Spatial Distribution of Flagellated Microalgae Chlamydomonas reinhardtii in a Quasi-Two-Dimensional Space
Abstract
:1. Introduction
2. Basis of Spatial Distribution of Cells
- (1)
- Cells occupy a finite space equal to their size and cannot occupy the same space multiple times. This is called the EV effect, or steric interaction, in which cells avoid each other, making VMR smaller. The larger the space occupied by a cell, the more pronounced this effect becomes.
- (2)
- If the space is not uniform and there is inhomogeneity, the distribution is spatially biased, which is an environmental factor that increases . Therefore, it was necessary to carefully check for this effect.
- (3)
- The purpose of this study is to determine whether interactions between cells, except for the excluded volume effect, tend to be attractive or repulsive.
3. Experimental Methods
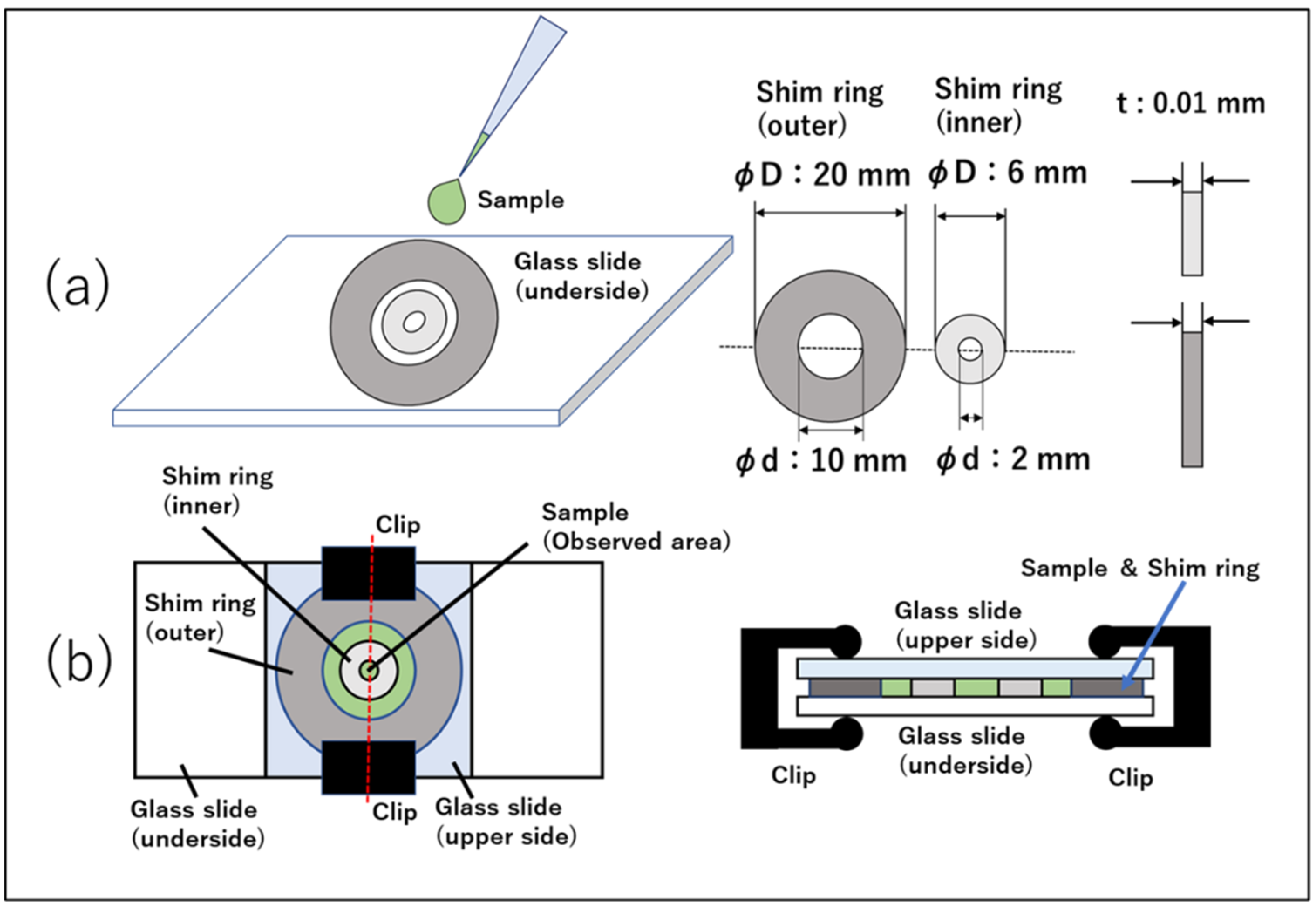
3.1. Sample Preparation
3.2. Preparation of Quasi-2D Spatial Device
3.3. Experimental Apparatus
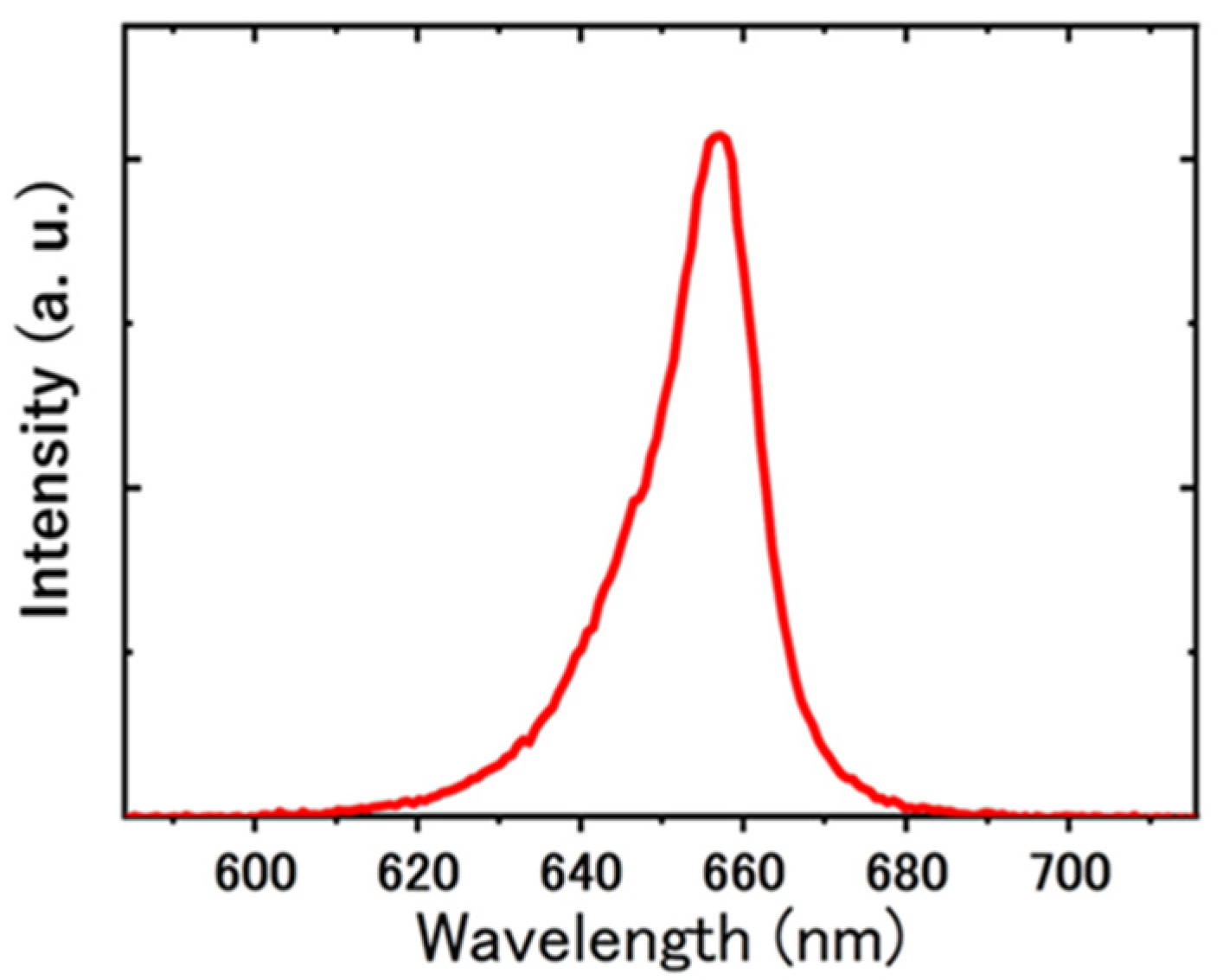
3.4. Image Analysis
3.4.1. Frame Interval
3.4.2. Binarization and Particle Tracking
3.4.3. Cell Counting
3.5. Calculation Method for Simulating the Spatial Distribution
- (1)
- Following the procedures (2) and (3) below, circular cells of diameter were distributed in a square 2-dimensional space of length on one side. The diameter was assumed to be the same for all the cells. The programming language used was Python.
- (2)
- The coordinates of the geometrical center of the cells were determined. First, a set of two-dimensional real random numbers between 0 and 1 was generated by the Monte Carlo method. Next, this random number was multiplied by the side length of the square area for observation, px (1.15 mm), to give each cell a random coordinate within the observation area. In addition, taking into account the EV effect of C. reinhardtii, the distance between two coordinates should not be smaller than the diameter 16.9 px (10 μm). That is, for the i-th and j-th coordinates,The cell of coordinates that did not satisfy this condition were not added, and the coordinates were again determined from random numbers.
- (3)
- The 2D space was divided into sections equally and the number of cells in each section were counted.
- (4)
- (2) and (3) were regarded as one frame and repeated 900 times to obtain the same number of frames as in the experiment.
- (5)
- A histogram was created with the horizontal axis as the number of cells in a section and with the vertical axis as the frequency of the occurrence of the section containing that number of cells.
3.6. Simulation including Stationary Cells
4. Results and Discussion
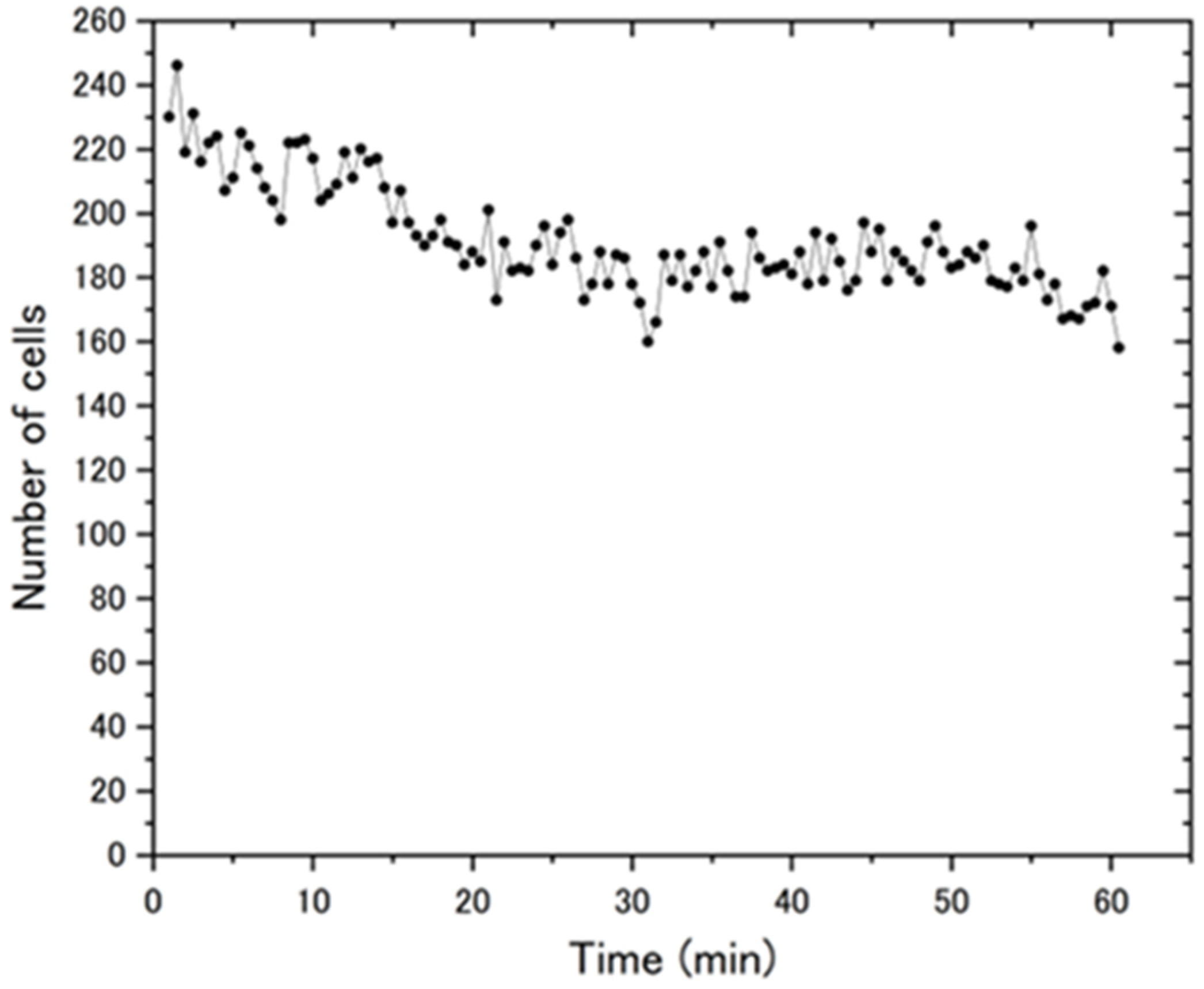
4.1. Change in the Number of Cells over Time
4.2. Partitioning and Cell Counting
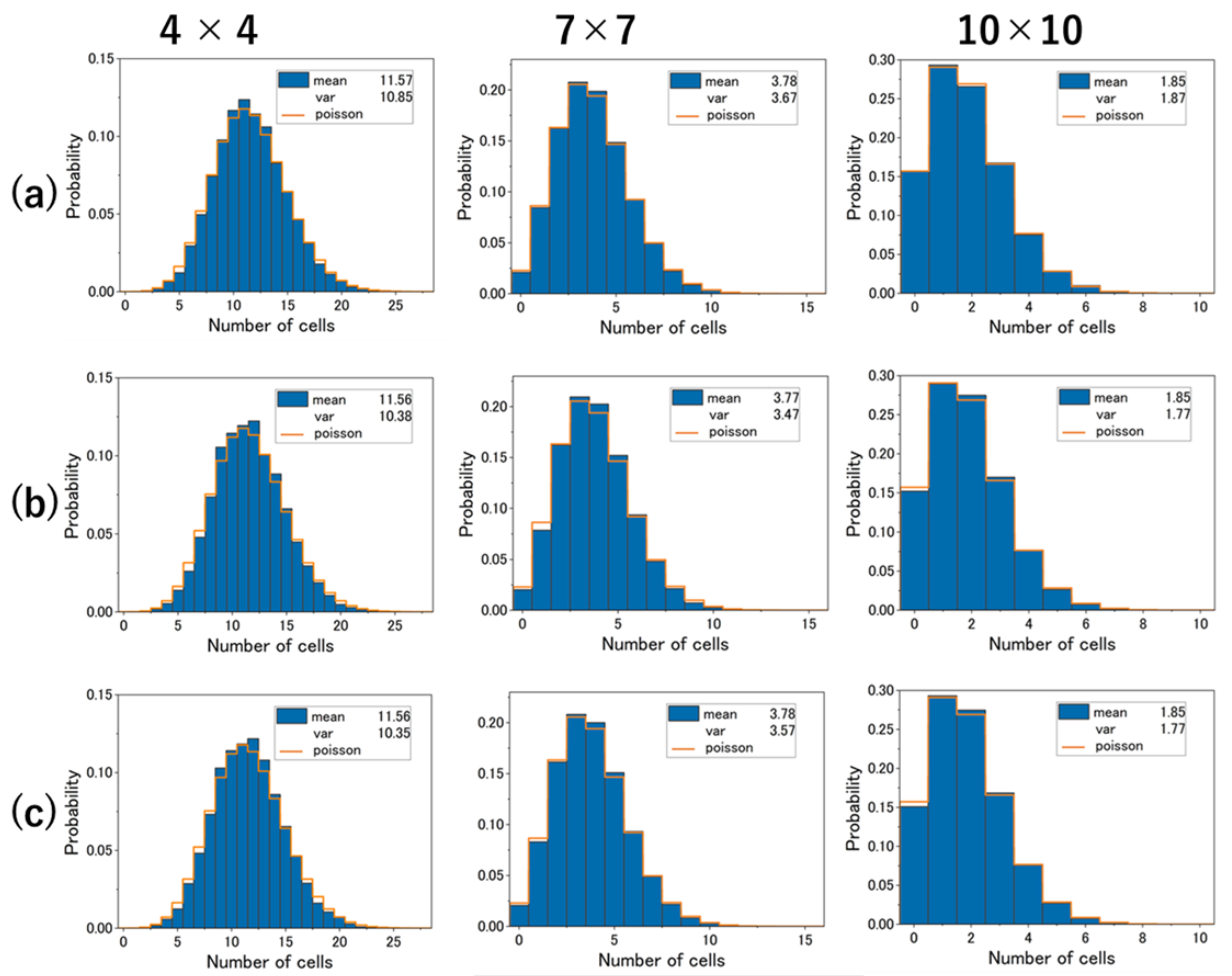
4.3. Experimental Results and Cell Distribution in Simulation
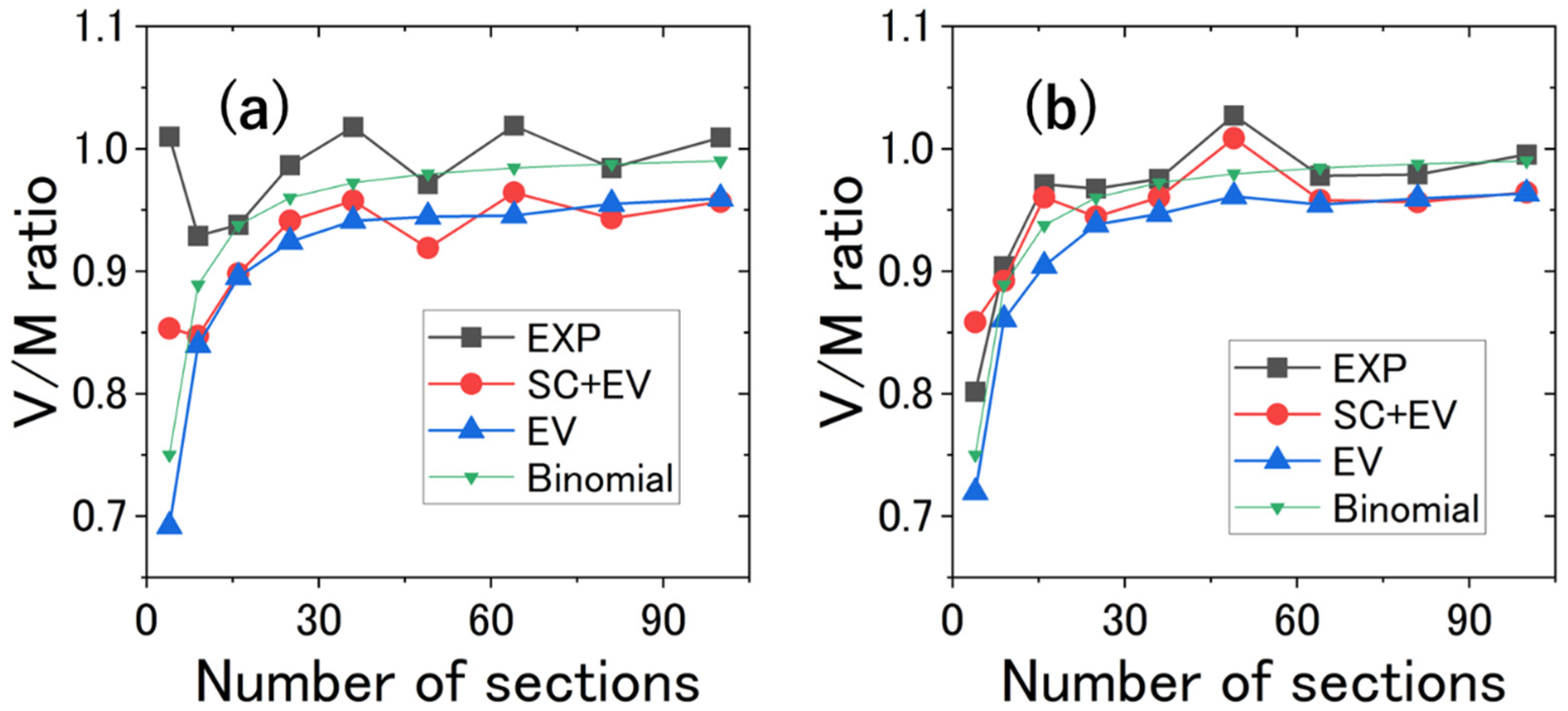
4.4. V/M Ratio vs. Number of Sections
5. Conclusions and Prospects
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fox, M.; Anthony, M. Quantum Optics: An Introduction; Oxford University Press: Oxford, UK, 2006. [Google Scholar]
- Koyama, K.; Hokunan, H.; Hasegawa, M.; Kawamura, S.; Koseki, S. Do Bacterial Cell Numbers Follow a Theoretical Poisson Distribution? Comparison of Experimentally Obtained Numbers of Single Cells with Random Number Generation via Computer Simulation. Food Microbiol. 2016, 60, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.T. Higher-Order Criteria for Nonclassical Effects in Photon Statistics. Phys. Rev. A 1990, 41, 1721–1723. [Google Scholar] [CrossRef] [PubMed]
- Bechinger, C.; Di Leonardo, R.; Löwen, H.; Reichhardt, C.; Volpe, G.; Volpe, G. Active Particles in Complex and Crowded Environments. arXiv 2016, arXiv:1602.00081. Available online: https://arxiv.org/abs/1602.00081v2 (accessed on 10 February 2023). [CrossRef]
- Reynolds, C.W. Flocks, Herds and Schools: A Distributed Behavioral Model. SIGGRAPH Comput. Graph. 1987, 21, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Couzin, I.D.; Krause, J.; James, R.; Ruxton, G.D.; Franks, N.R. Collective Memory and Spatial Sorting in Animal Groups. J. Theor. Biol. 2002, 218, 11. [Google Scholar] [CrossRef] [Green Version]
- Hemelrijk, C.K.; Hildenbrandt, H. Schools of Fish and Flocks of Birds: Their Shape and Internal Structure by Self-Organization. Interface Focus 2012, 2, 726–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, D.A.P.; Hildenbrandt, H.; Padding, J.T.; Hemelrijk, C.K. Fluid Dynamics of Moving Fish in a Two-Dimensional Multiparticle Collision Dynamics Model. Phys. Rev. E 2012, 85, 021901. [Google Scholar] [CrossRef] [Green Version]
- Cresswell, W. Flocking Is an Effective Anti-Predation Strategy in Redshanks, Tringa totanus. Anim. Behav. 1994, 47, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Krause, J.; Ruxton, G.D. Living in Groups; Oxford University Press: Oxford, UK, 2002. [Google Scholar]
- Cresswell, W.; Quinn, J.L. Predicting the Optimal Prey Group Size from Predator Hunting Behaviour. J. Anim. Ecol. 2011, 80, 310–319. [Google Scholar] [CrossRef]
- Moeur, M. Characterizing Spatial Patterns of Trees Using Stem-Mapped Data. For. Sci. 1993, 39, 756–775. [Google Scholar]
- Bees, M.A. Advances in Bioconvection. Annu. Rev. Fluid Mech. 2020, 52, 449–476. [Google Scholar] [CrossRef]
- Nonaka, Y.; Kikuchi, K.; Numayama-Tsuruta, K.; Kage, A.; Ueno, H.; Ishikawa, T. Inhomogeneous Distribution of Chlamydomonas in a Cylindrical Container with a Bubble Plume. Biol. Open 2016, 5, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Jánosi, I.M.; Czirók, A.; Silhavy, D.; Holczinger, A. Is Bioconvection Enhancing Bacterial Growth in Quiescent Environments? Environ. Microbiol. 2002, 4, 525–531. [Google Scholar] [CrossRef] [Green Version]
- Keller, E.F.; Segel, L.A. Initiation of Slime Mold Aggregation Viewed as an Instability. J. Theor. Biol. 1970, 26, 399–415. [Google Scholar] [CrossRef]
- Marée, A.F.M.; Hogeweg, P. How Amoeboids Self-Organize into a Fruiting Body: Multicellular Coordination in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 2001, 98, 3879–3883. [Google Scholar] [CrossRef] [Green Version]
- Scitable. Volvox, Chlamydomonas, Evolution of Multicellularity|Learn Science at Scitable. Available online: https://www.nature.com/scitable/topicpage/volvox-chlamydomonas-and-the-evolution-of-multicellularity-14433403/ (accessed on 10 February 2023).
- Vicsek, T.; Czirók, A.; Ben-Jacob, E.; Cohen, I.; Shochet, O. Novel Type of Phase Transition in a System of Self-Driven Particles. Phys. Rev. Lett. 1995, 75, 1226. Available online: https://journals.aps.org/prl/abstract/10.1103/PhysRevLett.75.1226 (accessed on 10 February 2023). [CrossRef] [Green Version]
- Lauga, E.; Powers, T.R. The Hydrodynamics of Swimming Microorganisms. Rep. Prog. Phys. 2009, 72, 096601. [Google Scholar] [CrossRef]
- Fragkopoulos, A.A.; Vachier, J.; Frey, J.; Le Menn, F.-M.; Mazza, M.G.; Wilczek, M.; Zwicker, D.; Bäumchen, O. Self-Generated Oxygen Gradients Control Collective Aggregation of Photosynthetic Microbes. J. R. Soc. Interface 2021, 18, 20210553. [Google Scholar] [CrossRef]
- Pooley, C.M.; Alexander, G.P.; Yeomans, J.M. Hydrodynamic Interaction between Two Swimmers at Low Reynolds Number. Phys. Rev. Lett. 2007, 99, 228103. [Google Scholar] [CrossRef]
- Knežević, M.; Welker, T.; Stark, H. Collective Motion of Active Particles Exhibiting Non-Reciprocal Orientational Interactions. Sci. Rep. 2022, 12, 19437. [Google Scholar] [CrossRef]
- Xiang, C.; Liu, J.; Ma, L.; Yang, M. Overexpressing Codon-Adapted Fusion Proteins of 4-Coumaroyl-CoA Ligase (4CL) and Stilbene Synthase (STS) for Resveratrol Production in Chlamydomonas reinhardtii. J. Appl. Phycol. 2020, 32, 1669–1676. [Google Scholar] [CrossRef]
- Bentley, S.A.; Laeverenz-Schlogelhofer, H.; Anagnostidis, V.; Cammann, J.; Mazza, M.G.; Gielen, F.; Wan, K.Y. Phenotyping Single-Cell Motility in Microfluidic Confinement. eLife 2022, 11, e76519. [Google Scholar] [CrossRef]
- Ostapenko, T.; Schwarzendahl, F.J.; Böddeker, T.J.; Kreis, C.T.; Cammann, J.; Mazza, M.G.; Bäumchen, O. Curvature-Guided Motility of Microalgae in Geometric Confinement. Phys. Rev. Lett. 2018, 120, 068002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cammann, J.; Schwarzendahl, F.J.; Ostapenko, T.; Lavrentovich, D.; Bäumchen, O.; Mazza, M.G. Emergent Probability Fluxes in Confined Microbial Navigation. Proc. Natl. Acad. Sci. USA 2021, 118, e2024752118. [Google Scholar] [CrossRef]
- Takemura, A. Modern Mathematical Statistics; Gakujutsu Tosho Shuppan: Tokyo, Japan, 2020. (In Japanese) [Google Scholar]
- Harris, E.H. The Chlamydomonas Sourcebook, 2nd ed.; Academic Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Kreis, C.T.; Le Blay, M.; Linne, C.; Makowski, M.M.; Bäumchen, O. Adhesion of Chlamydomonas Microalgae to Surfaces Is Switchable by Light. Nat. Phys. 2018, 14, 45–49. [Google Scholar] [CrossRef]
- Zhang, J.; Li, C.; Rahaman, M.M.; Yao, Y.; Ma, P.; Zhang, J.; Zhao, X.; Jiang, T.; Grzegorzek, M. A Comprehensive Review of Image Analysis Methods for Microorganism Counting: From Classical Image Processing to Deep Learning Approaches. Artif. Intell. Rev. 2022, 55, 2875–2944. [Google Scholar] [CrossRef] [PubMed]
- Fiji: ImageJ, with “Batteries Included”. Available online: https://fiji.sc/ (accessed on 10 February 2023).
- Dazzo, F.B.; Gross, C. CMEIAS Quadrat Maker: A Digital Software Tool to Optimize Grid Dimensions and Produce Quadrat Images for Landscape Ecology Spatial Analysis. J. Ecosyst. Ecogr. 2013, 3, 4. [Google Scholar]
- Pang, X.; Tong, Y.; Xue, W.; Yang, Y.-F.; Chen, X.; Liu, J.; Chen, D. Expression and Characterization of Recombinant Human Lactoferrin in Edible Alga Chlamydomonas reinhardtii. Biosci. Biotechnol. Biochem. 2019, 83, 851–859. [Google Scholar] [CrossRef]
- Melis, A.; Happe, T. Hydrogen Production. Green Algae as a Source of Energy. Plant Physiol. 2001, 127, 740–748. [Google Scholar]
- Yagi, T.; Yamashita, K.; Okada, N.; Isono, T.; Momose, D.; Mineki, S.; Tokunaga, E. Hydrogen Photoproduction in Green Algae Chlamydomonas reinhardtii Sustainable over 2 Weeks with the Original Cell Culture without Supply of Fresh Cells nor Exchange of the Whole Culture Medium. J. Plant Res. 2016, 129, 771–779. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aono, T.; Yamashita, K.; Hashimoto, M.; Ishikawa, Y.; Aizawa, K.; Tokunaga, E. Spatial Distribution of Flagellated Microalgae Chlamydomonas reinhardtii in a Quasi-Two-Dimensional Space. Micromachines 2023, 14, 813. https://doi.org/10.3390/mi14040813
Aono T, Yamashita K, Hashimoto M, Ishikawa Y, Aizawa K, Tokunaga E. Spatial Distribution of Flagellated Microalgae Chlamydomonas reinhardtii in a Quasi-Two-Dimensional Space. Micromachines. 2023; 14(4):813. https://doi.org/10.3390/mi14040813
Chicago/Turabian StyleAono, Tetsuo, Kyohei Yamashita, Masafumi Hashimoto, Yuji Ishikawa, Kentaro Aizawa, and Eiji Tokunaga. 2023. "Spatial Distribution of Flagellated Microalgae Chlamydomonas reinhardtii in a Quasi-Two-Dimensional Space" Micromachines 14, no. 4: 813. https://doi.org/10.3390/mi14040813
APA StyleAono, T., Yamashita, K., Hashimoto, M., Ishikawa, Y., Aizawa, K., & Tokunaga, E. (2023). Spatial Distribution of Flagellated Microalgae Chlamydomonas reinhardtii in a Quasi-Two-Dimensional Space. Micromachines, 14(4), 813. https://doi.org/10.3390/mi14040813




