410. [Google Scholar] [CrossRef] [Green Version]
- Ishizaki, T.; Saito, N.; Takai, O. Correlation of Cell Adhesive Behaviors on Superhydrophobic, Superhydrophilic, and Micropatterned Superhydrophobic/Superhydrophilic Surfaces to Their Surface Chemistry. Langmuir 2010, 26, 8147–8154. [Google Scholar] [CrossRef]
- Tu, C.; Huang, B.; Zhou, J.; Liang, Y.; Tian, J.; Ji, L.; Liang, X.; Ye, X. A Microfluidic Chip for Cell Patterning Utilizing Paired Microwells and Protein Patterns. Micromachines 2016, 8, 1. [Google Scholar] [CrossRef]
- Yang, T.; Gao, D.; Jin, F.; Jiang, Y.; Liu, H. Surface-Printed Microdot Array Chips Coupled with Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry for High-Throughput Single-Cell Patterning and Phospholipid Analysis: Surface-Printed Microdot Array Chips Coupled with MALDI-TOF MS. Rapid Commun. Mass Spectrom. 2016, 30, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Fu, W.; Sun, M.; Han, K.; Hu, R.; Liu, D.; Wang, J. Straightforward Neuron Micropatterning and Neuronal Network Construction on Cell-Repellent Polydimethylsiloxane Using Microfluidics-Guided Functionalized Pluronic Modification. Analyst 2021, 146, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Imanishi, Y.; Ito, Y. Effect of Protein and Cell Behavior on Pattern-Grafted Thermoresponsive Polymer. J. Biomed. Mater. Res. 1998, 42, 38–44. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Ikeda, K.; Suzuki, M.; Kiyohara, A.; Kudoh, S.N.; Shimizu, K.; Taira, T.; Ito, D.; Uchida, T.; Gohara, K. Cell Patterning Using a Template of Microstructured Organosilane Layer Fabricated by Vacuum Ultraviolet Light Lithography. Langmuir 2011, 27, 12521–12532. [Google Scholar] [CrossRef] [PubMed]
- Yamazoe, H.; Uemura, T.; Tanabe, T. Facile Cell Patterning on an Albumin-Coated Surface. Langmuir 2008, 24, 8402–8404. [Google Scholar] [CrossRef] [PubMed]
- Dabaghi, M.; Shahriari, S.; Saraei, N.; Da, K.; Chandiramohan, A.; Selvaganapathy, P.R.; Hirota, J.A. Surface Modification of PDMS-Based Microfluidic Devices with Collagen Using Polydopamine as a Spacer to Enhance Primary Human Bronchial Epithelial Cell Adhesion. Micromachines 2021, 12, 132. [Google Scholar] [CrossRef]
- Kang, G.; Lee, J.-H.; Lee, C.-S.; Nam, Y. Agarose Microwell Based Neuronal Micro-Circuit Arrays on Microelectrode Arrays for High Throughput Drug Testing. Lab Chip 2009, 9, 3236. [Google Scholar] [CrossRef]
- Männel, M.; Fischer, C.; Thiele, J. A Non-Cytotoxic Resin for Micro-Stereolithography for Cell Cultures of HUVECs. Micromachines 2020, 11, 246. [Google Scholar] [CrossRef] [Green Version]
- Richardson, S.A.; Rawlings, T.M.; Muter, J.; Walker, M.; Brosens, J.J.; Cameron, N.R.; Eissa, A.M. Covalent Attachment of Fibronectin onto Emulsion-Templated Porous Polymer Scaffolds Enhances Human Endometrial Stromal Cell Adhesion, Infiltration, and Function. Macromol. Biosci. 2019, 19, 1800351. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; He, W.; Huang, Q.; Zhang, R.; Feng, Q. Hydroxyapatite/Collagen Coating on PLGA Electrospun Fibers for Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells: Hydroxyapatite/Collagen Coating on PLGA Electrospun Fibers. J. Biomed. Mater. Res. 2018, 106, 2863–2870. [Google Scholar] [CrossRef]
- Kim, Y.H.; Baek, N.S.; Han, Y.H.; Chung, M.-A.; Jung, S.-D. Enhancement of Neuronal Cell Adhesion by Covalent Binding of Poly-D-lysine. J. Neurosci. Methods 2011, 202, 38–44. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Takasaki, Y.; Yamahira, S.; Nagamune, T. Photo-Cleavable Peptide-Poly(Ethylene Glycol) Conjugate Surfaces for Light-Guided Control of Cell Adhesion. Micromachines 2020, 11, 762. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Yamahira, S.; Kikuchi, K.; Sumaru, K.; Kanamori, T.; Nagamune, T. Photocontrollable Dynamic Micropatterning of Non-Adherent Mammalian Cells Using a Photocleavable Poly(Ethylene Glycol) Lipid. Angew. Chem. Int. Ed. 2012, 51, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Akagi, Y.; Matsumoto, S.; Yamamura, S. Control of Cell Adhesion and Detachment on a Nanostructured Scaffold Composed of Light-responsive Gas-generating Film. Sens. Mater. 2019, 31, 89–98. [Google Scholar]
- Akagi, Y.; Sugita, D.; Yamamoto, K. The Photoresponsive Micropump Tape Using Gas Generation Technology. Electron. Commun. Jpn. 2020, 103, 3–8. [Google Scholar] [CrossRef]
- Yatsushiro, S.; Yamamura, S.; Yamaguchi, Y.; Shinohara, Y.; Tamiya, E.; Horii, T.; Baba, Y.; Kataoka, M. Rapid and Highly Sensitive Detection of Malaria-Infected Erythrocytes Using a Cell Microarray Chip. PLoS ONE 2010, 5, e13179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamura, S.; Yatsushiro, S.; Yamaguchi, Y.; Abe, K.; Shinohara, Y.; Tamiya, E.; Baba, Y.; Kataoka, M. Accurate Detection of Carcinoma Cells by Use of a Cell Microarray Chip. PLoS ONE 2012, 7, e32370. [Google Scholar] [CrossRef]
- Yatsushiro, S.; Yamamoto, T.; Yamamura, S.; Abe, K.; Obana, E.; Nogami, T.; Hayashi, T.; Sesei, T.; Oka, H.; Okello-Onen, J.; et al. Application of a Cell Microarray Chip System for Accurate, Highly Sensitive and Rapid Diagnosis for Malaria in Uganda. Sci. Rep. 2016, 6, 30136. [Google Scholar] [CrossRef]
- Murali, M.; Yeo, S.H. Rapid Biocompatible Micro Device Fabrication by Micro Electro-Discharge Machining. Biomed. Microdevices 2004, 6, 41–45. [Google Scholar] [CrossRef]
- Bueeler, M.; Spoerl, E.; Seiler, T.; Mrochen, M. UV collagen cross-linking of the cornea-Safety aspects and design of a UV illumination system. In Proceedings of the SPIE-The International Society for Optical Engineering, San Jose, CA, USA, 25 February 2008. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ueno, H.; Akagi, Y.; Yamamura, S. Novel Quick Cell Patterning Using Light-Responsive Gas-Generating Polymer and Fluorescence Microscope. Micromachines 2022, 13, 320. https://doi.org/10.3390/mi13020320
Ueno H, Akagi Y, Yamamura S. Novel Quick Cell Patterning Using Light-Responsive Gas-Generating Polymer and Fluorescence Microscope. Micromachines. 2022; 13(2):320. https://doi.org/10.3390/mi13020320
Chicago/Turabian StyleUeno, Hidetaka, Yoshinori Akagi, and Shohei Yamamura. 2022. "Novel Quick Cell Patterning Using Light-Responsive Gas-Generating Polymer and Fluorescence Microscope" Micromachines 13, no. 2: 320. https://doi.org/10.3390/mi13020320
APA StyleUeno, H., Akagi, Y., & Yamamura, S. (2022). Novel Quick Cell Patterning Using Light-Responsive Gas-Generating Polymer and Fluorescence Microscope. Micromachines, 13(2), 320. https://doi.org/10.3390/mi13020320





