The Effect of Different System Parameters on the Movement of Microbial Cells Using Light-Induced Dielectrophoresis
Abstract
1. Introduction
2. Theoretical Background
3. Materials and Methods
3.1. Experimental Setup
3.2. Computational Model
3.3. Cell Culture and Sample Preparation
3.4. Experimental Protocol and Data Analysis
4. Results and Discussion
4.1. Physical Manipulation of Single Cells
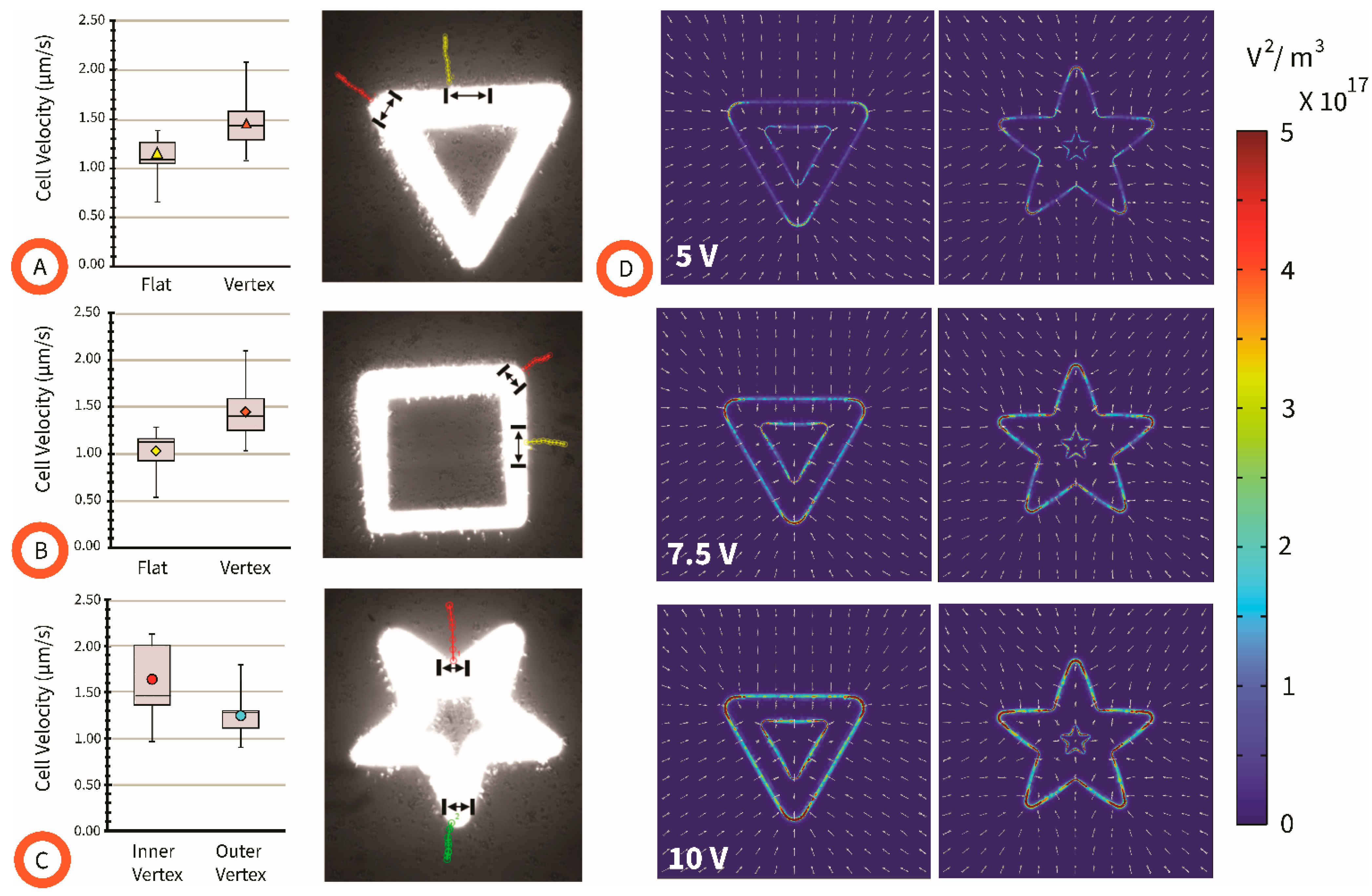
4.2. Effect of Pattern Shape on Cell Velocity
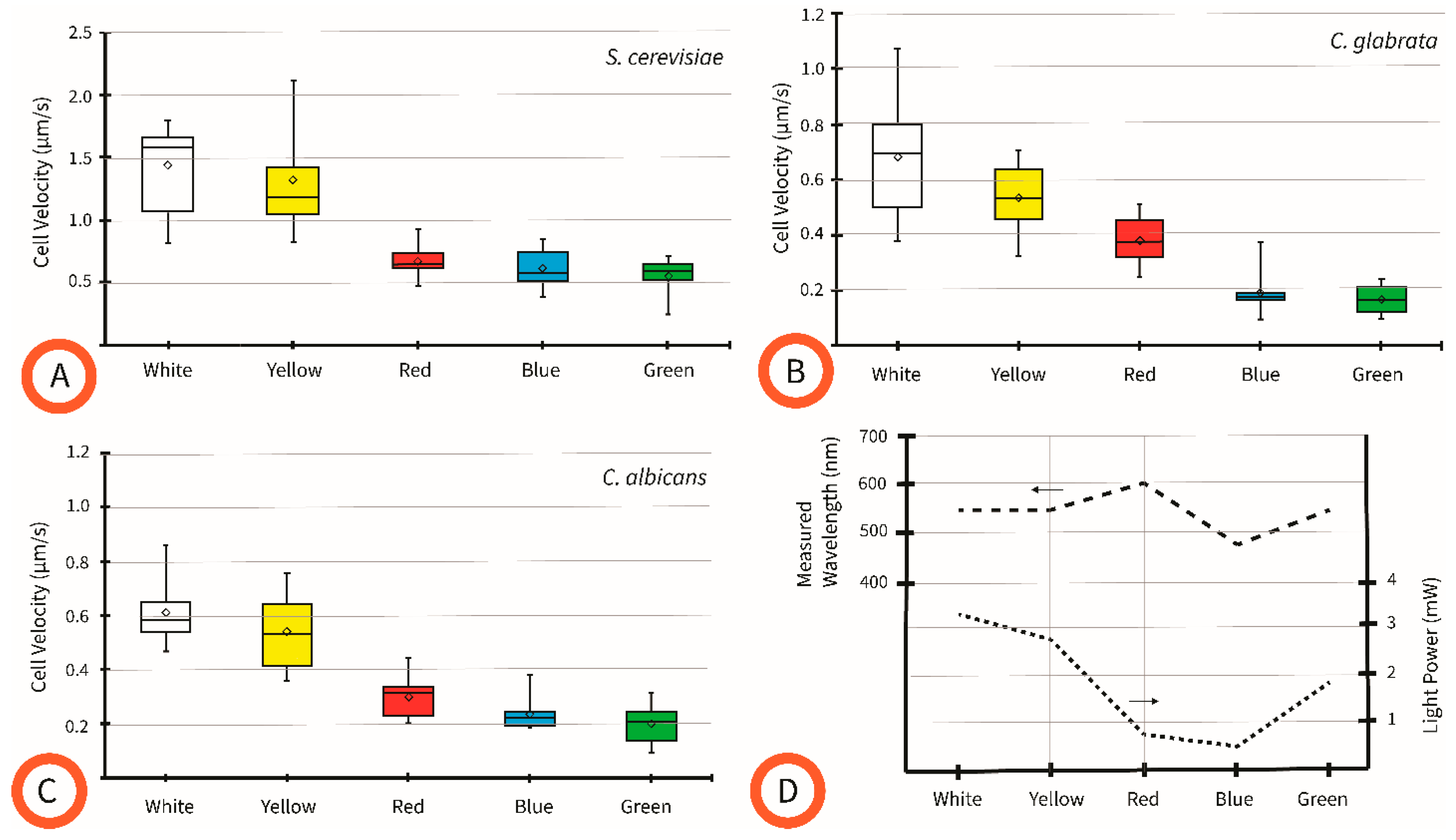
4.3. Effect of Pattern Color on Cell Velocity
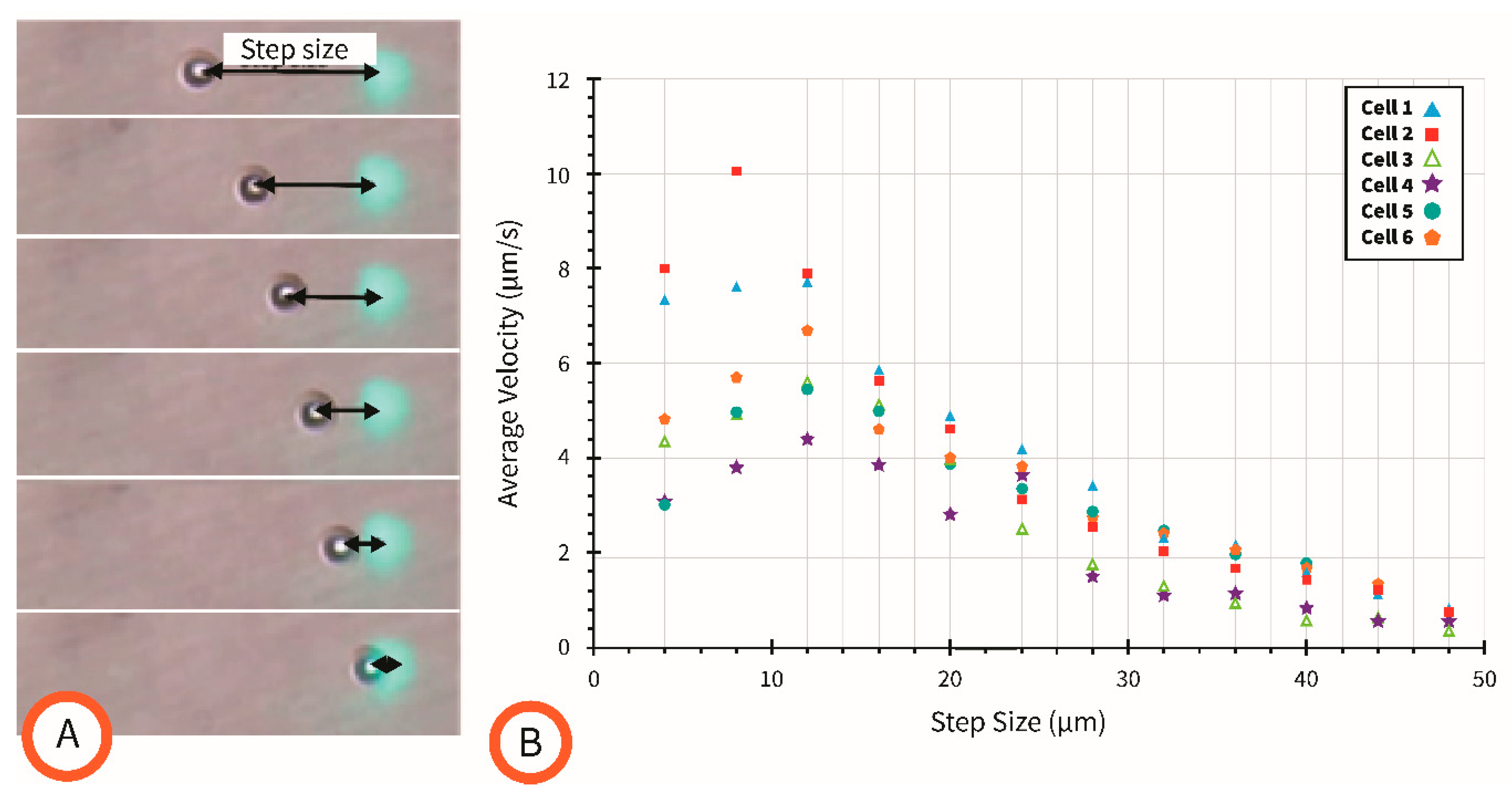
4.4. Effect of Movement Step Size on Cell Velocity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, W.; Wang, X.; Wang, Z.; Liang, W.; Ge, Z. Light-Powered Microrobots: Recent Progress and Future Challenges. Opt. Lasers Eng. 2023, 161, 107380. [Google Scholar] [CrossRef]
- Yu, H.; Fu, Y.; Zhang, X.; Chen, L.; Qi, D.; Shi, J.; Wang, W. Programmable Active Matter across Scales. Program. Mater. 2023, 1, e7. [Google Scholar] [CrossRef]
- Fu, Y.; Yu, H.; Zhang, X.; Malgaretti, P.; Kishore, V.; Wang, W. Microscopic Swarms: From Active Matter Physics to Biomedical and Environmental Applications. Micromachines 2022, 13, 295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Su, G.; Mei, J.; Li, J. A Review of Single-Cell Microrobots: Classification, Driving Methods and Applications. Micromachines 2023, 14, 1710. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, D.; Liang, S.; Dai, Y.; Bai, X.; Song, B.; Zhang, D.; Chen, H.; Feng, L. Recent Advances in Field-Controlled Micro–Nano Manipulations and Micro–Nano Robots. Adv. Intell. Syst. 2022, 4, 2100116. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, Z.; Zhu, B.; Zhang, Y.; Lin, J.; Wu, Y.; Wu, D. Design, Fabrication and Application of Magnetically Actuated Micro/Nanorobots: A Review. Nanotechnology 2022, 33, 152001. [Google Scholar] [CrossRef] [PubMed]
- Pethig, R. Dielectrophoresis: Theory, Methodology, and Biological Applications; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 245–300. [Google Scholar]
- Chou, W.P.; Wang, H.M.; Chang, J.H.; Chiu, T.K.; Hsieh, C.H.; Liao, C.J.; Wu, M.H. The Utilization of Optically-Induced-Dielectrophoresis (ODEP)-Based Virtual Cell Filters in a Microfluidic System for Continuous Isolation and Purification of Circulating Tumour Cells (CTCs) Based on Their Size Characteristics. Sens. Actuators B Chem. 2017, 241, 245–254. [Google Scholar] [CrossRef]
- Huang, S.B.; Wu, M.H.; Lin, Y.H.; Hsieh, C.H.; Yang, C.L.; Lin, H.C.; Tseng, C.P.; Lee, G. Bin High-Purity and Label-Free Isolation of Circulating Tumor Cells (CTCs) in a Microfluidic Platform by Using Optically-Induced-Dielectrophoretic (ODEP) Force. Lab Chip 2013, 13, 1371–1383. [Google Scholar] [CrossRef]
- Chiu, T.K.; Chou, W.P.; Huang, S.B.; Wang, H.M.; Lin, Y.C.; Hsieh, C.H.; Wu, M.H. Application of Optically-Induced-Dielectrophoresis in Microfluidic System for Purification of Circulating Tumour Cells for Gene Expression Analysis-Cancer Cell Line Model. Sci. Rep. 2016, 6, 32851. [Google Scholar] [CrossRef]
- Chu, P.Y.; Hsieh, C.H.; Lin, C.R.; Wu, M.H. The Effect of Optically Induced Dielectrophoresis (ODEP)-Based Cell Manipulation in a Microfluidic System on the Properties of Biological Cells. Biosensors 2020, 10, 65. [Google Scholar] [CrossRef]
- Liu, N.; Lin, Y.; Peng, Y.; Xin, L.; Yue, T.; Liu, Y.; Ru, C.; Xie, S.; Dong, L.; Pu, H.; et al. Automated Parallel Electrical Characterization of Cells Using Optically-Induced Dielectrophoresis. IEEE Trans. Autom. Sci. Eng. 2020, 17, 1084–1092. [Google Scholar] [CrossRef]
- Lu, Y.S.; Huang, Y.P.; Yeh, J.A.; Lee, C.; Chang, Y.H. Controllability of Non-Contact Cell Manipulation by Image Dielectrophoresis (IDEP). Opt. Quantum Electron. 2005, 37, 1385–1395. [Google Scholar] [CrossRef]
- Kremer, C.; Neale, S.; Menachery, A.; Barrett, M.; Cooper, J.M. Optoelectronic Tweezers for Medical Diagnostics. In Proceedings of the Frontiers in Biological Detection: From Nanosensors to Systems IV, San Francisco, CA, USA, 21–22 January 2012; SPIE: Washington, DC, USA, 2012; Volume 8212, p. 82120E. [Google Scholar]
- Ohta, A.T.; Chiou, P.Y.; Han, T.H.; Liao, J.C.; Bhardwaj, U.; McCabe, E.R.B.; Yu, F.; Sun, R.; Wu, M.C. Dynamic Cell and Microparticle Control via Optoelectronic Tweezers. J. Microelectromechanical Syst. 2007, 16, 491–499. [Google Scholar] [CrossRef]
- Yang, S.-M.; Yu, T.-M.; Huang, H.-P.; Ku, M.-Y.; Hsu, L.; Liu, C.-H. Dynamic Manipulation and Patterning of Microparticles and Cells by Using TiOPc-Based Optoelectronic Dielectrophoresis. Opt. Lett. 2010, 35, 1959–1961. [Google Scholar] [CrossRef]
- Chiou, P.Y.; Chang, Z.; Wu, M.C. A Novel Optoelectronic Tweezer Using Light Induced Dielectrophoresis. In Proceedings of the 2003 IEEE/LEOS International Conference on Optical MEMS, Waikoloa, HI, USA, 18–21 August 2003; pp. 8–9. [Google Scholar] [CrossRef]
- Chiou, P.Y.; Ohta, A.T.; Wu, M.C. Massively Parallel Manipulation of Single Cells and Microparticles Using Optical Images. Nature 2005, 436, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Hoeb, M.; Rädler, J.O.; Klein, S.; Stutzmann, M.; Brandt, M.S. Light-Induced Dielectrophoretic Manipulation of DNA. Biophys. J. 2007, 93, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Kalim, S.; Callahan, C.; Teitell, M.A.; Chiou, E.P.Y. A Light-Induced Dielectrophoretic Droplet Manipulation Platform. Lab Chip 2009, 9, 3228–3235. [Google Scholar] [CrossRef] [PubMed]
- Valley, J.K.; Neale, S.; Hsu, H.-Y.; Ohta, A.T.; Jamshidi, A.; Wu, M.C. Parallel Single-Cell Light-Induced Electroporation and Dielectrophoretic Manipulation. Lab Chip 2009, 9, 1714–1720. [Google Scholar] [CrossRef]
- Cheng, I.F.; Liu, S.L.; Chung, C.C.; Chang, H.C. Stepwise Gray-Scale Light-Induced Electric Field Gradient for Passive and Continuous Separation of Microparticles. Microfluid. Nanofluidics 2012, 12, 95–105. [Google Scholar] [CrossRef]
- Ohta, A.T.; Chiou, P.-Y.; Wu, M.C. Optically Controlled Manipulation of Live Cells Using Optoelectronic Tweezers. Opt. Trapp. Opt. Micromanipulation III 2006, 6326, 632617. [Google Scholar] [CrossRef]
- Martinez-Duarte, R. Microfabrication Technologies in Dielectrophoresis Applications—A Review. Electrophoresis 2012, 33, 3110–3132. [Google Scholar] [CrossRef] [PubMed]
- WHO Fungal Priority List 2022. Available online: https://www.who.int/publications/i/item/9789240060241 (accessed on 23 October 2023).
- Talapko, J.; Juzbašić, M.; Matijević, T.; Pustijanac, E.; Bekić, S.; Kotris, I.; Škrlec, I. Candida Albicans-the Virulence Factors and Clinical Manifestations of Infection. J. Fungi 2021, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.; Keck, D.; Gilmore, J.; Martinez-Duarte, R. Characterization of the Dielectrophoretic Response of Different Candida Strains Using 3d Carbon Microelectrodes. Micromachines 2020, 11, 255. [Google Scholar] [CrossRef] [PubMed]
- Natu, R.; Islam, M.; Martinez-Duarte, R. Nondimensional Streaming Dielectrophoresis Number for a System of Continuous Particle Separation. Anal. Chem. 2019, 91, 4357–4367. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Maltais, T.R.; Walter, T.M.; Wei, A.; Williams, S.J.; Wereley, S.T. Trapping and Viability of Swimming Bacteria in an Optoelectric Trap. Lab Chip 2016, 16, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Zanzucchi, P.J.; Wronski, C.R.; Carlson, D.E. Optical and Photoconductive Properties of Discharge-Produced Amorphous Silicon. J. Appl. Phys. 1977, 48, 5227–5236. [Google Scholar] [CrossRef]
- Liang, W.; Wang, S.; Dong, Z.; Lee, G.-B.; Li, W.J. Optical Spectrum and Electric Field Waveform dependent optically-induced dielectrophoretic (ODEP) micro-manipulation. Micromachines 2012, 3, 492–508. [Google Scholar] [CrossRef]
- Klis, F.M.; de Koster, C.G.; Brul, S. Cell Wall-Related Bionumbers and Bioestimates of Saccharomyces Cerevisiae and Candida Albicans. Eukaryot. Cell 2014, 13, 2–9. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, B.; Elsayed, M.; Nan, F.; Liang, W.; Valley, J.K.; Liu, L.; Huang, Q.; Wu, M.C.; Wheeler, A.R. Optoelectronic Tweezers: A versatile toolbox for nano-/micro-manipulations. Chem. Soc. Rev. 2022, 51, 9203–9242. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keck, D.; Ravi, S.; Yadav, S.; Martinez-Duarte, R. The Effect of Different System Parameters on the Movement of Microbial Cells Using Light-Induced Dielectrophoresis. Micromachines 2024, 15, 342. https://doi.org/10.3390/mi15030342
Keck D, Ravi S, Yadav S, Martinez-Duarte R. The Effect of Different System Parameters on the Movement of Microbial Cells Using Light-Induced Dielectrophoresis. Micromachines. 2024; 15(3):342. https://doi.org/10.3390/mi15030342
Chicago/Turabian StyleKeck, Devin, Suma Ravi, Shivam Yadav, and Rodrigo Martinez-Duarte. 2024. "The Effect of Different System Parameters on the Movement of Microbial Cells Using Light-Induced Dielectrophoresis" Micromachines 15, no. 3: 342. https://doi.org/10.3390/mi15030342
APA StyleKeck, D., Ravi, S., Yadav, S., & Martinez-Duarte, R. (2024). The Effect of Different System Parameters on the Movement of Microbial Cells Using Light-Induced Dielectrophoresis. Micromachines, 15(3), 342. https://doi.org/10.3390/mi15030342







