Design and Fabrication of a 3D-Printed Microfluidic Immunoarray for Ultrasensitive Multiplexed Protein Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
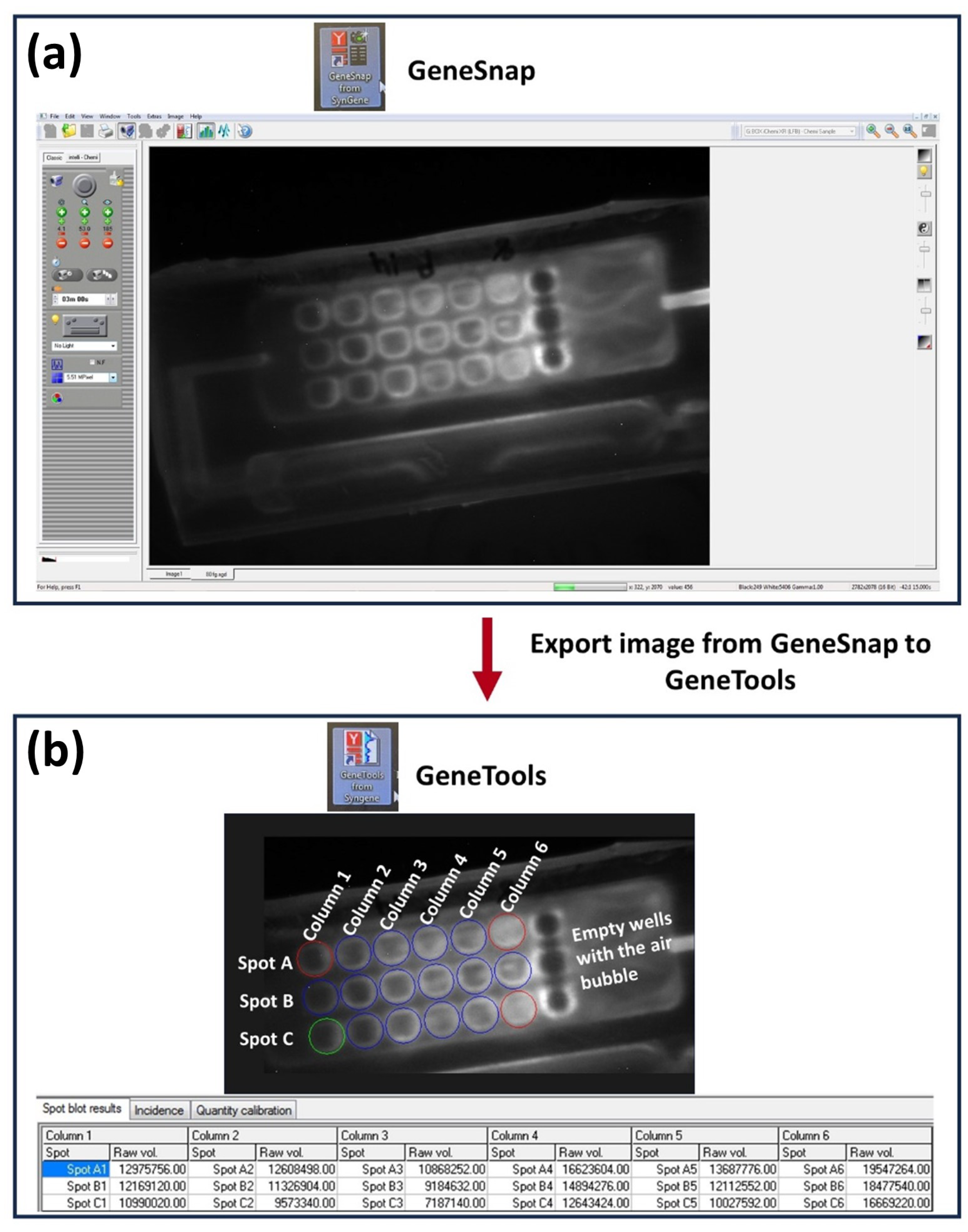
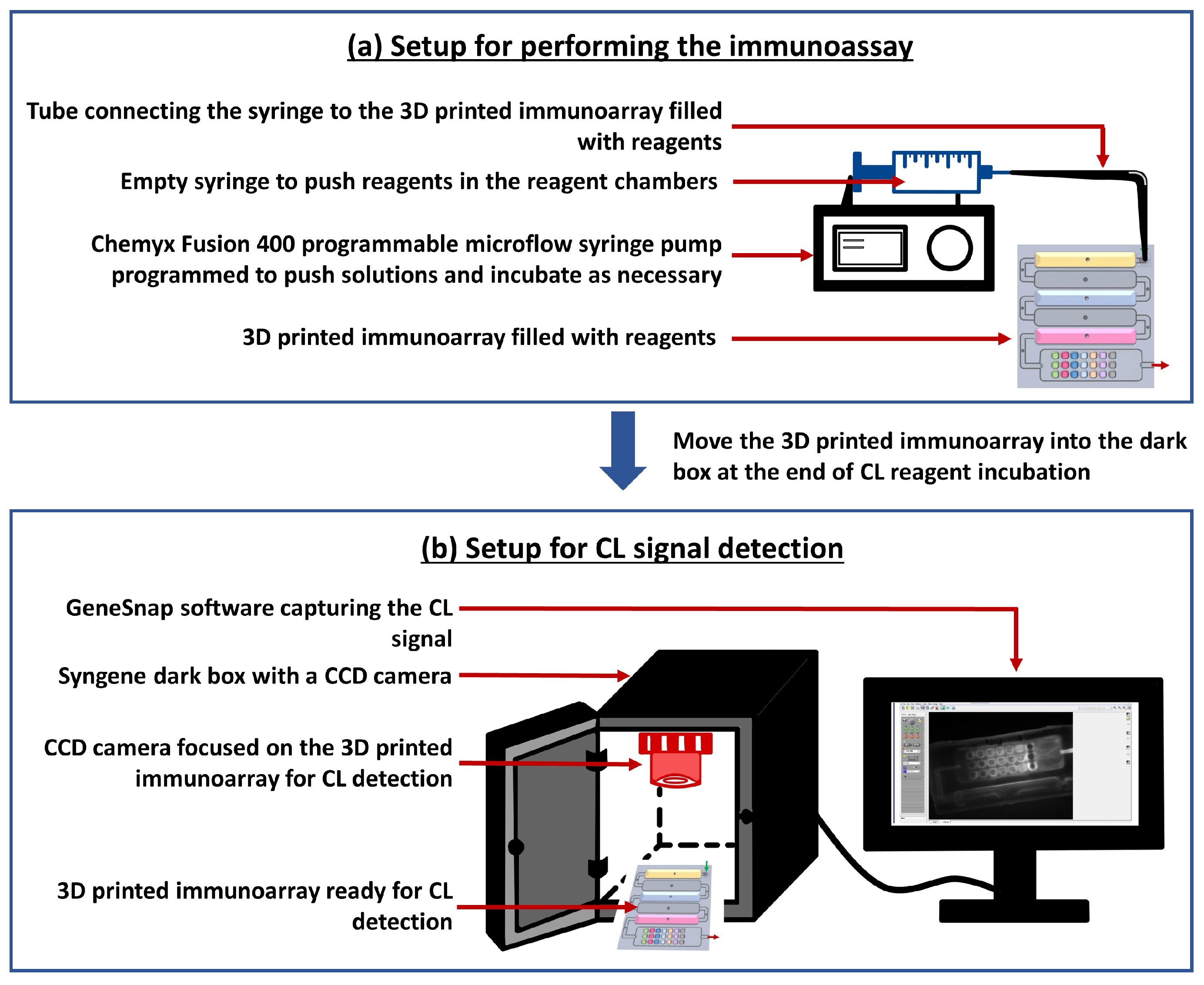
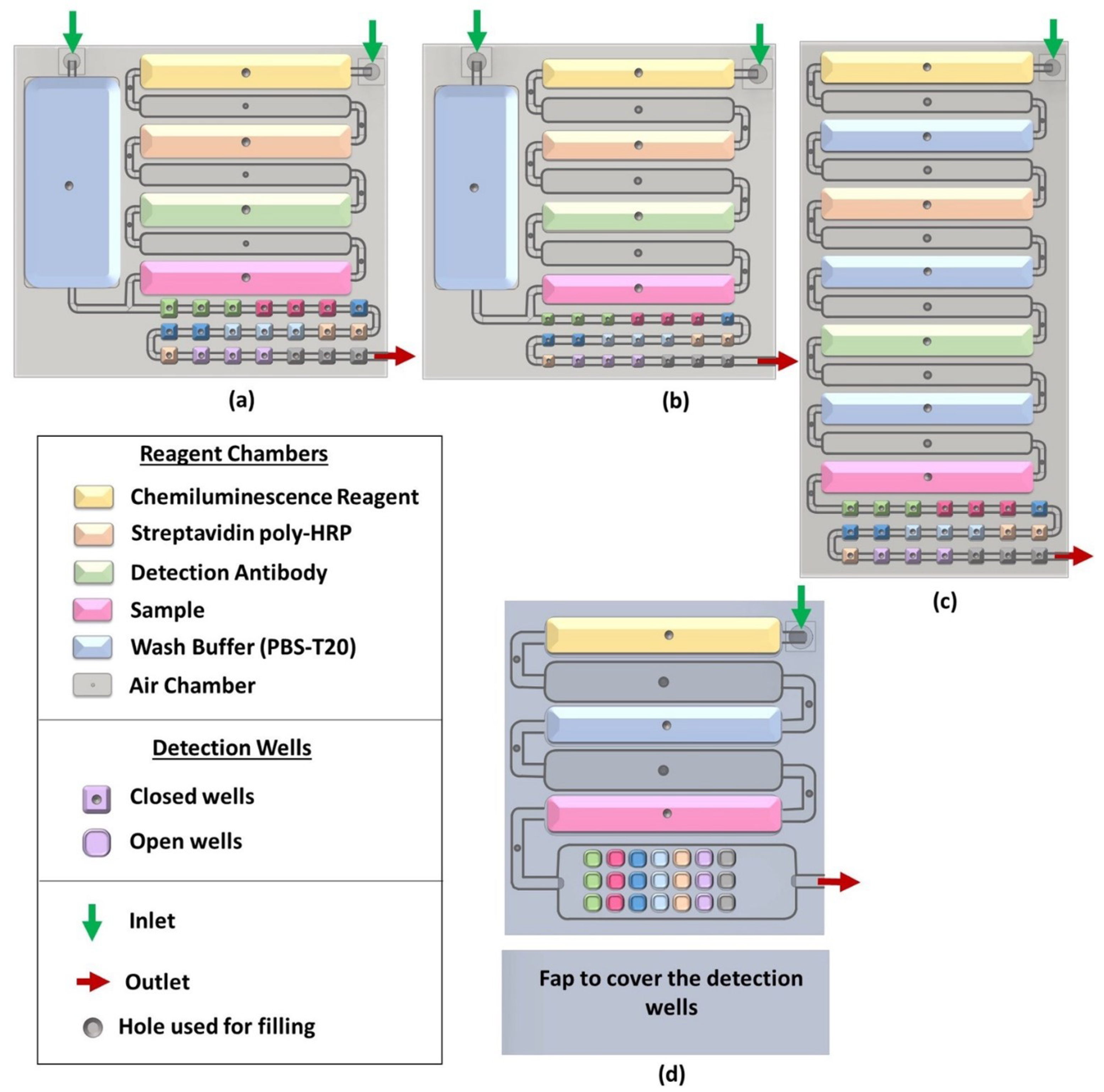
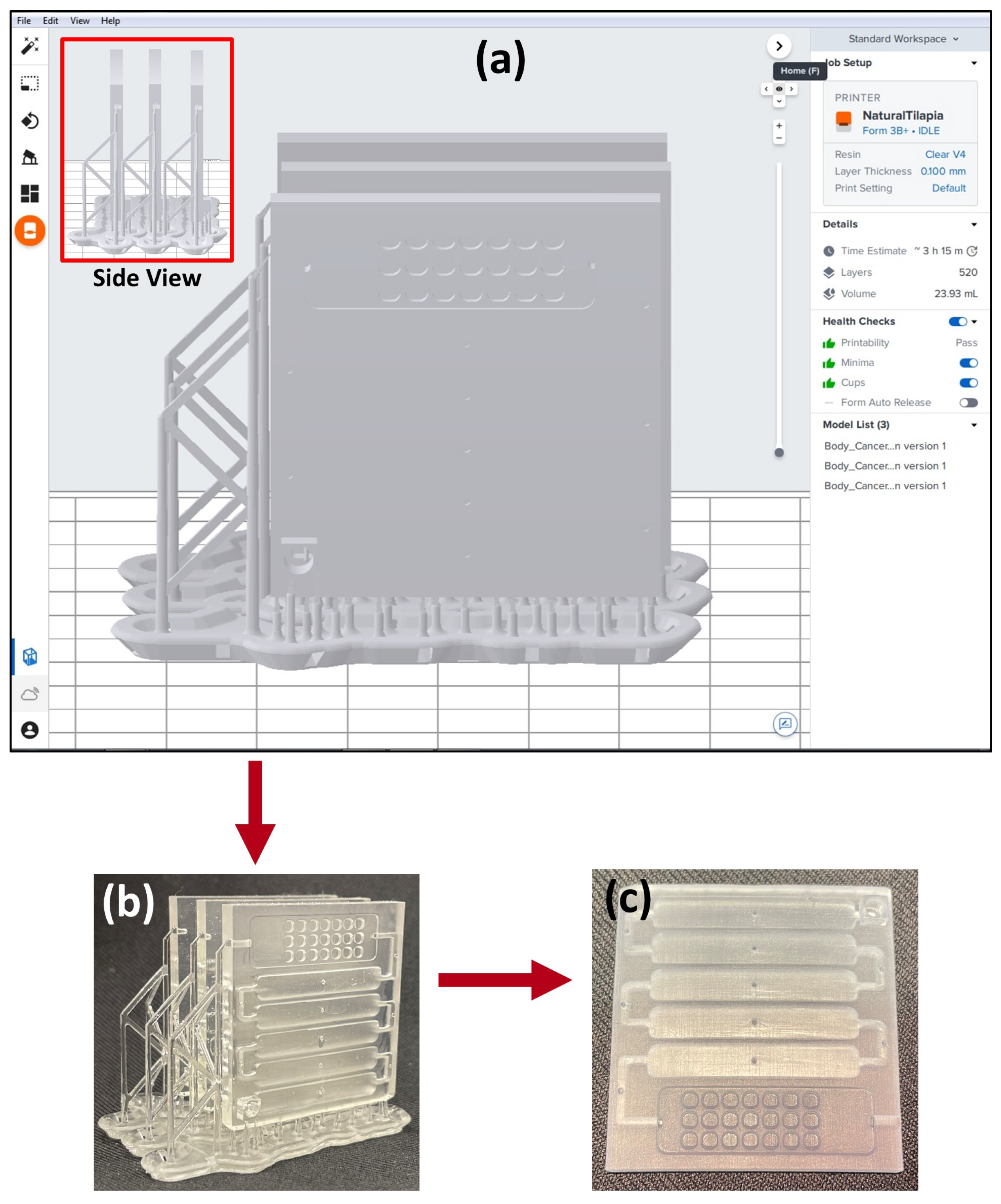
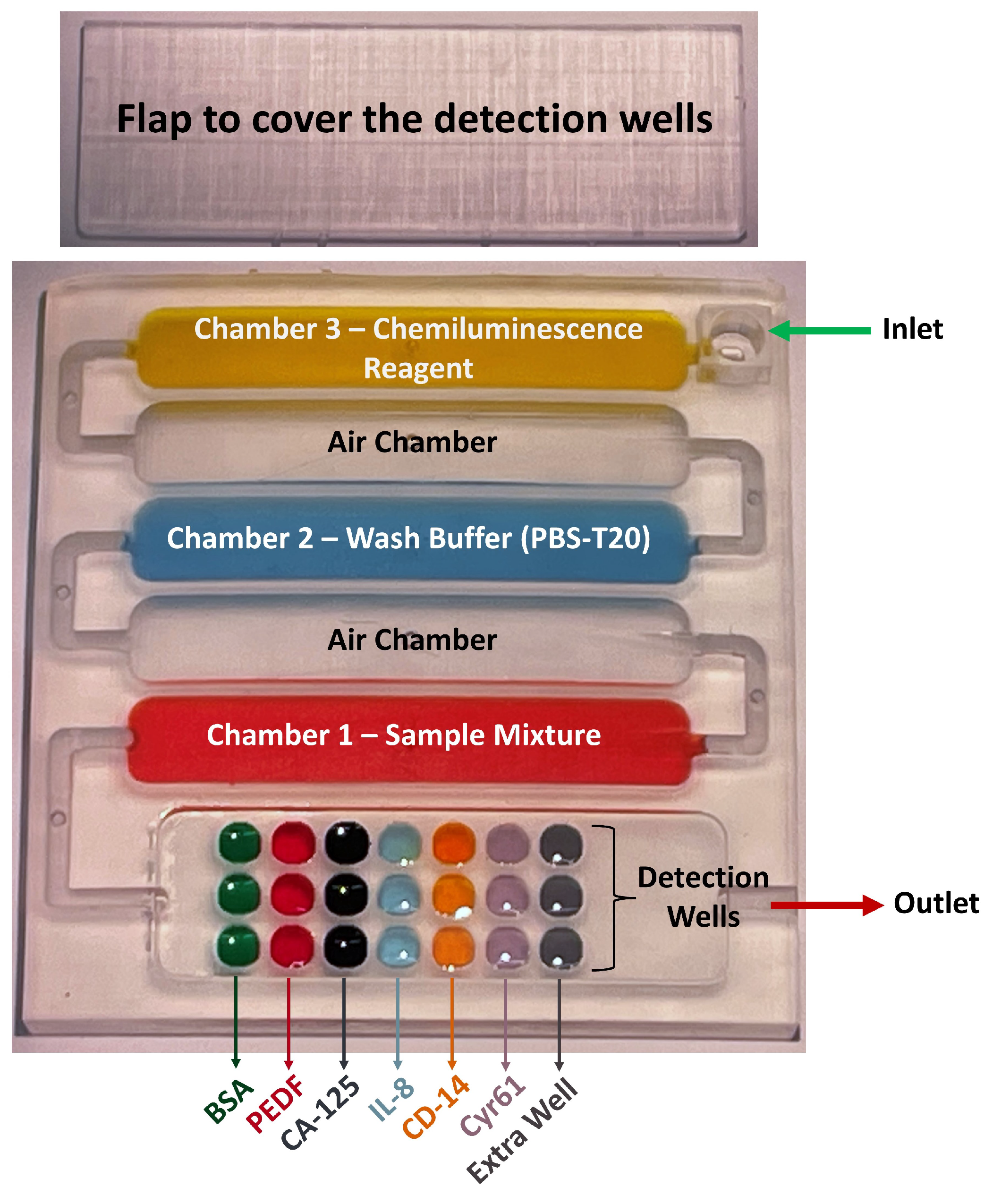
2.2.1. Designing and 3D Printing the Array
2.2.2. Biomarker Selection
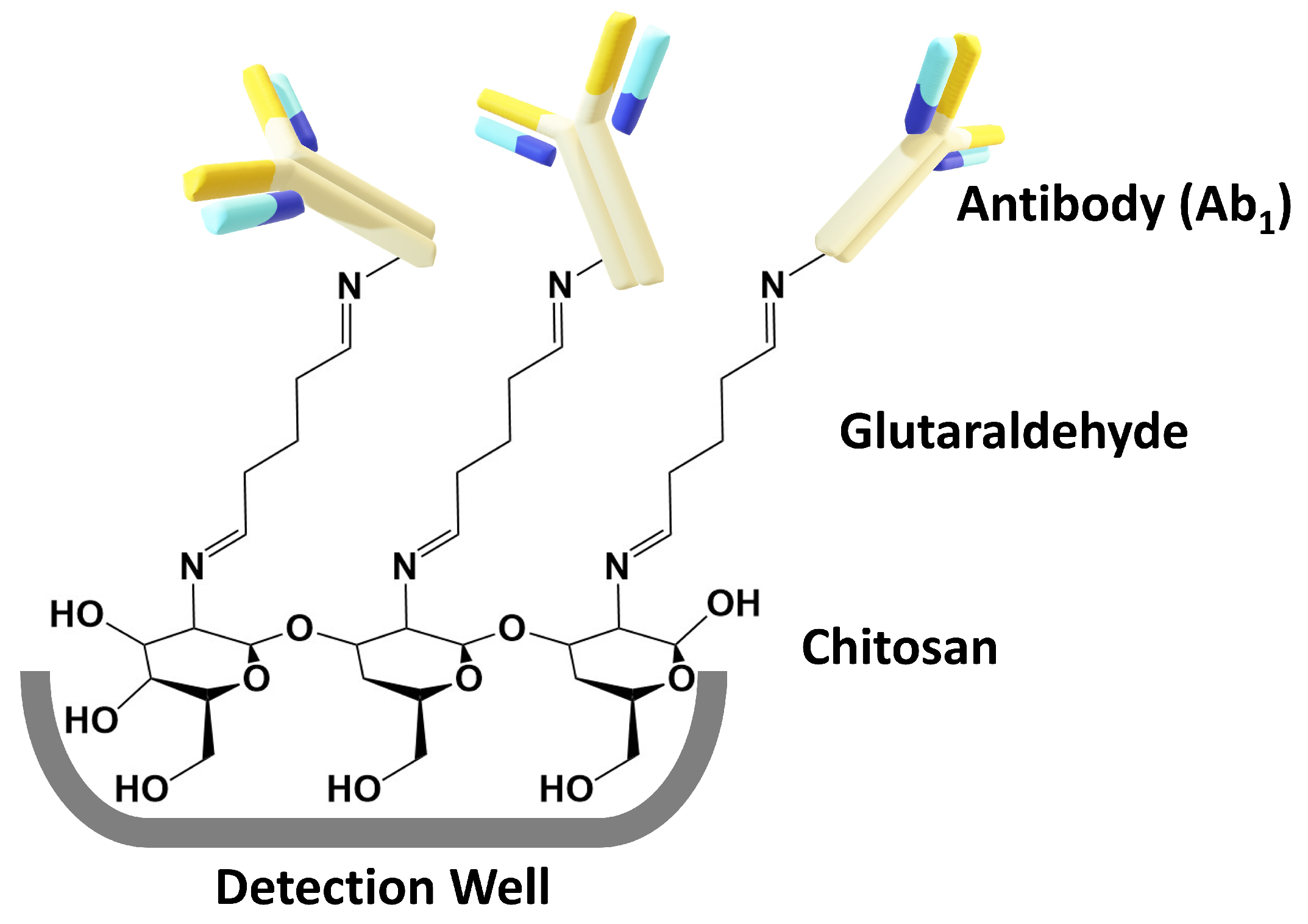
2.2.3. Immunoassay Development
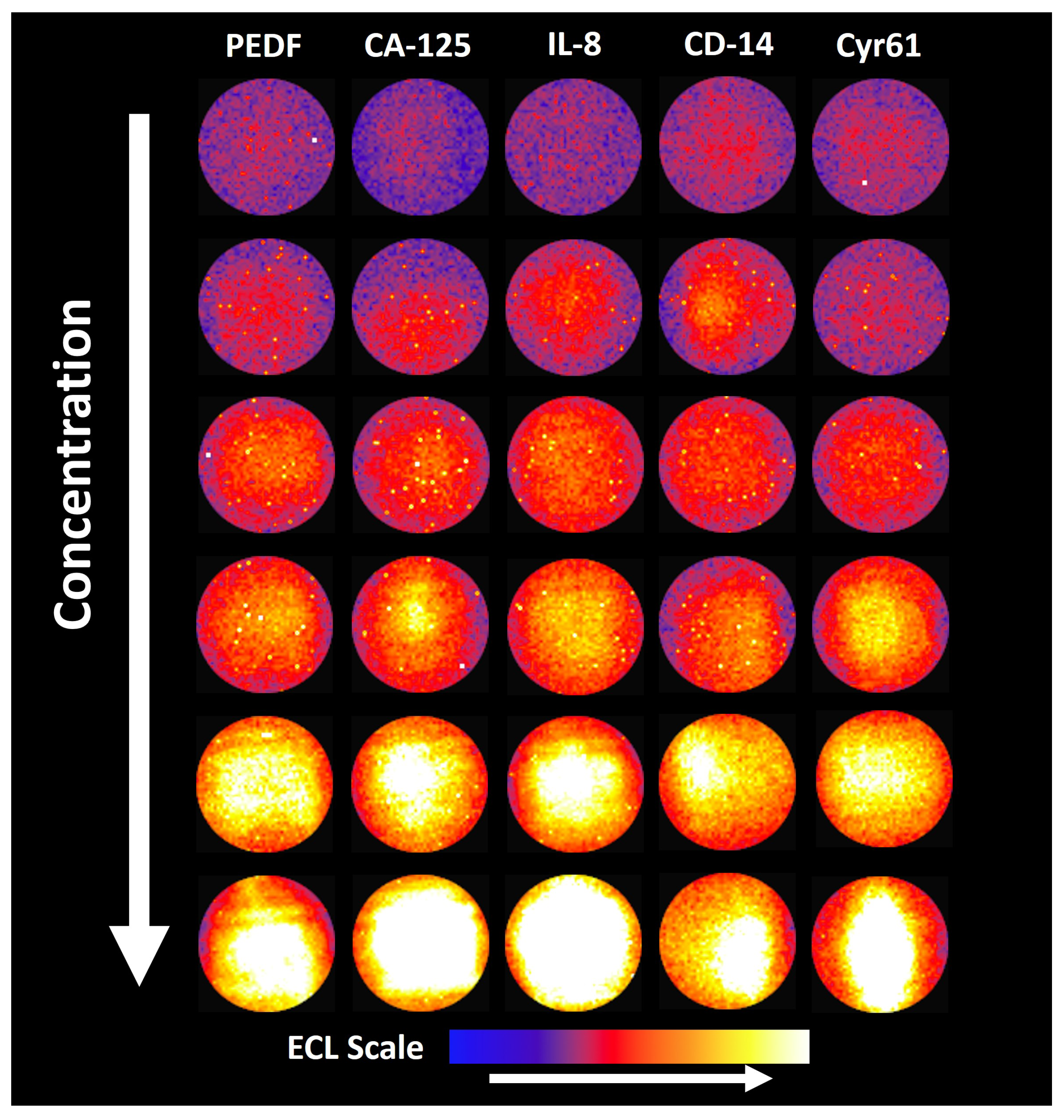
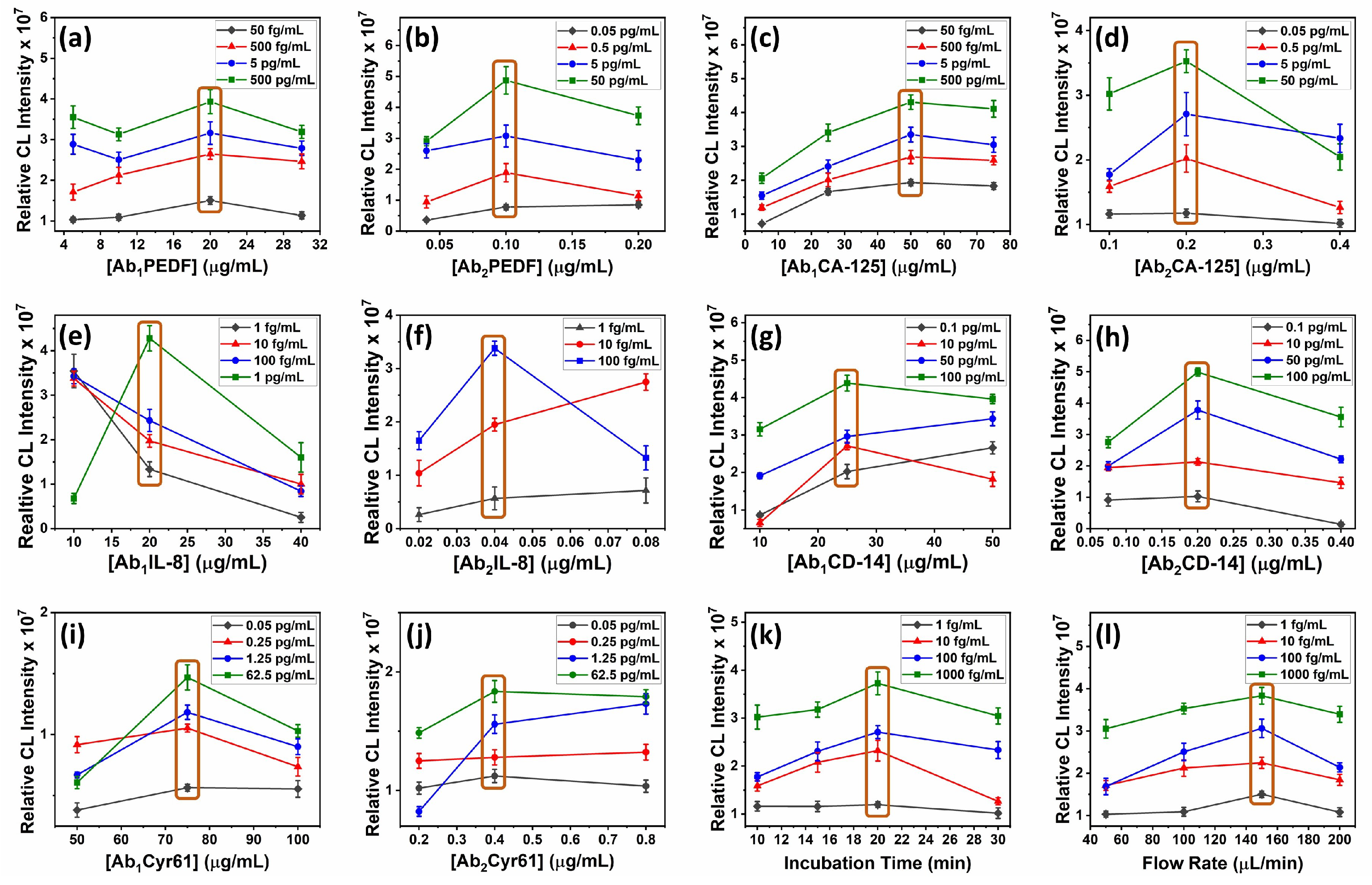
2.2.4. Optimization Steps
2.2.5. Assay Validation
3. Results
3.1. Biomarker Selection
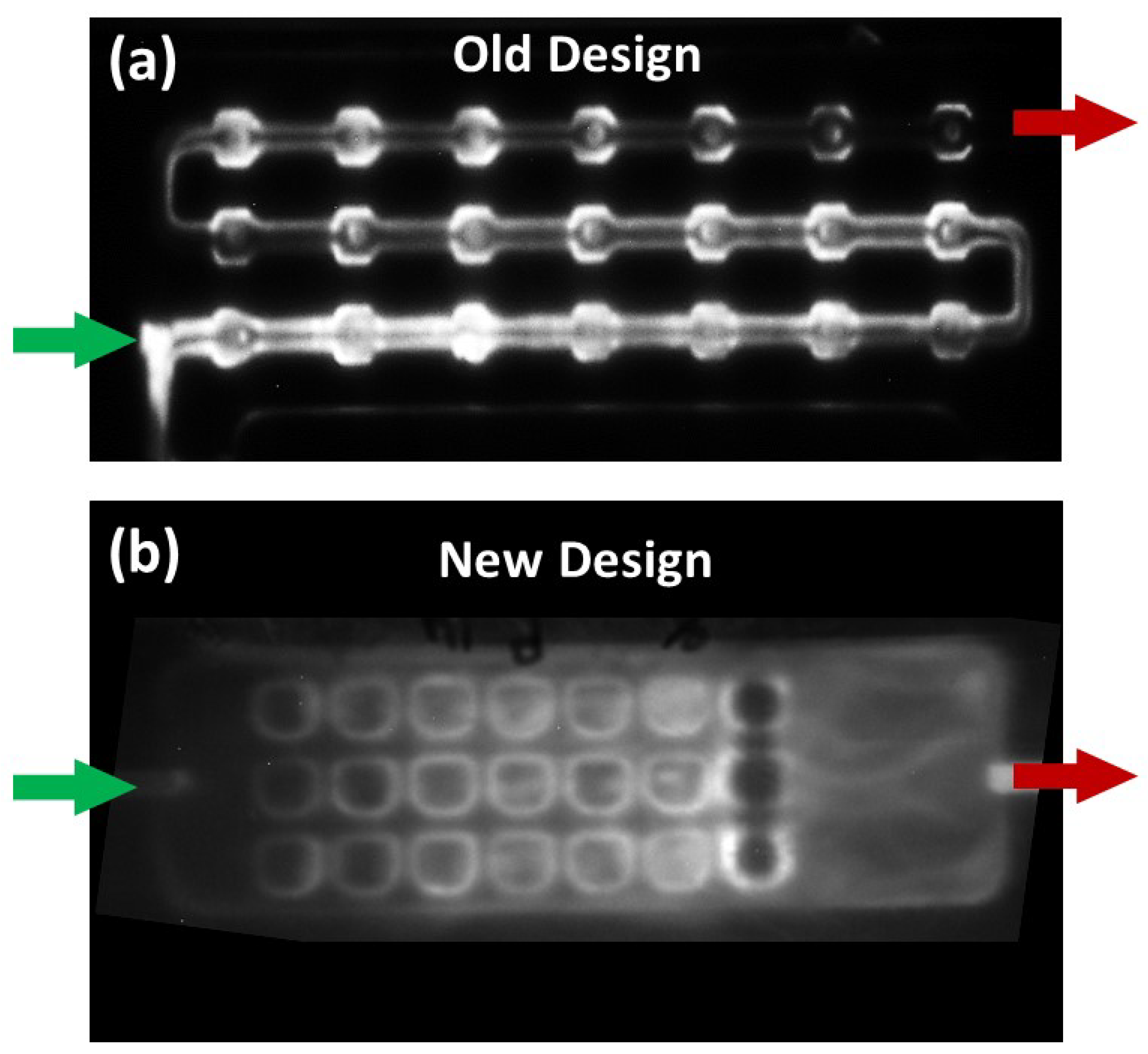
3.2. Designing and 3D Printing the Array
3.3. Optimization Steps
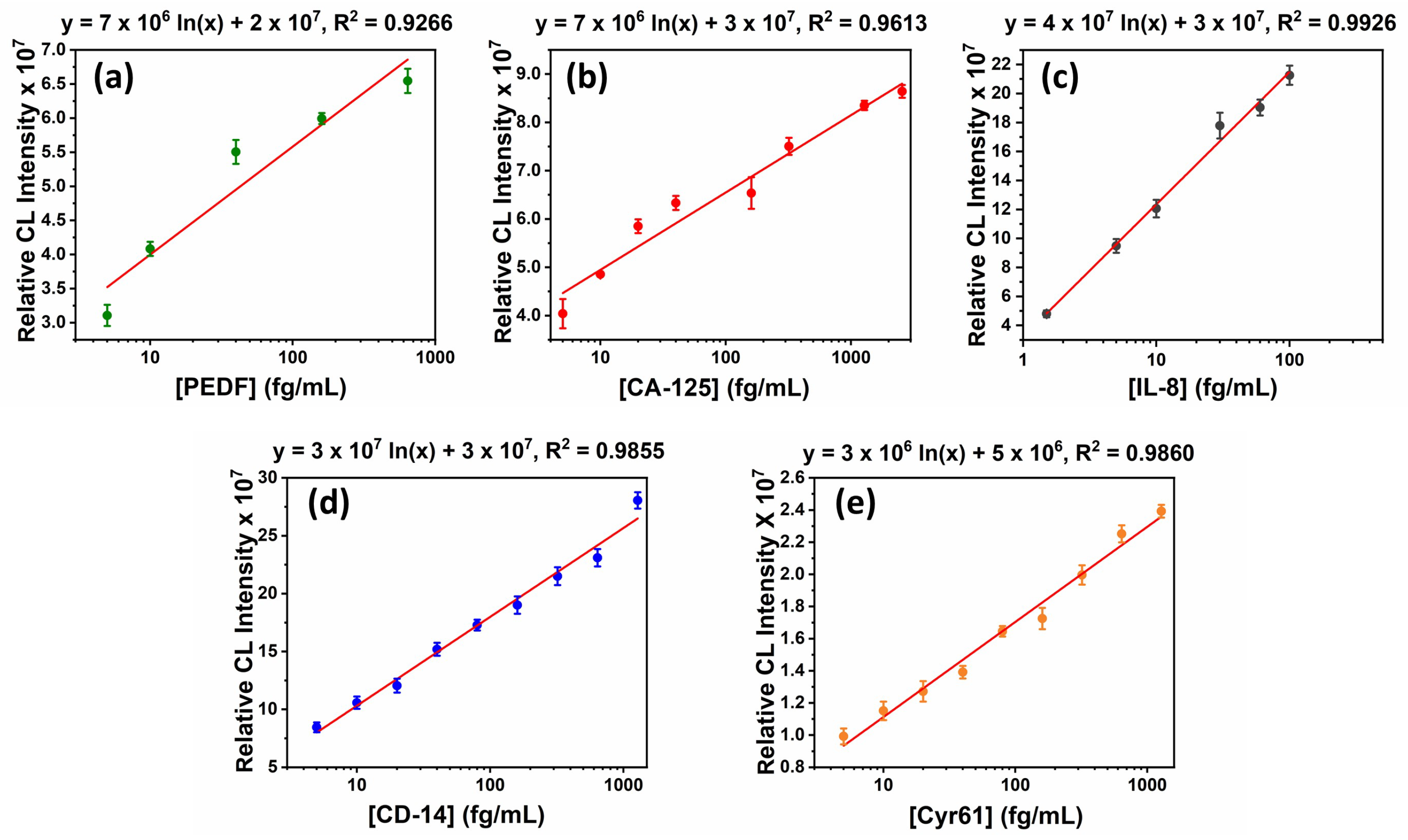
3.4. Assay Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LOD | Limit of Detection |
| 3D | Three Dimensional |
| CL | Chemiluminescence |
| PEDF | Human Pigment Epithelium-Derived Factor |
| CA-125 | Human Cancer Antigen 125 |
| IL-8 | Human Interleukin-8 |
| CD-14 | Human Cluster of Differentiation 14 |
| Cyr61 | Human Cysteine-rich Angiogenic Inducer 61 |
| BSA | Bovine Serum Albumin |
| CCD | Charge Coupled Device |
| PBS | Phosphate Buffer Saline |
| PBS-T20 | Phosphate Buffer Saline—Tween 20 |
| CFD | Computational Fluid Dynamic |
| ROC | Receiver Operating Characteristic |
| Ab | Biotinylated Detection/Secondary Antibody |
| ST-HRP | Streptavidin poly-Horseradish Peroxidase |
| Ab | Capture/Primary Antibody |
| RT | Room Temperature |
| SD | Standard Deviation |
Appendix A
| Biomarker | Cancer | Normal Con. | Conc. in Cancer Patients | Sensitivity (%) | Specificity (%) | AUC |
|---|---|---|---|---|---|---|
| PTHrp | Breast, prostate | <2.5 pM | >2.5 pM | 100 | 80.0 | 0.96 |
| CA 15-3 | Breast | <30 U/mL | >30 U/mL | 77.5 | 85.0 | 0.89 |
| CA-125 | Ovarian | <35 U/mL | >35 U/mL | 79.0 | 82.0 | 0.87 |
| IGFBP-4 | Ovarian | 400.9 ng/mL | 1344.09 ng/mL | 73.0 | 93.0 | 0.82 |
| IL-8 | Colorectal | 2.38–20.05 pg/mL | 0.61–32.86 pg/mL | 70.0 | 91.0 | 0.92 |
| sDC-SIGN & sDC-SIGNR | Colorectal | - | - | 98.7 | 98.8 | 0.99 |
| Cyr61 | Colorectal | <92.0 pg/mL | >92.0 pg/mL | 83.0 | 97.0 | 0.94 |
| CEACAM1 | Lung (NSCLC ) | 226.80–490.11 ng/mL | 381.30–968.13 ng/mL | 97.0 | 82.0 | 0.96 |
| ProGRP | Lung (SCLC ) | <50 pg/mL | >50 pg/mL | 84.0 | 95.0 | 0.72 |
| PEDF | Breast, prostate | 12.93 µg/mL | <9 µg/mL | 64.5 | 70.6 | 0.70 |
| CD-14 | Breast, prostate | 4.89 µg/mL | 6.69 µg/mL | 58.9 | 82.3 | 0.73 |
| Biomarker | Optimum Ab1 (µg/mL) | Optimum Ab2 (µg/mL) |
|---|---|---|
| PEDF | 20 | 0.1 |
| CA-125 | 50 | 0.2 |
| IL-8 | 20 | 0.04 |
| CD-14 | 25 | 0.2 |
| Cyr61 | 75 | 0.4 |
| Biomarker | Linear Range | Limit of Detection (fg/mL) | Normal Conc. in Serum | Conc. in Cancer Patient Serum |
|---|---|---|---|---|
| PEDF | 5–625 fg/mL | 3 | 12. 93 µg/mL | <9 µg/mL |
| CA-125 | 5 fg/mL–15.6 pg/mL | 1.5 | <35 U/mL | >35 U/mL |
| IL-8 | 1.5–100 fg/mL | 1.5 | 2.38–20.05 pg/mL | 0.61–32.86 pg/mL |
| CD-14 | 0.01 fg/mL–100 pg/mL | 0.01 | 4.89 µg/mL | 6.69 µg/mL |
| Cyr61 | 5–500 fg/mL | 4 | <92.0 pg/mL | >92.0 pg/mL |
References
- Niculescu, A.-G.; Chircov, C.; Bîrcă, A.C.; Grumezescu, A.M. Fabrication and Applications of Microfluidic Devices: A Review. Int. J. Mol. Sci. 2021, 22, 2011. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Prajapati, P.; Singh, V.; Pandya, A. An Introduction to Microfluidics and Their Applications. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2022; pp. 1–14. [Google Scholar] [CrossRef]
- Shrimal, P.; Jadeja, G.C.; Patel, S.R. A Review on Novel Methodologies for Drug Nanoparticle Preparation: Microfluidic Approach. Chem. Eng. Res. Des. 2020, 153, 728–756. [Google Scholar] [CrossRef]
- Song, Y.; Hormes, J.; Kumar, C.S.S.R. Microfluidic Synthesis of Nanomaterials. Small 2008, 4, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Tu, J.; Ma, H.; Yang, Y.; Tian, Z.; Pang, D.; Zhang, Z. Controllable Synthesis of Nanocrystals in Droplet Reactors. Lab Chip 2018, 18, 41–56. [Google Scholar] [CrossRef]
- Tokel, O.; İnci, F.; Demirci, U. Advances in Plasmonic Technologies for Point of Care Applications. Chem. Rev. 2014, 114, 5728–5752. [Google Scholar] [CrossRef]
- Zaman, M.A.; Padhy, P.; Wu, M.S.; Ren, W.; Jensen, M.A.; Davis, R.W.; Hesselink, L. Controlled Transport of Individual Microparticles Using Dielectrophoresis. Langmuir 2022, 39, 101–110. [Google Scholar] [CrossRef]
- Desagani, D.; Kleiman, S.; Zagardan, T.; Ben-Yoav, H. 3D-Printed Hydrodynamic Focusing Lab-on-a-Chip Device for Impedance Flow Particle Analysis. Chemosensors 2023, 11, 283. [Google Scholar] [CrossRef]
- Parolo, C.; Idili, A.; Heikenfeld, J.; Plaxco, K.W. Conformational-Switch Biosensors as Novel Tools to Support Continuous, Real-Time Molecular Monitoring in Lab-on-a-Chip Devices. Lab Chip 2023, 23, 1339–1348. [Google Scholar] [CrossRef]
- Da Silva, E.T.S.G.; Souto, D.E.P.; Barragan, J.T.C.; De Fátima Giarola, J.; De Moraes, A.C.M.; Kubota, L.T. Electrochemical Biosensors in Point-of-Care Devices: Recent Advances and Future Trends. ChemElectroChem 2017, 4, 778–794. [Google Scholar] [CrossRef]
- Craighead, H.G. Future Lab-on-a-Chip Technologies for Interrogating Individual Molecules. Nature 2006, 442, 387–393. [Google Scholar] [CrossRef]
- Wongkaew, N.; Simsek, M.; Griesche, C.; Baeumner, A.J. Functional Nanomaterials and Nanostructures Enhancing Electrochemical Biosensors and Lab-on-a-Chip Performances: Recent Progress, Applications, and Future Perspective. Chem. Rev. 2018, 119, 120–194. [Google Scholar] [CrossRef]
- Sengupta, P.; Khanra, K.; Chowdhury, A.R.; Datta, P. Lab-on-a-Chip Sensing Devices for Biomedical Applications. In Bioelectronics and Medical Devices; Elsevier: Amsterdam, The Netherlands, 2019; pp. 47–95. [Google Scholar] [CrossRef]
- Shi, H.; Nie, K.; Dong, B.; Long, M.; Xu, H.; Liu, Z. Recent Progress of Microfluidic Reactors for Biomedical Applications. Chem. Eng. J. 2019, 361, 635–650. [Google Scholar] [CrossRef]
- Moradi, E.; Jalili-Firoozinezhad, S.; Solati-Hashjin, M. Microfluidic Organ-on-a-Chip Models of Human Liver Tissue. Acta Biomater. 2020, 116, 67–83. [Google Scholar] [CrossRef]
- Sun, W.; Luo, Z.; Lee, J.; Kim, H.; Lee, K.; Tebon, P.; Feng, Y.; Dokmeci, M.R.; Sengupta, S.; Khademhosseini, A. Organ-on-a-Chip for Cancer and Immune Organs Modeling. Adv. Healthc. Mater. 2019, 8, 1801363. [Google Scholar] [CrossRef] [PubMed]
- Sharafeldin, M.; Kadimisetty, K.; Bhalerao, K.S.; Chen, T.; Rusling, J.F. 3D-Printed Immunosensor Arrays for Cancer Diagnostics. Sensors 2020, 20, 4514. [Google Scholar] [CrossRef] [PubMed]
- Rusling, J.F.; Forster, R.J. Biosensors Designed for Clinical Applications. Biomedicines 2021, 9, 702. [Google Scholar] [CrossRef]
- Podunavac, I.; Djocos, M.; Vejin, M.; Birgermajer, S.; Pavlović, Z.; Kojić, S.; Petrović, B.; Radonić, V. 3D-Printed Microfluidic Chip for Real-Time Glucose Monitoring in Liquid Analytes. Micromachines 2023, 14, 503. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Zhang, C.; Jiang, H. Advances in Microfluidic Nanobiosensors for the Detection of Foodborne Pathogens. LWT 2021, 151, 112172. [Google Scholar] [CrossRef]
- Sharafeldin, M.; Rusling, J.F. Multiplexed Electrochemical Assays for Clinical Applications. Curr. Opin. Electrochem. 2023, 39, 101256. [Google Scholar] [CrossRef]
- Sharafeldin, M.; Kadimisetty, K.; Bhalerao, K.R.; Bist, I.; Jones, A.; Chen, T.; Lee, N.H.; Rusling, J.F. Accessible Telemedicine Diagnostics with ELISA in a 3D Printed Pipette Tip. Anal. Chem. 2019, 91, 7394–7402. [Google Scholar] [CrossRef]
- Kadimisetty, K.; Malla, S.; Bhalerao, K.S.; Mosa, I.M.; Bhakta, S.; Lee, N.H.; Rusling, J.F. Automated 3D-Printed Microfluidic Array for Rapid Nanomaterial-Enhanced Detection of Multiple Proteins. Anal. Chem. 2018, 90, 7569–7577. [Google Scholar] [CrossRef] [PubMed]
- Rusling, J.F. Developing Microfluidic Sensing Devices Using 3D Printing. ACS Sens. 2018, 3, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Sharafeldin, M.; Jones, A.; Rusling, J.F. 3D-Printed Biosensor Arrays for Medical Diagnostics. Micromachines 2018, 9, 394. [Google Scholar] [CrossRef] [PubMed]
- Weerasuriya, D.R.K.; Hiniduma, K.; Bhakta, S.; Nigro, L.M.; Posada, L.F.; Tan, H.; Suib, S.L.; Kremer, R.; Rusling, J.F. COVID-19 Detection Using a 3D-Printed Micropipette Tip and a Smartphone. ACS Sens. 2023, 8, 848–857. [Google Scholar] [CrossRef]
- Darwish, I.A. Immunoassay Methods and Their Applications in Pharmaceutical Analysis: Basic Methodology and Recent Advances. Int. J. Biomed. Sci. IJBS 2006, 2, 217–235. [Google Scholar]
- Shi, Y.; Peng, Y.; Yang, K.; Meng, J.; Jie, G.; Pan, Z.; Qiaoge, B.; Zhao, W. Application of Microfluidics in Immunoassay: Recent Advancements. J. Healthc. Eng. 2021, 2021, 1–24. [Google Scholar] [CrossRef]
- Hristov, D.R.; Rodriguez-Quijada, C.; Gómez-Márquez, J.; Hamad-Schifferli, K. Designing Paper-Based Immunoassays for Biomedical Applications. Sensors 2019, 19, 554. [Google Scholar] [CrossRef]
- Giljohann, D.A.; Mirkin, C.A. Drivers of Biodiagnostic Development. Nature 2009, 462, 461–464. [Google Scholar] [CrossRef]
- Prabhakar, P.; Sen, R.K.; Dwivedi, N.; Khan, R.; Solanki, P.R.; Srivastava, A.K.; Dhand, C. 3D-Printed Microfluidics and Potential Biomedical Applications. Front. Nanotechnol. 2021, 3. [Google Scholar] [CrossRef]
- Zhang, Y. Three-Dimensional-Printing for Microfluidics or the Other Way Around? Int. J. Bioprint. 2019, 5, 192. [Google Scholar] [CrossRef]
- Fusion 360 Help. Available online: https://help.autodesk.com/view/fusion360/ENU/courses/ (accessed on 11 October 2023).
- Amini, Y.; Ghazanfari, V.; Heydari, M.; Shadman, M.M.; Khamseh, A.G.; Khani, M.H.; Hassanvand, A. Computational Fluid Dynamics Simulation of Two-Phase Flow Patterns in a Serpentine Microfluidic Device. Sci. Rep. 2023, 13, 9483. [Google Scholar] [CrossRef] [PubMed]
- Zweig, M.H.; Campbell, G. Receiver-Operating Characteristic (ROC) Plots: A Fundamental Evaluation Tool in Clinical Medicine. Clin. Chem. 1993, 39, 561–577. [Google Scholar] [CrossRef]
- Schoonjans, F. ROC Curves in MedCalc; MedCalc: Ostend, Belgium, 2023; Available online: https://www.medcalc.org/features/roccurves.php (accessed on 30 September 2023).
- Wikipedia Contributors. Aptamer; Wikipedia: 2023. Available online: https://en.wikipedia.org/wiki/Aptamer (accessed on 30 September 2023).
- Sharafeldin, M.; Chen, T.; Ozkaya, G.U.; Choudhary, D.; Molinolo, A.A.; Gutkind, J.S.; Rusling, J.F. Detecting Cancer Metastasis and Accompanying Protein Biomarkers at Single Cell Levels Using a 3D-Printed Microfluidic Immunoarray. Biosens. Bioelectron. 2021, 171, 112681. [Google Scholar] [CrossRef] [PubMed]
- Kadimisetty, K.; Andrew, P.; Bhalerao, K.S.; Sharafeldin, M.; Mosa, I.M.; Lee, N.H.; Rusling, J.F. Automated 4-Sample Protein Immunoassays Using 3D-Printed Microfluidics. Anal. Methods 2018, 10, 4000–4006. [Google Scholar] [CrossRef] [PubMed]
- Otieno, B.; Krause, C.E.; Jones, A.; Kremer, R.; Rusling, J.F. Cancer Diagnostics via Ultrasensitive Multiplexed Detection of Parathyroid Hormone-Related Peptides with a Microfluidic Immunoarray. Anal. Chem. 2016, 88, 9269–9275. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, L.; Shen, L.; Shen, X.; Ju, S.; Cong, H. Diagnostic Value of Serum Concentration and Integrity of Circulating Cell-Free DNA in Breast Cancer: A Comparative Study with CEA and CA15-3. Labmedicine 2018, 49, 323–332. [Google Scholar] [CrossRef]
- Wang, J.; Jia, G.; Yao, H.; Wu, Z.; Wang, M.; Qi, J. Diagnostic Accuracy of Serum HE4, CA125 and ROMA in Patients with Ovarian Cancer: A Meta-Analysis. Tumor Biol. 2014, 35, 6127–6138. [Google Scholar] [CrossRef]
- Mosig, R.A.; Lobl, M.; Senturk, E.; Shah, H.; Cohen, S.; Chudin, E.; Fruscio, R.; Marchini, S.; D’Incalci, M.; Sachidanandam, R.; et al. IGFBP-4 Tumor and Serum Levels Are Increased across All Stages of Epithelial Ovarian Cancer. J. Ovarian Res. 2012, 5, 3. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Okamura, S.; Yamaji, T.; Iwasaki, M.; Tsugane, S.; Shetty, V.; Koizumi, T. Plasma Cytokine Levels and the Presence of Colorectal Cancer. PLoS ONE 2019, 14, e0213602. [Google Scholar] [CrossRef]
- Xia, W.; Chen, W.; Zhang, Z.; Wu, D.; Wu, P.; Chen, Z.; Li, C.; Huang, J. Prognostic Value, Clinicopathologic Features and Diagnostic Accuracy of Interleukin-8 in Colorectal Cancer: A Meta-Analysis. PLoS ONE 2015, 10, e0123484. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, C.; Chen, K.; Chen, Z.; Sun, Z.; Zhang, Z.; Ding, D.; Ren, S.; Zuo, Y. The Clinical Significance of DC-SIGN and DC-SIGNR, Which Are Novel Markers Expressed in Human Colon Cancer. PLoS ONE 2014, 9, e114748. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Xu, Z.; Zhu, X.; Xue, T.; Liu, J.L.; Gao, F.; Wu, C.; Song, B.; Qi, L. Serum Cyr61 as a Potential Biomarker for Diagnosis of Colorectal Cancer. Clin. Transl. Oncol. 2016, 19, 519–524. [Google Scholar] [CrossRef]
- Zhou, M.; Du, Y.; Liu, Y.; Wang, Y.-Z.; He, Y.; Yang, C.; Wang, W.; Gao, F. Clinical and Experimental Studies Regarding the Expression and Diagnostic Value of Carcinoembryonic Antigen-Related Cell Adhesion Molecule 1 in Non-Small-Cell Lung Cancer. BMC Cancer 2013, 13, 359. [Google Scholar] [CrossRef]
- Oremek, G.M.; Sauer-Eppel, H.; Bruzdziak, T. Value of Tumour and Inflammatory Markers in Lung Cancer. Anticancer Res. 2007, 27, 1911–1915. [Google Scholar] [PubMed]
- Jones, A.; Dhanapala, L.; Baldo, T.A.; Sharafeldin, M.; Krause, C.E.; Shen, M.; Moghaddam, S.; Faria, R.C.; Dey, D.K.; Watson, R.W.G.; et al. Prostate Cancer Diagnosis in the Clinic Using an 8-Protein Biomarker Panel. Anal. Chem. 2020, 93, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Mou, L.; Jiang, X. Materials for Microfluidic Immunoassays: A Review. Adv. Healthc. Mater. 2017, 6, 1601403. [Google Scholar] [CrossRef] [PubMed]
- Dixit, C.K.; Kadimisetty, K.; Otieno, B.; Tang, C.K.; Malla, S.; Krause, C.E.; Rusling, J.F. Electrochemistry-Based Approaches to Low Cost, High Sensitivity, Automated, Multiplexed Protein Immunoassays for Cancer Diagnostics. Analyst 2016, 141, 536–547. [Google Scholar] [CrossRef]
- Li, X.; Pan, X.; Lu, J.; Zhou, Y.; Gong, J. Dual-Modal Visual/Photoelectrochemical All-in-One Bioassay for Rapid Detection of AFP Using 3D Printed Microreactor Device. Biosens. Bioelectron. 2020, 158, 112158. [Google Scholar] [CrossRef]
- Longhitano, G.A.; Nunes, G.B.; Candido, G.; Da Silva, J.V.L. The Role of 3D Printing during COVID-19 Pandemic: A Review. Prog. Addit. Manuf. 2020, 6, 19–37. [Google Scholar] [CrossRef]
- Ordutowski, H.; Parra-Cabrera, C.; Achille, C.; Dochy, R.; Dosso, F.D.; Lammertyn, J.; Ameloot, R.; Spasić, D. A Multistep Immunoassay on a 3D-Printed Capillarity-Driven Microfluidic Device for Point-of-Care Diagnostics. Appl. Mater. Today 2023, 32, 101788. [Google Scholar] [CrossRef]
- Yin, B.; Qian, C.; Wan, X.; Sohan, A.S.M.M.F.; Lin, X. Tape Integrated Self-Designed Microfluidic Chip for Point-of-Care Immunoassays Simultaneous Detection of Disease Biomarkers with Tunable Detection Range. Biosens. Bioelectron. 2022, 212, 114429. [Google Scholar] [CrossRef]
- Yang, Y.; Griffin, K.; Li, X.; Sharp, E.; Young, L.; Garcia, L.; Griswold, J.; Pappas, D. Combined CD25, CD64, and CD69 Biomarker in 3D-Printed Multizone Millifluidic Device for Sepsis Detection in Clinical Samples. Anal. Chem. 2023, 95, 12819–12825. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.B.; Ng, S.H.; Yoon, Y.J. A Review on 3D Printed Bioimplants. Int. J. Precis. Eng. Manuf. 2015, 16, 1035–1046. [Google Scholar] [CrossRef]
- Gross, B.C.; Erkal, J.L.; Lockwood, S.Y.; Chen, C.; Spence, D.M. Evaluation of 3D Printing and Its Potential Impact on Biotechnology and the Chemical Sciences. Anal. Chem. 2014, 86, 3240–3253. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.B.; Ng, S.H.; Li, K.H.H.; Yoon, Y. 3D Printed Microfluidics for Biological Applications. Lab Chip 2015, 15, 3627–3637. [Google Scholar] [CrossRef]
- Cox, K.L. Immunoassay Methods. Assay Guidance Manual—NCBI Bookshelf. Available online: https://www.ncbi.nlm.nih.gov/books/NBK92434/ (accessed on 10 October 2023).
- Minić, R.; Živković, I. Optimization, Validation and Standardization of ELISA. In Norovirus; IntechOpen: Rijeka, Croatia, 2021. [Google Scholar] [CrossRef]
- Shah, V.P.; Midha, K.K.; Findlay, J.W.A.; Hill, H.; Hulse, J.D.; McGilveray, I.J.; McKay, G.; Miller, K.J.; Patnaik, R.; Powell, M.L.; et al. Bioanalytical Method Validation—A Revisit with a Decade of Progress. Pharm. Res. 2000, 17, 1551–1557. [Google Scholar] [CrossRef]
| Spiked Conc: (fg/mL) | |||||
|---|---|---|---|---|---|
| 15 | 75 | 200 | 500 | ||
| PEDF | Found Concentration (fg/mL) | 17 (±8) | 66 (±4) | 177 (±19) | 580 (±31) |
| Percent Recovery (%) | 111 (±10) | 88 (±13) | 89 (±16) | 116 (±14) | |
| CA-125 | Found Concentration (fg/mL) | 14 (±2) | 83 (±11) | 190 (±15) | 522 (±25) |
| Percent Recovery (%) | 92 (±6) | 111 (±14) | 95 (±7) | 104 (±11) | |
| IL-8 | Found Concentration (fg/mL) | 17 (±5) | 65 (±8) | 169 (±10) | 569 (±24) |
| Percent Recovery (%) | 110 (±14) | 86 (±8) | 84 (±16) | 114 (±17) | |
| CD-14 | Found Concentration (fg/mL) | 17 (±3) | 63 (±4) | 164 (±12) | 470 (±18) |
| Percent Recovery (%) | 115 (±15) | 84 (±9) | 82 (±13) | 94 (±12) | |
| Cyr61 | Found Concentration (fg/mL) | 16 (±5) | 86 (±15) | 211 (±15) | 586 (±21) |
| Percent Recovery (%) | 106 (±11) | 115 (±13) | 106 (±19) | 117 (±14) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiniduma, K.; Bhalerao, K.S.; De Silva, P.I.T.; Chen, T.; Rusling, J.F. Design and Fabrication of a 3D-Printed Microfluidic Immunoarray for Ultrasensitive Multiplexed Protein Detection. Micromachines 2023, 14, 2187. https://doi.org/10.3390/mi14122187
Hiniduma K, Bhalerao KS, De Silva PIT, Chen T, Rusling JF. Design and Fabrication of a 3D-Printed Microfluidic Immunoarray for Ultrasensitive Multiplexed Protein Detection. Micromachines. 2023; 14(12):2187. https://doi.org/10.3390/mi14122187
Chicago/Turabian StyleHiniduma, Keshani, Ketki S. Bhalerao, Peyahandi I. Thilini De Silva, Tianqi Chen, and James F. Rusling. 2023. "Design and Fabrication of a 3D-Printed Microfluidic Immunoarray for Ultrasensitive Multiplexed Protein Detection" Micromachines 14, no. 12: 2187. https://doi.org/10.3390/mi14122187
APA StyleHiniduma, K., Bhalerao, K. S., De Silva, P. I. T., Chen, T., & Rusling, J. F. (2023). Design and Fabrication of a 3D-Printed Microfluidic Immunoarray for Ultrasensitive Multiplexed Protein Detection. Micromachines, 14(12), 2187. https://doi.org/10.3390/mi14122187







