Nanoscale Extracellular Vesicle-Enabled Liquid Biopsy: Advances and Challenges for Lung Cancer Detection
Abstract
1. Introduction
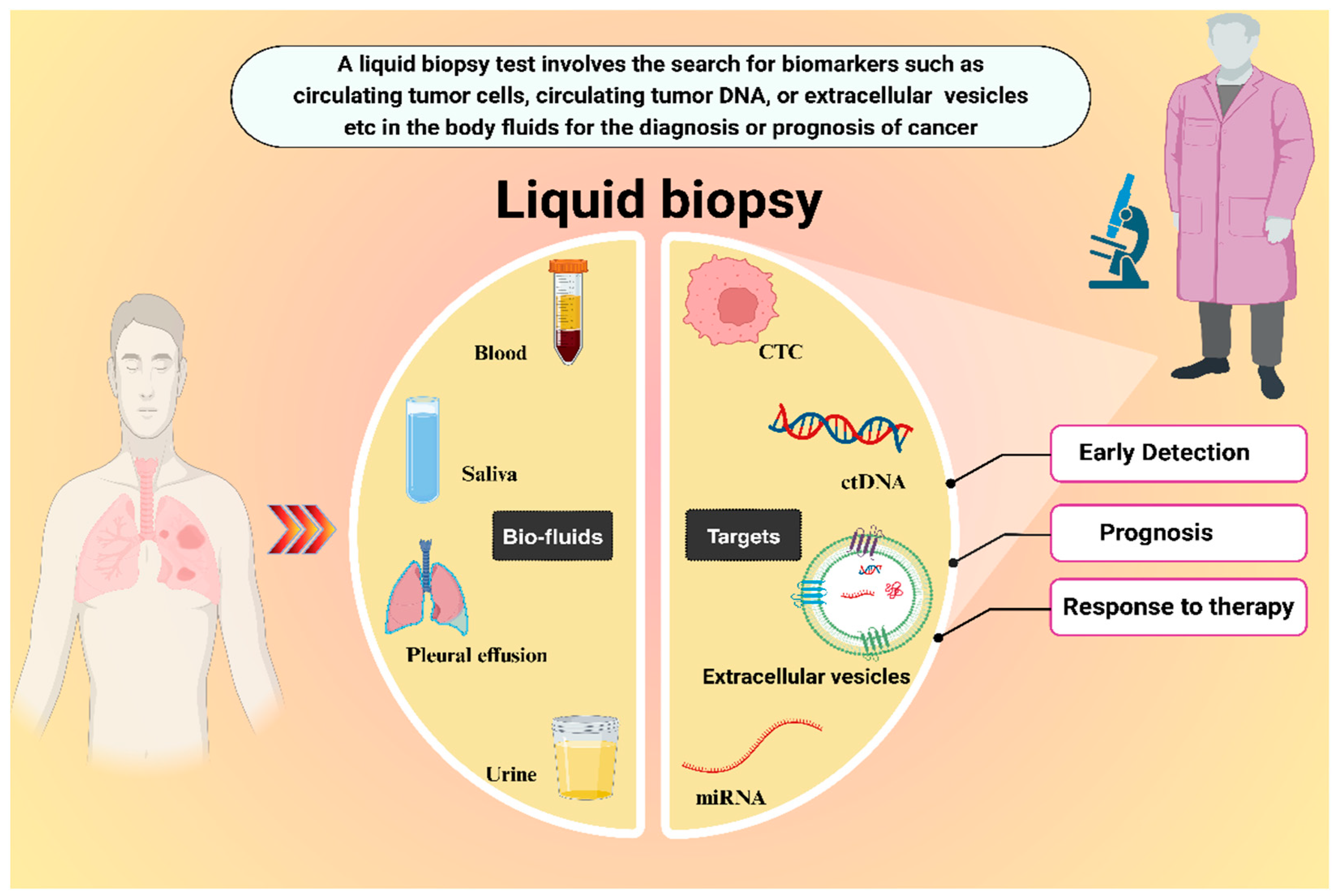
2. Liquid Biopsy Approach
3. Extracellular Vesicles (EVs)
4. Current State of EV-Based Liquid Biopsy for Lung Cancer
5. Two Critical Factors for EV-Based Liquid Biopsies
5.1. Isolation Techniques for EVs
| Isolation Technique | Strengths | Weaknesses |
|---|---|---|
| Ultracentrifugation |
|
|
| Density Gradient Centrifugation |
|
|
| Size Exclusion Chromatography |
|
|
| Ultrafiltration |
|
|
| Precipitation |
|
|
| Asymmetrical Flow Field-Flow Fractionation |
|
|
| Tangential Flow Filtration |
|
|
| Anion Exchange Chromatography |
|
|
| Immunoaffinity |
|
|
| Microfluidic Platform |
|
|
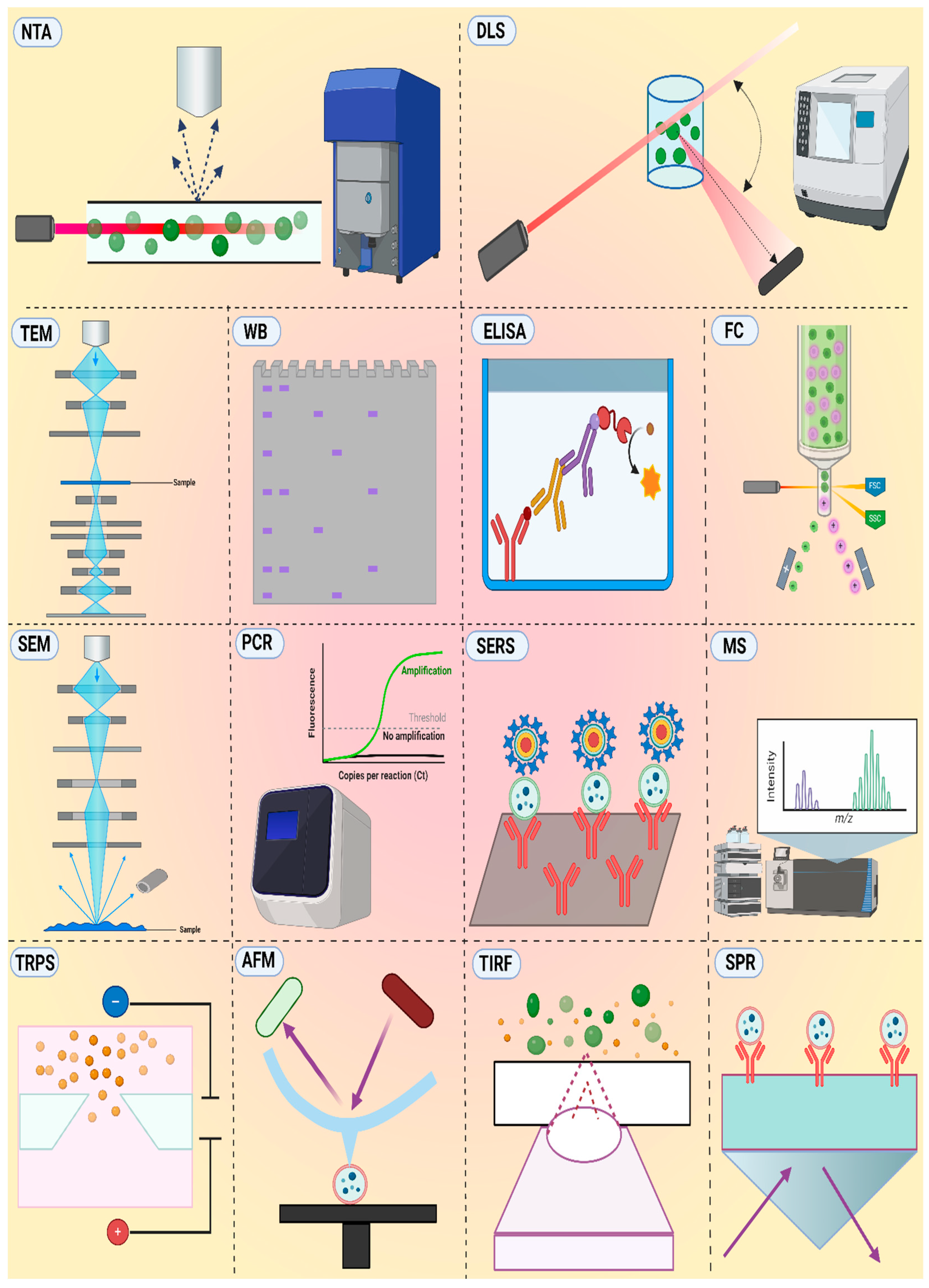
5.2. Analysis Techniques for EVs
| Technique | Strengths | Weaknesses |
|---|---|---|
| TEM |
|
|
| SEM |
|
|
| AFM |
|
|
| NTA |
|
|
| DLS |
|
|
| TRPS |
|
|
| WB |
|
|
| ELISA |
|
|
| MS |
|
|
| PCR |
|
|
| TIRF |
|
|
| FC |
|
|
| SPR |
|
|
| SERS |
|
|
6. Challenges
7. Future Prospects
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Qu, H.; Zhu, M.; Shan, C.; Ji, X.; Ji, G.; Zhang, W.; Zhang, H.; Chen, B. Prevalence, diagnosis, and treatment of chronic obstructive pulmonary disease in a hospitalized lung cancer population: A single center study. J. Thorac. Dis. 2023, 15, 4182. [Google Scholar] [CrossRef] [PubMed]
- Leiter, A.; Veluswamy, R.R.; Wisnivesky, J.P. The global burden of lung cancer: Current status and future trends. Nat. Rev. Clin. Oncol. 2023, 20, 624–639. [Google Scholar] [CrossRef] [PubMed]
- Elshoeibi, A.M.; Elsayed, B.; Kaleem, M.Z.; Elhadary, M.R.; Abu-Haweeleh, M.N.; Haithm, Y.; Krzyslak, H.; Vranic, S.; Pedersen, S. Proteomic Profiling of Small-Cell Lung Cancer: A Systematic Review. Cancers 2023, 15, 5005. [Google Scholar] [CrossRef]
- Mehta, A.; Barreto, G. Non-invasive approaches for lung cancer diagnosis. Indian J. Thorac. Cardiovasc. Surg. 2018, 34, 11–19. [Google Scholar] [CrossRef]
- Meriggi, F. Second-Line Treatment Options for Small-Cell Lung Cancer: A Light at The End of the Tunnel. Cancers 2024, 16, 255. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Zhao, D.; Lu, J.; Zeng, W.; Zhang, C.; You, Y. Changing trends in disease burden of lung cancer in China from 1990–2019 and following 15-year prediction. Curr. Probl. Cancer 2024, 48, 101036. [Google Scholar] [CrossRef]
- Sharma, R. Mapping of global, regional and national incidence, mortality and mortality-to-incidence ratio of lung cancer in 2020 and 2050. Int. J. Clin. Oncol. 2022, 27, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, M.; Alhusseini, N.; Samhan, L.; Samhan, S.; Abbad, T. Tobacco control policies implementation and future lung cancer incidence in Saudi Arabia. A population-based study. Prev. Med. Rep. 2023, 36, 102439. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, S.; Zhou, Z.; Lei, C.; Yu, H.; Zeng, C.; Xia, X.; Qiao, G.; Shi, Q. Unpleasant symptoms of immunotherapy for people with lung cancer: A mixed-method study. Int. J. Nurs. Stud. 2023, 139, 104430. [Google Scholar] [CrossRef]
- Kops, S.E.; Heus, P.; Korevaar, D.A.; Damen, J.A.; Idema, D.L.; Verhoeven, R.L.; Annema, J.T.; Hooft, L.; van der Heijden, E.H. Diagnostic yield and safety of navigation bronchoscopy: A systematic review and meta-analysis. Lung Cancer 2023, 180, 107196. [Google Scholar] [CrossRef] [PubMed]
- Visser, E.; Genet, S.A.A.M.; de Kock, R.P.P.A.; van den Borne, B.E.E.M.; Youssef-El Soud, M.; Belderbos, H.N.A.; Stege, G.; de Saegher, M.E.A.; van’t Westeinde, S.C.; Brunsveld, L.; et al. Liquid biopsy-based decision support algorithms for diagnosis and subtyping of lung cancer. Lung Cancer 2023, 178, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Chinnappan, R.; Mir, T.A.; Alsalameh, S.; Makhzoum, T.; Alzhrani, A.; Alnajjar, K.; Adeeb, S.; Al Eman, N.; Ahmed, Z.; Shakir, I. Emerging Biosensing Methods to Monitor Lung Cancer Biomarkers in Biological Samples: A Comprehensive Review. Cancers 2023, 15, 3414. [Google Scholar] [CrossRef] [PubMed]
- Jalal, A.H.; Sikder, A.K.; Alam, F.; Samin, S.; Rahman, S.S.; Khan, M.M.A.; Siddiquee, M.R. Early diagnosis with alternative approaches: Innovation in lung cancer care. Shanghai Chest 2021, 5, 1–14. [Google Scholar] [CrossRef]
- Silvestri, G.A.; Goldman, L.; Tanner, N.T.; Burleson, J.; Gould, M.; Kazerooni, E.A.; Mazzone, P.J.; Rivera, M.P.; Doria-Rose, V.P.; Rosenthal, L.S. Outcomes from more than 1 million people screened for lung cancer with low-dose CT imaging. Chest 2023, 164, 241–251. [Google Scholar] [CrossRef]
- Ledda, R.E.; Funk, G.-C.; Sverzellati, N. The pros and cons of lung cancer screening. Eur. Radiol. 2024, 1–9. [Google Scholar] [CrossRef]
- Zarinshenas, R.; Amini, A.; Mambetsariev, I.; Abuali, T.; Fricke, J.; Ladbury, C.; Salgia, R. Assessment of barriers and challenges to screening, diagnosis, and biomarker testing in early-stage lung cancer. Cancers 2023, 15, 1595. [Google Scholar] [CrossRef]
- De Margerie-Mellon, C.; Chassagnon, G. Artificial intelligence: A critical review of applications for lung nodule and lung cancer. Diagn. Interv. Imaging 2023, 104, 11–17. [Google Scholar] [CrossRef]
- Medhin, L.B.; Beasley, A.B.; Warburton, L.; Amanuel, B.; Gray, E.S. Extracellular vesicles as a liquid biopsy for melanoma: Are we there yet? Semin. Cancer Biol. 2023, 89, 92–98. [Google Scholar] [CrossRef]
- Li, L.; Jiang, H.; Zeng, B.; Wang, X.; Bao, Y.; Chen, C.; Ma, L.; Yuan, J. Liquid biopsy in lung cancer. Clin. Chim. Acta 2024, 554, 117757. [Google Scholar] [CrossRef]
- Bamankar, S.; Londhe, V.Y. The rise of extracellular vesicles as new age biomarkers in cancer diagnosis: Promises and pitfalls. Technol. Cancer Res. Treat. 2023, 22, 15330338221149266. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Niu, X.; Zhang, Z.; O’Byrne, K.; Kulasinghe, A.; Fielding, D.; Möller, A.; Wuethrich, A.; Lobb, R.J.; Trau, M. Glycan Profiling in Small Extracellular Vesicles with a SERS Microfluidic Biosensor Identifies Early Malignant Development in Lung Cancer. Adv. Sci. 2024, 11, 2401818. [Google Scholar] [CrossRef]
- Ilié, M.; Hofman, P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl. Lung Cancer Res. 2016, 5, 420–423. [Google Scholar] [CrossRef]
- Thenuwara, G.; Curtin, J.; Tian, F. Advances in diagnostic tools and therapeutic approaches for gliomas: A comprehensive review. Sensors 2023, 23, 9842. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, S.; Gaude, E.; Ruparel, M.; Van Der Schee, M.; Janes, S.; Rintoul, R.; Group, L.R. The potential of breath analysis to improve outcome for patients with lung cancer. J. Breath Res. 2019, 13, 034002. [Google Scholar] [CrossRef]
- Mahuron, K.M.; Fong, Y. Applications of liquid biopsy for surgical patients with cancer: A review. JAMA Surg. 2024, 159, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Shegekar, T.; Vodithala, S.; Juganavar, A. The emerging role of liquid biopsies in revolutionising cancer diagnosis and therapy. Cureus 2023, 15, e43650. [Google Scholar] [CrossRef]
- Cui, S.; Cheng, Z.; Qin, W.; Jiang, L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer 2018, 116, 46–54. [Google Scholar] [CrossRef]
- Davidson, S.M.; Boulanger, C.M.; Aikawa, E.; Badimon, L.; Barile, L.; Binder, C.J.; Brisson, A.; Buzas, E.; Emanueli, C.; Jansen, F. Methods for the identification and characterization of extracellular vesicles in cardiovascular studies: From exosomes to microvesicles. Cardiovasc. Res. 2023, 119, 45–63. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Boysen, J.; Nelson, M.; Magzoub, G.; Maiti, G.P.; Sinha, S.; Goswami, M.; Vesely, S.K.; Shanafelt, T.D.; Kay, N.E.; Ghosh, A.K. Dynamics of microvesicle generation in B-cell chronic lymphocytic leukemia: Implication in disease progression. Leukemia 2017, 31, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Chen, T.; Dahlman, J.; Eniola-Adefeso, L.; Ghiran, I.C.; Kurre, P.; Lam, W.A.; Lang, J.K.; Marbán, E.; Martín, P.; et al. Current challenges and future directions for engineering extracellular vesicles for heart, lung, blood and sleep diseases. J. Extracell. Vesicles 2023, 12, 12305. [Google Scholar] [CrossRef]
- Greening, D.W.; Simpson, R.J. Understanding extracellular vesicle diversity–current status. Expert Rev. Proteom. 2018, 15, 887–910. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, L.; Meng, Y.; Li, B.; Yang, Y.; Gao, F. Migrasome: A new functional extracellular vesicle. Cell Death Discov. 2023, 9, 381. [Google Scholar] [CrossRef]
- Shishido, S.N.; Lin, E.; Nissen, N.; Courcoubetis, G.; Suresh, D.; Mason, J.; Osipov, A.; Hendifar, A.E.; Lewis, M.; Gaddam, S. Cancer-related cells and oncosomes in the liquid biopsy of pancreatic cancer patients undergoing surgery. NPJ Precis. Oncol. 2024, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jeppesen, D.K.; Higginbotham, J.N.; Franklin, J.L.; Coffey, R.J. Comprehensive isolation of extracellular vesicles and nanoparticles. Nat. Protoc. 2023, 18, 1462–1487. [Google Scholar] [CrossRef]
- Liu, C.; Xiang, X.; Han, S.; Lim, H.Y.; Li, L.; Zhang, X.; Ma, Z.; Yang, L.; Guo, S.; Soo, R. Blood-based liquid biopsy: Insights into early detection and clinical management of lung cancer. Cancer Lett. 2022, 524, 91–102. [Google Scholar] [CrossRef]
- Ruzycka-Ayoush, M.; Prochorec-Sobieszek, M.; Cieszanowski, A.; Glogowski, M.; Szumera-Cieckiewicz, A.; Podgorska, J.; Targonska, A.; Sobczak, K.; Mosieniak, G.; Grudzinski, I.P. Extracellular Vesicles as Next-Generation Biomarkers in Lung Cancer Patients: A Case Report on Adenocarcinoma and Squamous Cell Carcinoma. Life 2024, 14, 408. [Google Scholar] [CrossRef]
- Rahimian, S.; Najafi, H.; Afzali, B.; Doroudian, M. Extracellular Vesicles and Exosomes: Novel Insights and Perspectives on Lung Cancer from Early Detection to Targeted Treatment. Biomedicines 2024, 12, 123. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, C.; Zeng, X.; Xiong, Y.; Li, K.; Huang, K.; Chen, P. Tandem hybridization chain reaction and selective coordination enable fluorescence detection of exosomes in lung cancer. Sens. Actuators B Chem. 2024, 410, 135722. [Google Scholar] [CrossRef]
- Lim, S.; Ha, Y.; Lee, B.; Shin, J.; Rhim, T. Calnexin as a dual-role biomarker: Antibody-based diagnosis and therapeutic targeting in lung cancer. BMB Rep. 2024, 57, 155. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Luo, S.; Lu, P.; Cai, C.; Li, W.; Li, C. Composition, functions, and applications of exosomal membrane proteins. Front. Immunol. 2024, 15, 1408415. [Google Scholar] [CrossRef]
- Sandfeld-Paulsen, B.; Aggerholm-Pedersen, N.; Bæk, R.; Jakobsen, K.R.; Meldgaard, P.; Folkersen, B.H.; Rasmussen, T.R.; Varming, K.; Jørgensen, M.M.; Sorensen, B.S. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 2016, 10, 1595–1602. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Di, K.; Khan, H.; He, N.; Li, Z. Rapid Capturing and Chemiluminescent Sensing of Programmed Death Ligand-1 Expressing Extracellular Vesicles. Biosensors 2022, 12, 281. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Qiu, F.; Qiu, Z. Proteomic identification of exosomal LRG1: A potential urinary biomarker for detecting NSCLC. Electrophoresis 2011, 32, 1976–1983. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Lee, J.-E.; Sung, K.J.; Sung, Y.H.; Pack, C.-G.; Jung, M.-k.; Han, B. Exosomal PD-L1 promotes tumor growth through immune escape in non-small cell lung cancer. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Yoh, K.E.; Lowe, C.J.; Mahajan, S.; Suttmann, R.; Nguy, T.; Reichelt, M.; Yang, J.; Melendez, R.; Li, Y.; Molinero, L. Enrichment of circulating tumor-derived extracellular vesicles from human plasma. J. Immunol. Methods 2021, 490, 112936. [Google Scholar] [CrossRef]
- Yamashita, T.; Kamada, H.; Kanasaki, S.; Maeda, Y.; Nagano, K.; Abe, Y.; Inoue, M.; Yoshioka, Y.; Tsutsumi, Y.; Katayama, S.; et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 2013, 68, 969–973. [Google Scholar]
- Sandfeld-Paulsen, B.; Jakobsen, K.R.; Bæk, R.; Folkersen, B.H.; Rasmussen, T.R.; Meldgaard, P.; Varming, K.; Jørgensen, M.M.; Sorensen, B.S. Exosomal proteins as diagnostic biomarkers in lung cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. [Google Scholar] [CrossRef]
- Zhou, X.; Wen, W.; Shan, X.; Zhu, W.; Xu, J.; Guo, R.; Cheng, W.; Wang, F.; Qi, L.-W.; Chen, Y. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget 2017, 8, 6513. [Google Scholar] [CrossRef] [PubMed]
- Dejima, H.; Iinuma, H.; Kanaoka, R.; Matsutani, N.; Kawamura, M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol. Lett. 2017, 13, 1256–1263. [Google Scholar] [CrossRef]
- Hydbring, P.; De Petris, L.; Zhang, Y.; Brandén, E.; Koyi, H.; Novak, M.; Kanter, L.; Hååg, P.; Hurley, J.; Tadigotla, V. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer 2018, 124, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yu, X.; Bu, J.; Xiao, Q.; Ma, S.; Chen, N.; Qu, C. Comparative analyses of salivary exosomal miRNAs for patients with or without lung cancer. Front. Genet. 2023, 14, 1249678. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Lu, F.; Wang, J.; Wang, K.; Liu, B.; Li, N.; Tang, B. A portable point-of-care testing system to diagnose lung cancer through the detection of exosomal miRNA in urine and saliva. Chem. Commun. 2020, 56, 8968–8971. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, Y.; Song, X.; Xie, L.; Zhao, S.; Song, X. Tumor-derived exosomal miRNAs as diagnostic biomarkers in non-small cell lung cancer. Front. Oncol. 2020, 10, 560025. [Google Scholar] [CrossRef]
- Rolfo, C.; Chacartegui, J.; Giallombardo, M.; Alessandro, R.; Peeters, M. 7P Exosomes isolated in plasma of non-small cell lung cancer patients contain microRNA related to the EGFR pathway: Proof of concept. J. Thorac. Oncol. 2016, 11, S85. [Google Scholar] [CrossRef]
- Sha, L.; Bo, B.; Li, J.; Liu, Q.; Cao, Y.; Zhao, J. Precise assessment of lung cancer-derived exosomes based on dual-labelled membrane interface. Chin. Chem. Lett. 2024, 110109. [Google Scholar] [CrossRef]
- Luo, S.; Wu, Y.; Pan, W.; Zhong, G.; Situ, B.; Li, B.; Ye, X.; Jiang, X.; Li, W.; Zhang, Y. An integrated magneto-fluorescent nanosensor for rapid and sensitive detection of tumor-derived exosomes. Sens. Actuators B Chem. 2023, 374, 132792. [Google Scholar] [CrossRef]
- Feng, J.; Jia, L.; Pan, W.; Fan, Y.; Guo, J.; Luo, T.; Liu, C.; Wang, W.; Zheng, L.; Li, B. Rapid and efficient fluorescent aptasensor for PD-L1 positive extracellular vesicles isolation and analysis: EV-ANCHOR. Chem. Eng. J. 2023, 465, 142811. [Google Scholar] [CrossRef]
- Ren, F.; Fei, Q.; Qiu, K.; Zhang, Y.; Zhang, H.; Sun, L. Liquid biopsy techniques and lung cancer: Diagnosis, monitoring and evaluation. J Exp Clin Cancer Res. 2024, 43, 96. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.-Y.; Huang, D.-F.; Si, M.-Y.; Lu, H.; Tang, Z.-X.; Zhang, Z.-X. Role of exosomes in non-small cell lung cancer and EGFR-mutated lung cancer. Front. Immunol. 2023, 14, 1142539. [Google Scholar] [CrossRef] [PubMed]
- Kan, C.F.K.; Unis, G.D.; Li, L.Z.; Gunn, S.; Li, L.; Soyer, H.P.; Stark, M.S. Circulating Biomarkers for Early Stage Non-Small Cell Lung Carcinoma Detection: Supplementation to Low-Dose Computed Tomography. Front. Oncol. 2021, 11, 555331. [Google Scholar] [CrossRef]
- Zhang, Q.; Qin, S.; Peng, C.; Liu, Y.; Huang, Y.; Ju, S. Circulating circular RNA hsa_circ_0023179 acts as a diagnostic biomarker for non-small-cell lung cancer detection. J. Cancer Res. Clin. Oncol. 2023, 149, 3649–3660. [Google Scholar] [CrossRef]
- Cazzoli, R.; Buttitta, F.; Di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef]
- Yang, B.; Xin, X.; Cao, X.; Nasifu, L.; Nie, Z.; He, B. The diagnostic and prognostic value of exosomal microRNAs in lung cancer: A systematic review. Clin. Transl. Oncol. 2024, 26, 1921–1933. [Google Scholar] [CrossRef]
- Liu, S.; Wu, X.; Wang, Y.; Chen, Y. Exosomal circ_0000735 contributes to non-small lung cancer malignant progression. J. Biochem. Mol. Toxicol. 2024, 38, e23700. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, S.; Hååg, P.; Sahu, S.S.; Berisha, L.; Kaminskyy, V.O.; Ekman, S.; Lewensohn, R.; Linnros, J.; Viktorsson, K.; Dev, A. Multiplexed electrokinetic sensor for detection and therapy monitoring of extracellular vesicles from liquid biopsies of non-small-cell lung cancer patients. Biosens. Bioelectron. 2021, 193, 113568. [Google Scholar] [CrossRef]
- Yu, Q.; Zhao, Q.; Wang, S.; Zhao, S.; Zhang, S.; Yin, Y.; Dong, Y. Development of a lateral flow aptamer assay strip for facile identification of theranostic exosomes isolated from human lung carcinoma cells. Anal. Biochem. 2020, 594, 113591. [Google Scholar] [CrossRef]
- Fan, Y.; Duan, X.; Zhao, M.; Wei, X.; Wu, J.; Chen, W.; Liu, P.; Cheng, W.; Cheng, Q.; Ding, S. High-sensitive and multiplex biosensing assay of NSCLC-derived exosomes via different recognition sites based on SPRi array. Biosens. Bioelectron. 2020, 154, 112066. [Google Scholar] [CrossRef]
- Conteduca, D.; Brunetti, G.; Barth, I.; Quinn, S.D.; Ciminelli, C.; Krauss, T.F. Multiplexed Near-Field Optical Trapping Exploiting Anapole States. ACS Nano 2023, 17, 16695–16702. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, X.; Kong, Q.; He, H.; Sun, J.; Qiu, W.; Zhang, L.; Yang, M. Extracellular Vesicle Preparation and Analysis: A State-of-the-Art Review. Adv. Sci. 2024, 11, 2401069. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wijerathne, H.; Godwin, A.K.; Soper, S.A. Isolation and analysis methods of extracellular vesicles (EVs). Extracell. Vesicles Circ. Nucleic Acids 2021, 2, 80. [Google Scholar]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Clos-Sansalvador, M.; Monguió-Tortajada, M.; Roura, S.; Franquesa, M.; Borras, F.E. Commonly used methods for extracellular vesicles’ enrichment: Implications in downstream analyses and use. Eur. J. Cell Biol. 2022, 101, 151227. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q. Progress, opportunity, and perspective on exosome isolation-efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684. [Google Scholar] [CrossRef]
- Macías, M.; Rebmann, V.; Mateos, B.; Varo, N.; Perez-Gracia, J.L.; Alegre, E.; González, Á. Comparison of six commercial serum exosome isolation methods suitable for clinical laboratories. Effect in cytokine analysis. Clin. Chem. Lab. Med. (CCLM) 2019, 57, 1539–1545. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhu, D.; Wang, J.; Wu, X. A highly efficient method for isolating urinary exosomes. Int. J. Mol. Med. 2019, 43, 83–90. [Google Scholar] [CrossRef]
- Pirolli, N.H.; Reus, L.S.C.; Mamczarz, Z.; Khan, S.; Bentley, W.E.; Jay, S.M. High performance anion exchange chromatography purification of probiotic bacterial extracellular vesicles enhances purity and anti-inflammatory efficacy. Biotechnol. Bioeng. 2023, 120, 3368–3380. [Google Scholar] [CrossRef]
- Bian, J.; Gobalasingham, N.; Purchel, A.; Lin, J. The power of field-flow fractionation in characterization of nanoparticles in drug delivery. Molecules 2023, 28, 4169. [Google Scholar] [CrossRef]
- Ströhle, G.; Gan, J.; Li, H. Affinity-based isolation of extracellular vesicles and the effects on downstream molecular analysis. Anal. Bioanal. Chem. 2022, 414, 7051–7067. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, K.P.; Rossi, I.; Abdullahi, M.; Ramirez, M.I.; Stratton, D.; Inal, J.M. Isolation and characterization of extracellular vesicles and future directions in diagnosis and therapy. WIREs Nanomed. Nanobiotechnol. 2023, 15, e1835. [Google Scholar] [CrossRef] [PubMed]
- Visan, K.S.; Wu, L.-Y.; Voss, S.; Wuethrich, A.; Möller, A. Status quo of Extracellular Vesicle isolation and detection methods for clinical utility. Semin. Cancer Biol. 2023, 88, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.; Fernandez-Rhodes, M.; Law, A.; Peacock, B.; Lewis, M.P.; Davies, O.G. Comparison of extracellular vesicle isolation processes for therapeutic applications. J. Tissue Eng. 2023, 14, 20417314231174609. [Google Scholar] [CrossRef] [PubMed]
- Pallares-Rusiñol, A.; Bernuz, M.; Moura, S.L.; Fernández-Senac, C.; Rossi, R.; Martí, M.; Pividori, M.I. Chapter Two-Advances in exosome analysis. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2023; Volume 112, pp. 69–117. [Google Scholar]
- Havers, M.; Broman, A.; Lenshof, A.; Laurell, T. Advancement and obstacles in microfluidics-based isolation of extracellular vesicles. Anal. Bioanal. Chem. 2023, 415, 1265–1285. [Google Scholar] [CrossRef]
- Omrani, M.; Beyrampour-Basmenj, H.; Jahanban-Esfahlan, R.; Talebi, M.; Raeisi, M.; Serej, Z.A.; Akbar-Gharalari, N.; Khodakarimi, S.; Wu, J.; Ebrahimi-Kalan, A. Global trend in exosome isolation and application: An update concept in management of diseases. Mol. Cell. Biochem. 2024, 479, 679–691. [Google Scholar] [CrossRef]
- Bachurski, D.; Schuldner, M.; Nguyen, P.-H.; Malz, A.; Reiners, K.S.; Grenzi, P.C.; Babatz, F.; Schauss, A.C.; Hansen, H.P.; Hallek, M. Extracellular vesicle measurements with nanoparticle tracking analysis–An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles 2019, 8, 1596016. [Google Scholar] [CrossRef]
- Parisse, P.; Rago, I.; Ulloa Severino, L.; Perissinotto, F.; Ambrosetti, E.; Paoletti, P.; Ricci, M.; Beltrami, A.P.; Cesselli, D.; Casalis, L. Atomic force microscopy analysis of extracellular vesicles. Eur. Biophys. J. 2017, 46, 813–820. [Google Scholar] [CrossRef]
- Maas, S.L.; Broekman, M.L.; de Vrij, J. Tunable resistive pulse sensing for the characterization of extracellular vesicles. Exosomes Microvesicles Methods Protoc. 2017, 1545, 21–33. [Google Scholar]
- Pang, Y.; Shi, J.; Yang, X.; Wang, C.; Sun, Z.; Xiao, R. Personalized detection of circling exosomal PD-L1 based on Fe3O4@ TiO2 isolation and SERS immunoassay. Biosens. Bioelectron. 2020, 148, 111800. [Google Scholar] [CrossRef]
- Palmieri, V.; Lucchetti, D.; Gatto, I.; Maiorana, A.; Marcantoni, M.; Maulucci, G.; Papi, M.; Pola, R.; De Spirito, M.; Sgambato, A. Dynamic light scattering for the characterization and counting of extracellular vesicles: A powerful noninvasive tool. J. Nanoparticle Res. 2014, 16, 1–8. [Google Scholar] [CrossRef]
- Jimenez, L.; Yu, H.; McKenzie, A.J.; Franklin, J.L.; Patton, J.G.; Liu, Q.; Weaver, A.M. Quantitative proteomic analysis of small and large extracellular vesicles (EVs) reveals enrichment of adhesion proteins in small EVs. J. Proteome Res. 2019, 18, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Chen, K.; Wang, Z.; Wang, Y.; Liu, J.; Lin, L.; Shao, Y.; Gao, L.; Yin, H.; Cui, C. DNA in serum extracellular vesicles is stable under different storage conditions. BMC Cancer 2016, 16, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.A.; Van Der Pol, E.; Arkesteijn, G.J.; Bremer, M.; Brisson, A.; Coumans, F.; Dignat-George, F.; Duggan, E.; Ghiran, I.; Giebel, B. MIFlowCyt-EV: A framework for standardized reporting of extracellular vesicle flow cytometry experiments. J. Extracell. Vesicles 2020, 9, 1713526. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Phan, M.M.; Sun, Y.; Koerber, J.T.; Ho, H.; Chen, Y.; Yang, J. Development of an SPR-based binding assay for characterization of anti-CD20 antibodies to CD20 expressed on extracellular vesicles. Anal. Biochem. 2022, 646, 114635. [Google Scholar] [CrossRef]
- Martín-Cófreces, N.B.; Torralba, D.; Lozano-Prieto, M.; Fernández-Gallego, N.; Sánchez-Madrid, F. TIRF microscopy as a tool to determine exosome composition. Stem Cell Renew. Cell-Cell Commun. Methods Protoc. 2021, 2346, 91–104. [Google Scholar]
- Lee, J.; Kim, H.; Heo, Y.; Yoo, Y.K.; Han, S.I.; Kim, C.; Hur, D.; Kim, H.; Kang, J.Y.; Lee, J.H. Enhanced paper-based ELISA for simultaneous EVs/exosome isolation and detection using streptavidin agarose-based immobilization. Analyst 2020, 145, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Altıntaş, O.; Saylan, Y. Exploring the versatility of exosomes: A review on isolation, characterization, detection methods, and diverse applications. Anal. Chem. 2023, 95, 16029–16048. [Google Scholar] [CrossRef] [PubMed]
- Sonbhadra, S.; Mehak; Pandey, L.M. Biogenesis, isolation, and detection of exosomes and their potential in therapeutics and Diagnostics. Biosensors 2023, 13, 802. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, H.; Liu, Q.; Zeng, N.; Fu, G.; Qiu, Y.; Yang, Y.; Yuan, H.; Wang, W.; Li, B. Exosomes as Powerful Biomarkers in Cancer: Recent Advances in Isolation and Detection Techniques. Int. J. Nanomed. 2024, 19, 1923–1949. [Google Scholar] [CrossRef]
- Palmieri, M.; Frullanti, E. Different Liquid Biopsies for the Management of Non-Small Cell Lung Cancer in the Mutational Oncology Era. Med. Sci. 2023, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Di Capua, D.; Bracken-Clarke, D.; Ronan, K.; Baird, A.-M.; Finn, S. The Liquid Biopsy for Lung Cancer: State of the Art, Limitations and Future Developments. Cancers 2021, 13, 3923. [Google Scholar] [CrossRef] [PubMed]
- Ospina, A.V. Overview of the Role of Liquid Biopsy in Non-small Cell Lung Cancer (NSCLC). Clin. Oncol. 2024, 36, e371–e380. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Amin Mahdian, S.M.; Ebrahimi, M.S.; Taghizadieh, M.; Vosough, M.; Sadri Nahand, J.; Hosseindoost, S.; Vousooghi, N.; Javar, H.A.; Larijani, B.; et al. Microfluidics for detection of exosomes and microRNAs in cancer: State of the art. Mol. Ther.-Nucleic Acids 2022, 28, 758–791. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.A.; Chan, L.K.W.; Hung, L.C.; Phoebe, L.K.W.; Park, Y.; Yi, K.-H. Clinical Applications of Exosomes: A Critical Review. Int. J. Mol. Sci. 2024, 25, 7794. [Google Scholar] [CrossRef]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.K.; Enderle, D.; Noerholm, M.; Breakefield, X.O.; Skog, J.K. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef]
- Yakubovich, E.; Polischouk, A.; Evtushenko, V. Principles and problems of exosome isolation from biological fluids. Biochem. Suppl. Ser. A Membr. Cell Biol. 2022, 16, 115–126. [Google Scholar] [CrossRef]
- Malapelle, U.; Pisapia, P.; Addeo, A.; Arrieta, O.; Bellosillo, B.; Cardona, A.F.; Cristofanilli, M.; De Miguel-Perez, D.; Denninghoff, V.; Durán, I. Liquid biopsy from research to clinical practice: Focus on non-small cell lung cancer. Expert Rev. Mol. Diagn. 2021, 21, 1165–1178. [Google Scholar] [CrossRef]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in exosome isolation and analysis in health and disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef]
- Martins, T.S.; Vaz, M.; Henriques, A.G. A review on comparative studies addressing exosome isolation methods from body fluids. Anal. Bioanal. Chem. 2023, 415, 1239–1263. [Google Scholar] [CrossRef]

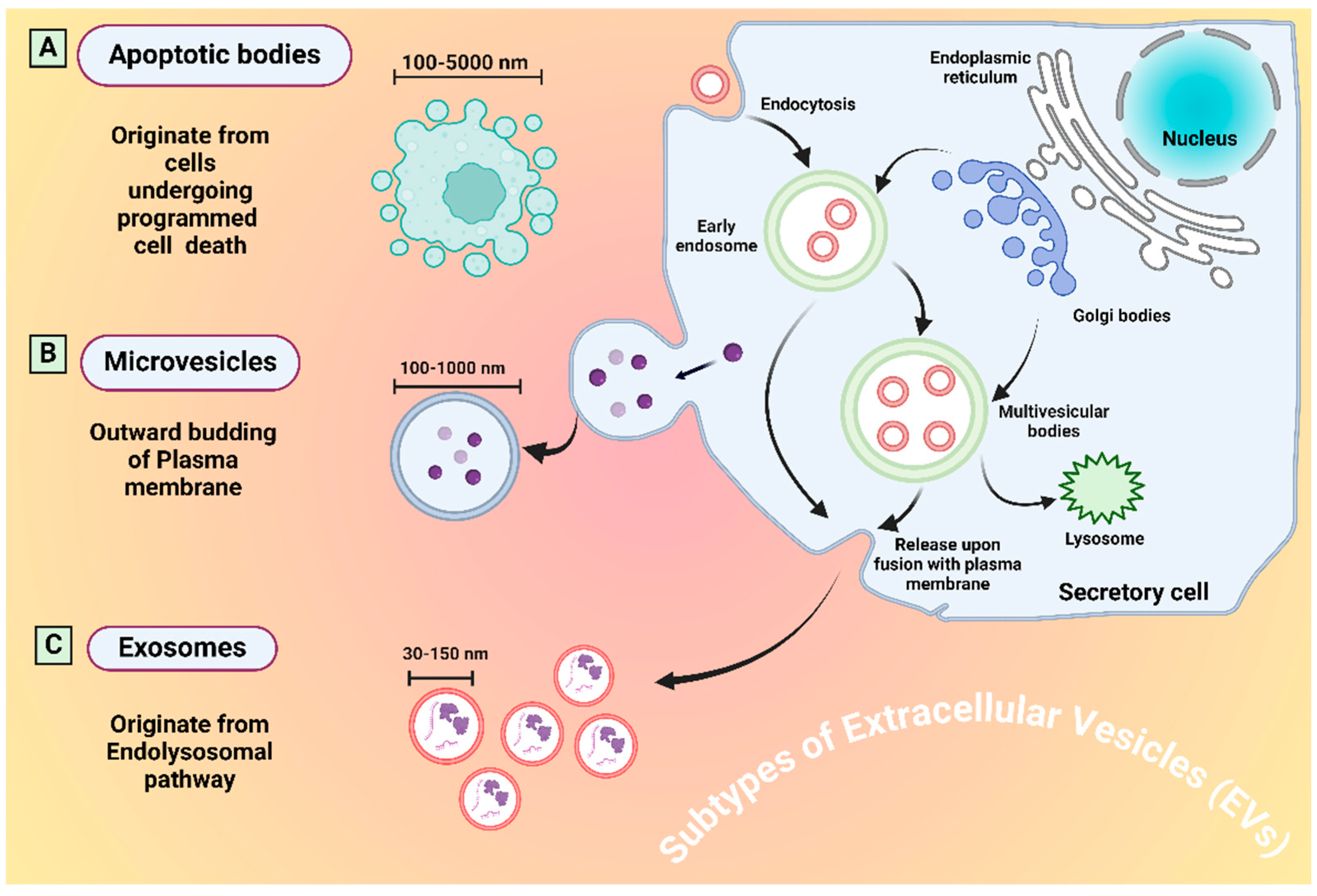

| Subtype | Size | Origination | Markers | Ref. |
|---|---|---|---|---|
| Exosomes | 30–150 nm | Late endosomal through MVBs | CD9, CD63, Tsg101, CD81, ALIX, HSP70 | [31] |
| Microvesicles | 100–1000 nm | Directly through plasma membrane budding | Integrins, Selectins, CD40, tissue factor | [32] |
| Apoptotic bodies | 100–5000 nm | Through programmed cell death | Annexin V, C3b, thrombospondin, Annexin A1, histone coagulation factor | [33] |
| Exomeres | ≤50 nm | Through cleavage of cytoplasmic extension | TGFBI, ENO1 and GPC1 | [34] |
| Migrasomes | 500–3000 | Through bifurcation of retraction fibers during cell migration | Tspan4, CD63, Annexin A1 | [35] |
| Oncosomes | 1000–10,000 | Through cancer cell amoeboid movement | Cav-1 or ADP ribosylation factor 6 | [36] |
| Supermeres | 35–50 | Not explored | TGFBI, ACE2, PCSK9, miR-1246, MET, GPC1 and AGO2. | [37] |
| Source | EV-Based Biomarker | Utility | Ref. |
|---|---|---|---|
| Plasma | NY-ESO-1 | Prognostic | [44] |
| Serum | PD-L1 | Diagnosis | [45] |
| Urine | LRG1 | Diagnosis | [46] |
| Serum | PD-L1 | Prognosis | [47] |
| Plasma | EpCam | Diagnosis and prognosis | [48] |
| Serum | EGFR | Diagnosis | [49] |
| plasma | CD151, CD171, and tetraspanin 8 | Diagnosis | [44] |
| Plasma | CD63, CD9, CD81 | Diagnosis and prognosis | [50] |
| Plasma | miR-19-3p, miR-221-3p, and miR-21-5p | Diagnosis | [51] |
| Plasma | miR-21 and miR-4257 | Prognosis | [52] |
| Pleural effusion | miR-200 | Diagnosis | [53] |
| Saliva | miR-92b-5p | Diagnosis | [54] |
| Urine and saliva | miRNA-205 | Diagnosis | [55] |
| Serum | miR-125b-5p | Diagnosis | [56] |
| Plasma | miR-30b/30c | Diagnosis | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, A.; Raza, F.; He, N. Nanoscale Extracellular Vesicle-Enabled Liquid Biopsy: Advances and Challenges for Lung Cancer Detection. Micromachines 2024, 15, 1181. https://doi.org/10.3390/mi15101181
Khan A, Raza F, He N. Nanoscale Extracellular Vesicle-Enabled Liquid Biopsy: Advances and Challenges for Lung Cancer Detection. Micromachines. 2024; 15(10):1181. https://doi.org/10.3390/mi15101181
Chicago/Turabian StyleKhan, Adeel, Faisal Raza, and Nongyue He. 2024. "Nanoscale Extracellular Vesicle-Enabled Liquid Biopsy: Advances and Challenges for Lung Cancer Detection" Micromachines 15, no. 10: 1181. https://doi.org/10.3390/mi15101181
APA StyleKhan, A., Raza, F., & He, N. (2024). Nanoscale Extracellular Vesicle-Enabled Liquid Biopsy: Advances and Challenges for Lung Cancer Detection. Micromachines, 15(10), 1181. https://doi.org/10.3390/mi15101181









