Biochemical and Structural Characterization of Cross-Linked Enzyme Aggregates (CLEAs) of Organic Solvent Tolerant Protease
Abstract
:1. Introduction
2. Results
2.1. Effect of Glutaraldehyde Concentration on CLEA–Elastase Formation
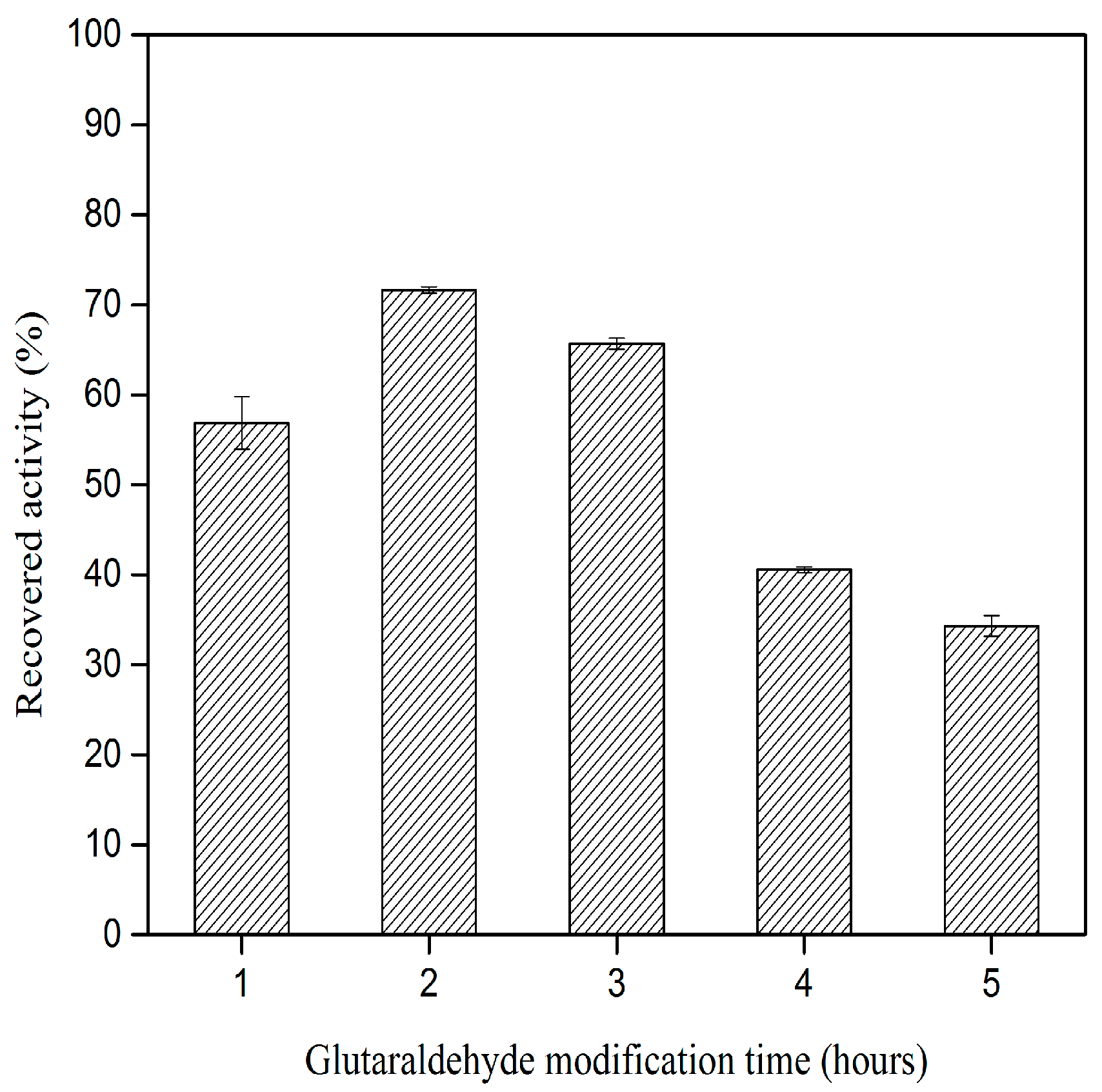
2.2. Effect of Glutaraldehyde Modification Time on CLEA–Elastase Formation
2.3. Effect of Different Co–Aggregants on CLEA–Elastase Formation
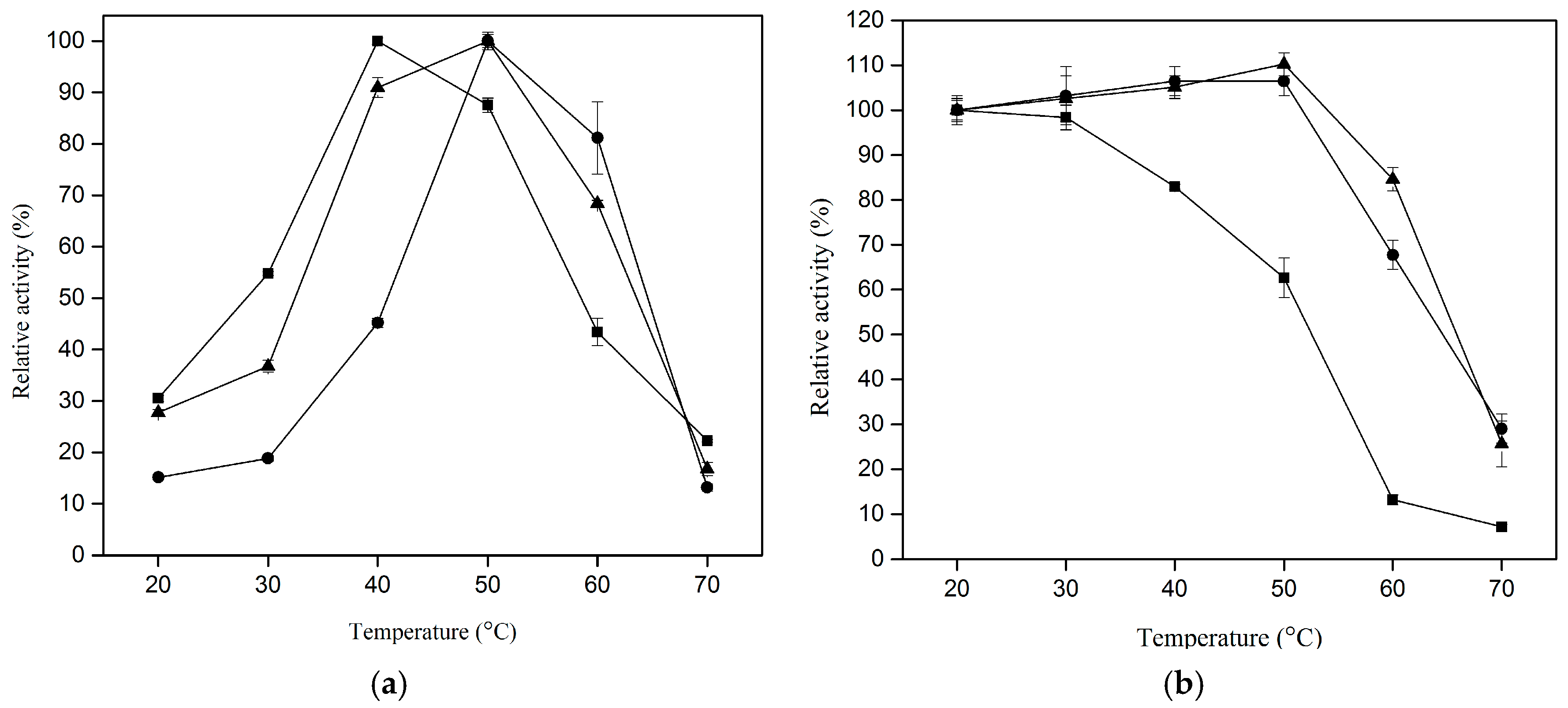
2.4. Effect of Temperature on Proteolytic Activity of CLEA–Elastase
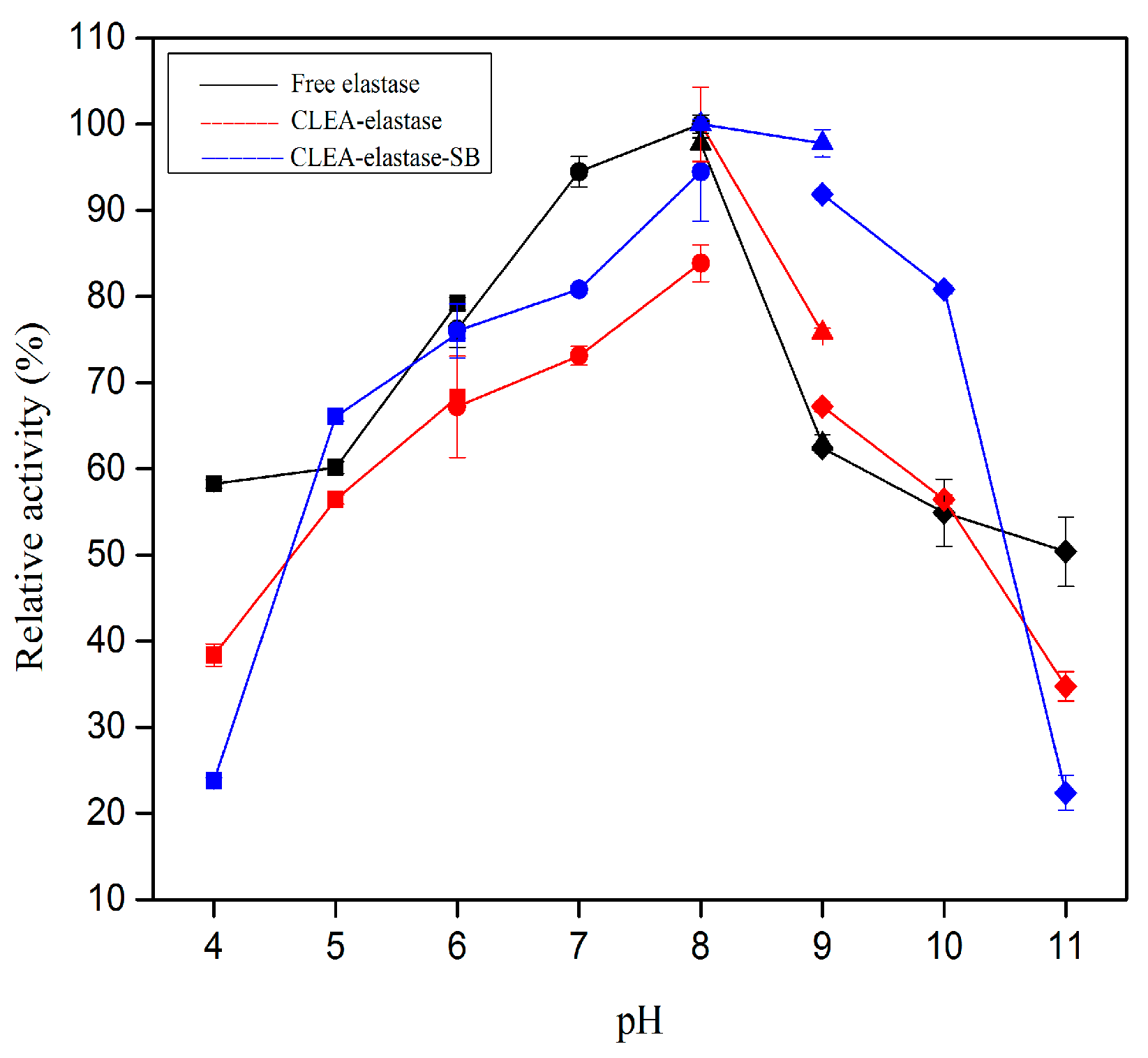
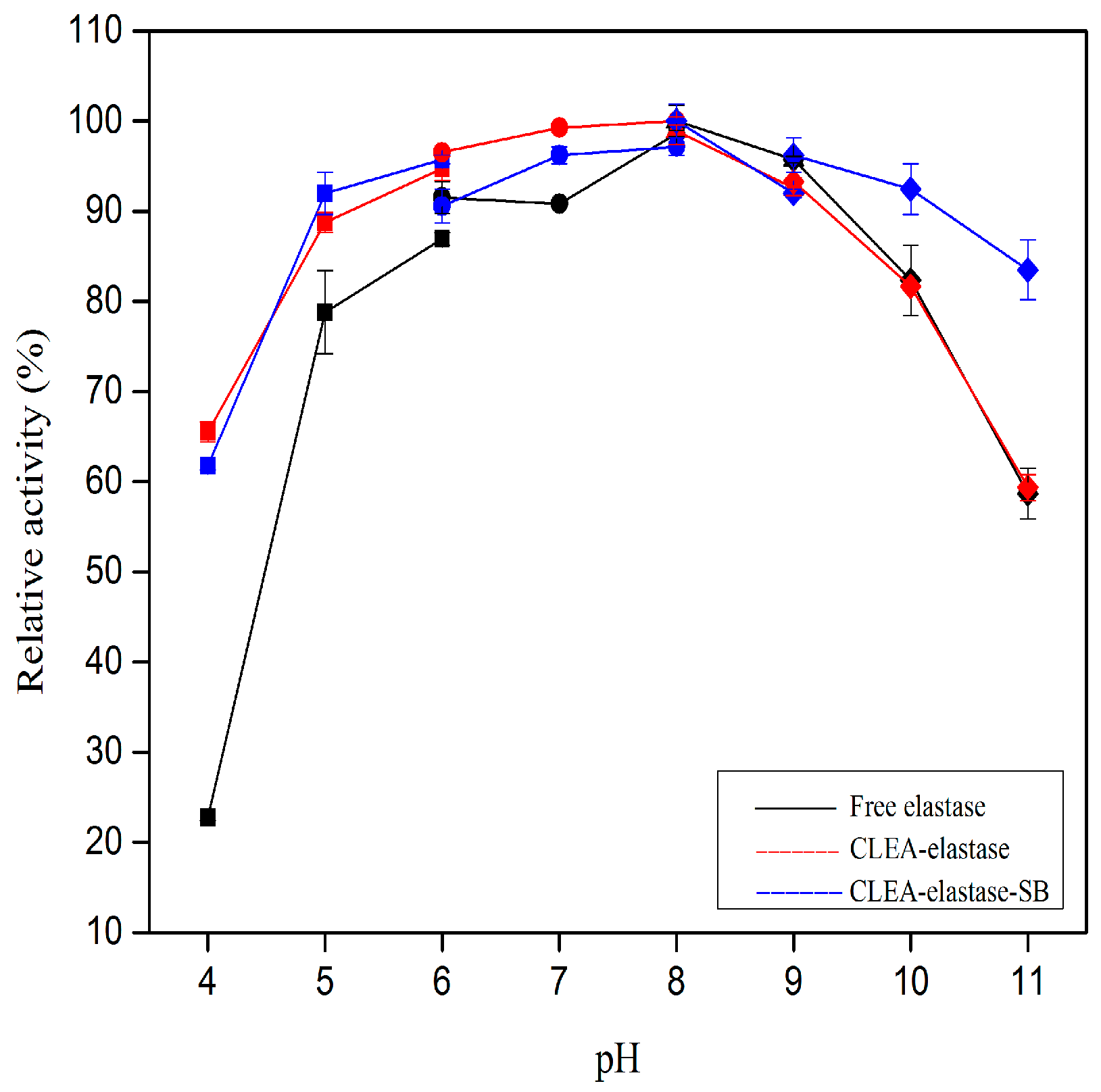
2.5. Effect of pH on Proteolytic Activity of CLEA–Elastase
2.6. pH Stability of CLEA–Elastase
2.7. Stability of CLEA–Elastase in Organic Solvents
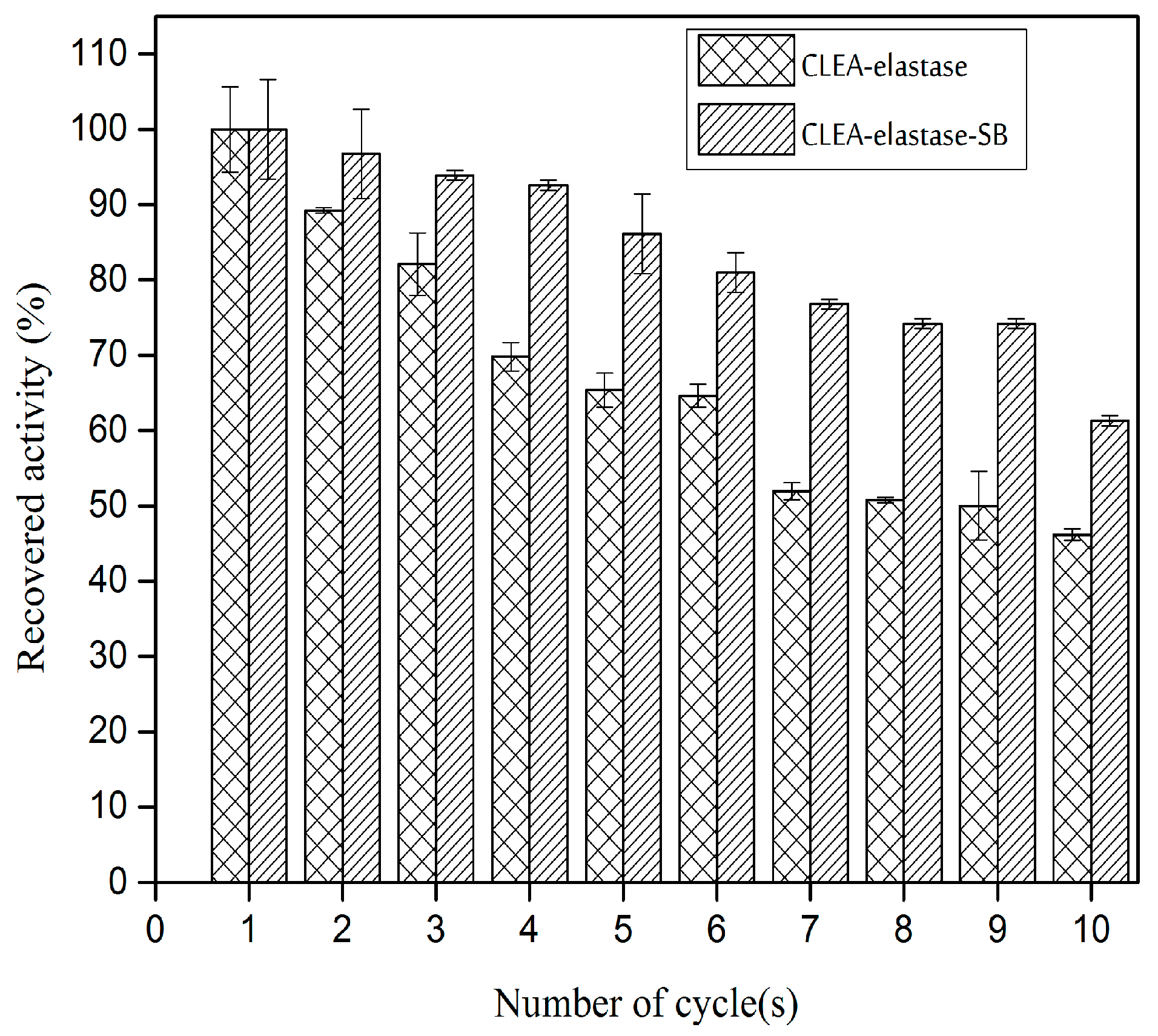
2.8. Reusability of CLEA–Elastase
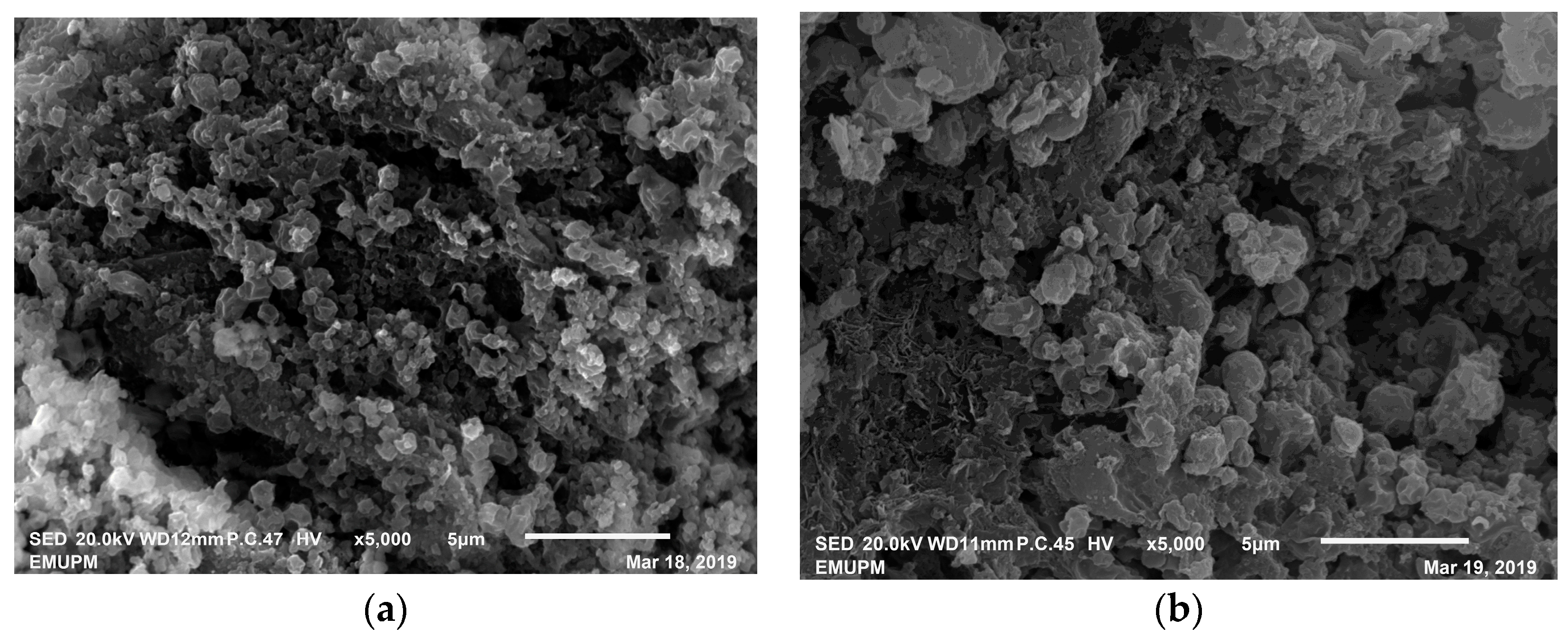
2.9. Scanning Electron Microscopy of CLEA–Elastase
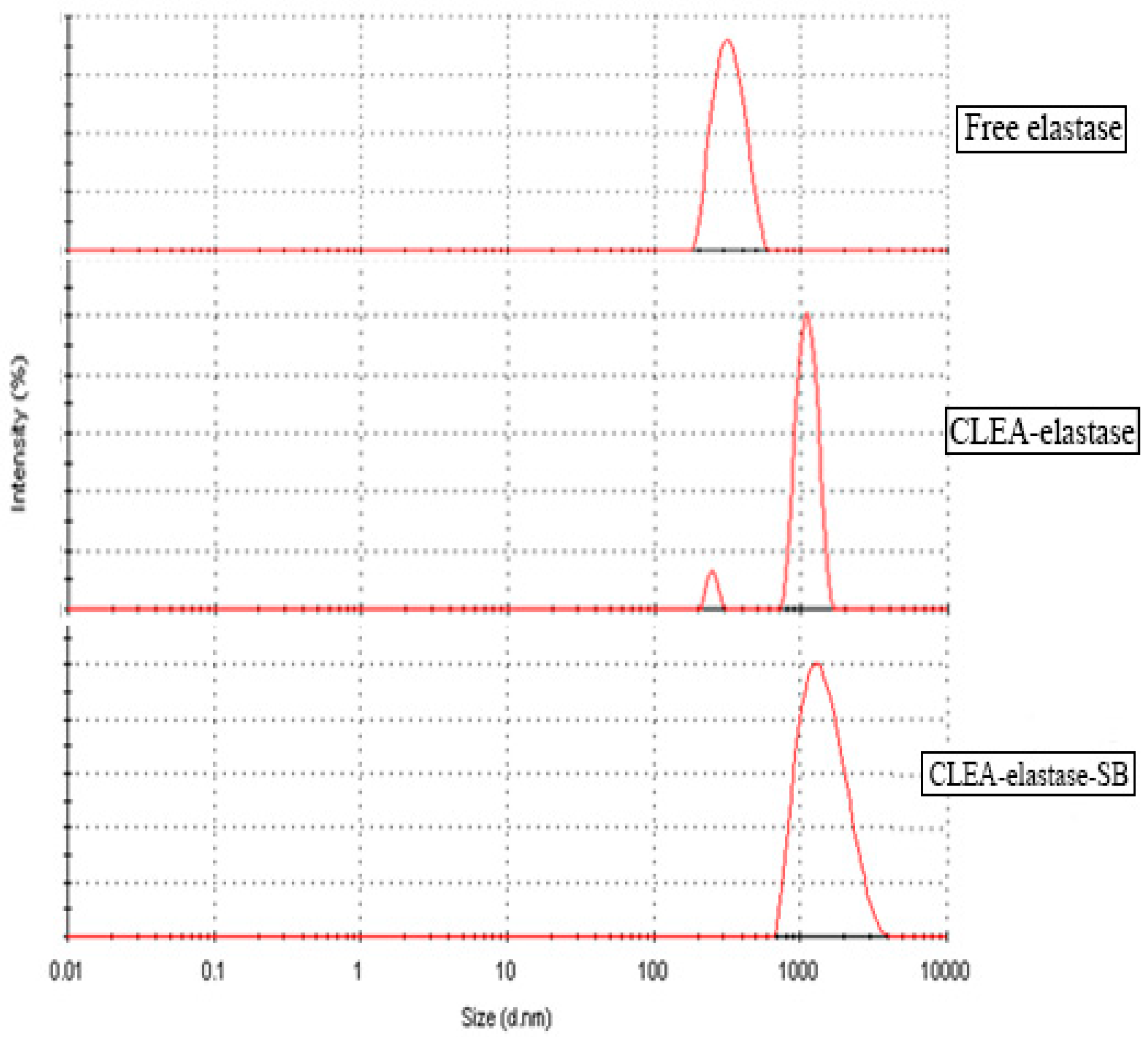
2.10. Dynamic Light Scattering (DLS) of CLEA–Elastase
3. Discussion
4. Materials and Methods
4.1. Bacterial and Enzyme Source
4.2. Assay of Proteolytic Activity
4.3. Preparation of CLEA–Elastase
4.4. Optimum Temperature and Thermal Stability of CLEA–Elastase
4.5. Optimum pH and pH Stability of CLEA–Elastase
4.6. Stability of CLEA–Elastase in Organic Solvent
4.7. Reusability of CLEA–Elastase
4.8. Structural Analysis of CLEA–Elastase
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sheldon, R.A. Cross–Linked Enzyme Aggregates as Industrial Biocatalysts. Org. Process Res. Dev. 2011, 15, 213–223. [Google Scholar] [CrossRef]
- Walsh, G. Industrial Enzymes: Proteases and Carbohydrases. Proteins Biochem. Biotechnol. 2015, 327–369. [Google Scholar] [CrossRef]
- Li, Q.; Yi, L.; Iverson, B.L. Commercial proteases: Present and future. FEBS Lett. 2013, 587, 1155–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawant, R.; Nagendran, S. Protease: An enzyme with multiple industrial applications. World J. Pharm. Pharm. Sci. 2014, 3, 568–579. [Google Scholar]
- Ahmad, R.; Sardar, M. Enzyme Immobilization: An Overview on Nanoparticles as Immobilization Matrix. Biochem. Anal. Biochem. 2015, 4, 1. [Google Scholar]
- Jesionowski, T.; Zdarta, J. Enzyme immobilization by adsorption: A review. Adsorption 2014, 20, 801–821. [Google Scholar] [CrossRef] [Green Version]
- Laane, C.; Boeren, S.; Vos, K.; Veeger, C. Rules for Optimization of Biocatalysis in Organic Solvents. Biotechnol. Bioeng. 2009, 102, 1–8. [Google Scholar] [CrossRef]
- Ivanov, I.P.; Kasche, V.; Petkov, D.D. Thermodynamics of Enzymic Peptide Synthesis in Biphasic Aqueous–Organic Systems: Coupling of the Chemical and Phase Equilibria. Biocatal. Biotransform. 1996, 14, 181–194. [Google Scholar] [CrossRef]
- Kasche, V.; Michaelis, G.; Galunsky, B. Binding of Organic Solvent Molecules Influences the P1’–P2’ Stereo–and Sequence–Specificity of α–Chymotrypsin in Kinetically Controlled Peptide Synthesis. Biotechnol. Lett. 1991, 13, 75–80. [Google Scholar] [CrossRef]
- Doukyu, N.; Ogino, H. Organic solvent–tolerant enzymes. Biochem. Eng. J. 2010, 48, 270–282. [Google Scholar] [CrossRef]
- Gu, Z.; Lai, J.; Hang, J.; Zhang, C.; Wang, S.; Jiao, Y.; Liu, S.; Fang, Y. Theoretical and experimental studies on the conformational changes of organic solvent–tolerant protease from Bacillus sphaericus DS11 in methanol/water mixture. Int. J. Biol. Macromol. 2019, 128, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.S.; Simair, A.A.; Ali, C.H.; Khushk, I.; Khokhar, J.A.; Ahmad, A.; Danish, M.; Lu, C. Production, Purification and Partial Characterization of Organo–Solvent Tolerant Protease from Newly Isolated Bacillus sp. BBXS–2. Ferment. Technol. 2018, 7, 2. [Google Scholar] [CrossRef]
- Ogino, H.; Yasui, K.; Shiotani, T.; Ishihara, T.; Ishikawa, H. Organic Solvent–Tolerant Bacterium Which Secretes an Organic Solvent–Stable Proteolytic Enzyme. Appl. Environ. Microbiol. 1995, 61, 4258–4262. [Google Scholar] [PubMed]
- Rahman, R.N.Z.A.; Geok, L.P.; Basri, M.; Salleh, A.B. Physical factors affecting the production of organic solvent–tolerant protease by Pseudomonas aeruginosa strain K. Bioresour. Technol. 2005, 96, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Rahman, R.N.Z.A.; Geok, L.P.; Basri, M.; Salleh, A.B. An organic solvent–stable alkaline protease from Pseudomonas aeruginosa strain K: Enzyme purification and characterization. Enzyme Microb. Technol. 2006, 39, 1484–1491. [Google Scholar] [CrossRef]
- Rahman, R.N.Z.A.; Salleh, A.B.; Basri, M.; Wong, C.F. Role of α–Helical Structure of Organic Solvent–Activated Homodimer of Elastase Strain, K. Int. J. Mol. Sci. 2011, 12, 5797–5814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zdarta, J.; Meyer, A.S.; Jesionowski, T.; Pinelo, M. A General Overview of Support Materials for Enzyme Immobilization: Characteristics, Properties, Practical Utility. Catalysts 2018, 8, 92. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.C.; Ortiz, C.; Berenguer–Murcia, A.; Torres, R.; Fernandez–Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290. [Google Scholar] [CrossRef]
- Garcia–Galan, C.; Berenguer–Murcia, A.; Fernandez–Lafuente, R.; Rodrigues, R.C. Potential of Diferent Enzyme Immobilization Strategies to Improve Enzyme Performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Sheldon, R.A. Cross–linked enzyme aggregates (CLEA®s): Stable and recyclable biocatalysts. Biochem. Soc. Trans. 2007, 35, 1583–1587. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, O.; Ortiz, C.; Berenguer–Murcia, A.; Torres, R.; Rodrigues, R.C.; Fernandez–Lafuente, R. Glutaraldehyde in bio–catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, R.A.; Schoevaart, R.; Van Langen, L.M. Cross–linked enzyme aggragates (CLEAs): A novel and versatile method for enzyme immobilization (a review). Biocatal. Biotransform. 2005, 23, 141–147. [Google Scholar] [CrossRef]
- Rojas, M.J.; Amaral–Fonseca, M.; Zanin, G.M.; Fernandez–Lafuente, R.; Giordano, R.D.L.C.; Tardioli, P.W. Preparation of Crosslinked Enzyme Aggregates of a Thermostable Cyclodextrin Glucosyltransferase from Thermoanaerobacter sp. Critical Effect of the Cross–linking Agent. Catalysts 2019, 9, 120. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.D.; Sun, L.M.; Li, L.L. A simple Technique of Preparing Stable CLEAs of Phenylalanine Ammonia Lyase Using Co–aggregation with Starch and Bovine Serum Albumin. Appl. Biochem. Biotechnol. 2013, 170, 1827–1837. [Google Scholar] [CrossRef]
- Shah, S.; Sharma, A.; Gupta, M.N. Preparation of cross–linked enzyme aggregates by using bovine serum albumin as a proteic feeder. Anal. Biochem. 2006, 351, 207–213. [Google Scholar] [CrossRef]
- Nadar, S.S.; Rathod, V.K. Magnetic macromolecular cross linked enzyme aggregates (CLEAs) of glucoamylase. Enzyme Microb. Technol. 2016, 83, 78–87. [Google Scholar] [CrossRef]
- Schoevaart, R.; Wolbers, M.W.; Golubovic, M.; Ottens, M.; Kieboom, A.P.G.; van Rantwijk, F.; van der Wielen, L.A.M.; Sheldon, R.A. Preparation, Optimization, and Structures of Cross–Linked Enzyme Aggregates (CLEAs). Biotechnol. Bioeng. 2004, 87, 754–762. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Seow, N.; Yang, K.L. Hollow cross–linked enzyme aggregates (h–CLEA) of laccase with high uniformity and activity. Colloids Surf. B Biointerfaces 2017, 151, 88–94. [Google Scholar] [CrossRef]
- Talekar, S.; Shah, V.; Patil, S.; Nimbalkar, M. Porous cross linked enzyme aggregates (p–CLEAs) of Saccharomyces cerevisiae invertase. Catal. Sci. Technol. 2012, 2, 1575–1579. [Google Scholar] [CrossRef]
- Galvis, M.; Barbosa, O.; Ruiz, M.; Cruz, J.; Ortiz, C.; Torres, R.; Fernandez-Lafuente, R. Chemical amination of lipase B from Candida antarctica is an efficient solution for the preparation of crosslinked enzyme aggregates. Process Biochem. 2012, 47, 2373–2378. [Google Scholar] [CrossRef]
- Tutar, H.; Yilmaz, E.; Pehlivan, E.; Yilmaz, M. Immobilization of Candida rugose lipase on sporopollenin from Lycopodium clavatum. Int. J. Biol. Macromol. 2009, 45, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Talekar, S.; Pandharbale, A.; Ladole, M.; Nadar, S.; Mulla, M.; Japhalekar, K.; Pattankude, K.; Arage, D. Carrier free co–immobilization of alpha amylase, glucoamylase and pullulanase as combined cross–linked enzyme aggregates (combi–CLEAs): A tri–enzyme biocatalyst with one pot starch hydrolytic activity. Bioresour. Technol. 2013, 147, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Bott, S.; Huo, Q. Techniques for Accurate Sizing of Gold Nanoparticles Using Dynamic Light Scattering with Particular Application to Chemical and Biological Sensing Based on Aggregate Formation. ACS Appl. Mater. Interfaces 2016, 8, 21585–21594. [Google Scholar] [CrossRef] [PubMed]
- Asgher, M.; Bashir, F.; Iqbal, H.M.N. Protease–based cross–linked enzyme aggregates with improved catalytic stability, silver removal, and dehairing potentials. Int. J. Biol. Macromol. 2018, 118, 1247–1256. [Google Scholar] [CrossRef]
- Mahmod, S.S.; Yusof, F.; Jami, M.S.; Khanahmadi, S.; Shah, H. Development of an immobilized biocatalyst with lipase and protease activities as a multipurpose cross–linked enzyme aggregates (multi–CLEA). Process Biochem. 2015, 50, 2144–2157. [Google Scholar] [CrossRef]
- Sheldon, R.A. Characteristic features and biotechnological applications of cross–linked enzyme aggregates (CLEAs). Appl. Microbiol. Biotechnol. 2011, 92, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme cross–linking. Biotechniques 2004, 37, 790–802. [Google Scholar] [CrossRef]
- Talekar, S.; Joshi, A.; Joshi, G.; Kamat, P.; Haripurkar, R.; Kambale, S. Parameters in preparation and characterization of cross linked enzyme aggregates (CLEAs). RSC Adv. 2013, 3, 12485–12511. [Google Scholar] [CrossRef]
- Easa, M.N.; Yusof, F.; Halim, A.A. Characterization of Cross–Linked Enzyme Aggregates (CLEA)–amylase from Zophobas morio. Int. Food Res. J. 2017, 24, 335–339. [Google Scholar]
- Li, X.D.; Wu, J.; Jia, D.C.; Wan, Y.H.; Yang, N.; Qiao, M. Preparation of Cross–Linked Glucoamylase Aggregates Immobilization by Using Dextrin and Xanthan Gum as Protecting Agents. Catalysts 2016, 6, 77. [Google Scholar] [CrossRef] [Green Version]
- Talekar, S.; Waingade, S.; Gaikwad, V.; Patil, S.; Nagavekar, N. Preparation and characterization of cross linked enzyme aggregates (CLEAs) of Bacillus amyloloquefaciens alpha amylase. J. Biochem. Technol. 2008, 43, 314–319. [Google Scholar]
- Rehman, S.; Bhatti, H.N.; Bilal, M.; Asgher, M. Cross–linked enzyme aggregates (CLEAs) of Pencilluim notatum lipase enzyme with improved activity, stability and reusability characteristics. Int. J. Biol. Macromol. 2016, 91, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.Y.; Yang, Z. Cross–linked tyrosinase aggregates for elimination of phenolic compounds from wastewater. Chemosphere 2013, 92, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Said, Z.S.A.M.; Arifi, F.A.M.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Leow, A.L.T.; Latip, W.; Ali, M.S.M. Unravelling protein–organic solvent interaction of organic solvent tolerant elastase from Pseudomonas aeruginosa strain K crystal structure. Int. J. Biol. Macromol. 2019, 127, 575–584. [Google Scholar] [CrossRef]
- Zerva, A.; Antonopoulou, I.; Enman, J.; Iancu, L.; Rova, U.; Christakopoulos, P. Cross–Linked Enzyme Aggregates of Feruloyl Esterase Preparations from Thermothelomyces thermophilia and Talaromyces wortmannii. Calalysts 2018, 8, 208. [Google Scholar] [CrossRef] [Green Version]

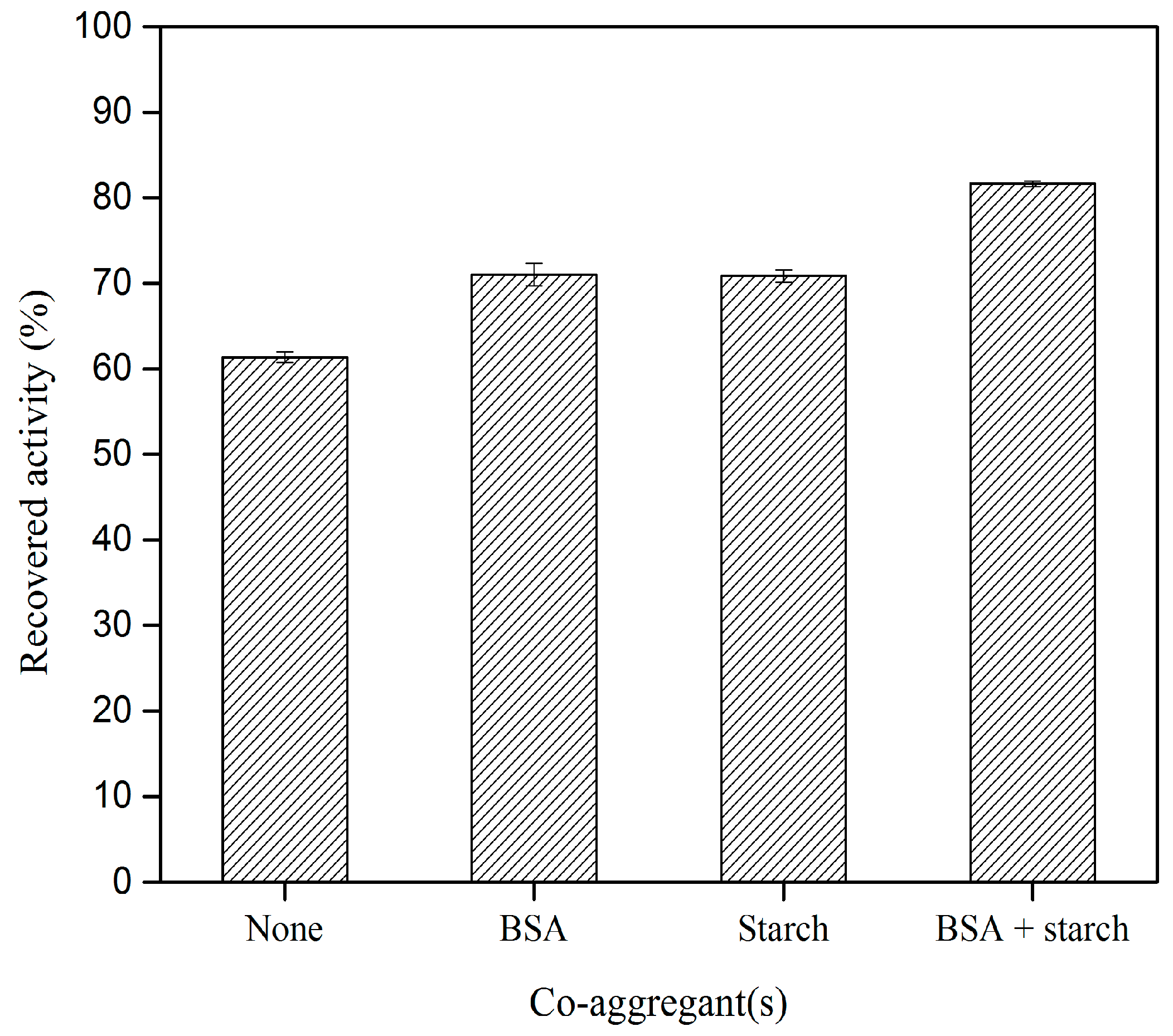
| Organic Solvents | Log Po/w | Free Elastase | CLEA–Elastase | CLEA–Elastase–SB |
|---|---|---|---|---|
| Control | – | 100 | 100 | 100 |
| DMSO | −1.30 | 102.39 ± 1.64 | 104.55 ± 3.20 | 47.59 ± 2.06 |
| Methanol | −0.78 | 87.29 ± 1.06 | 111.36 ± 3.18 | 101.87 ± 1.64 |
| Acetonitrile | −0.33 | 82.81 ± 0.71 | 164.55 ± 1.15 | 100.80 ± 1.13 |
| Ethanol | −0.24 | 79.22 ± 1.91 | 172.73 ± 3.57 | 101.34 ± 0.87 |
| 1–propanol | 0.28 | 31.39 ± 1.21 | 111.36 ± 2.16 | 94.12 ± 5.81 |
| Benzene | 2.13 | 79.52 ± 1.74 | 152.73 ± 1.22 | 98.40 ± 4.57 |
| Toluene | 2.50 | 86.70 ± 0.77 | 96.36 ± 2.87 | 79.14 ± 0.34 |
| Xylene | 3.15 | 82.96 ± 0.62 | 133.18 ± 0.84 | 89.84 ± 0.59 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razib, M.S.M.; Rahman, R.N.Z.R.A.; Shariff, F.M.; Ali, M.S.M. Biochemical and Structural Characterization of Cross-Linked Enzyme Aggregates (CLEAs) of Organic Solvent Tolerant Protease. Catalysts 2020, 10, 55. https://doi.org/10.3390/catal10010055
Razib MSM, Rahman RNZRA, Shariff FM, Ali MSM. Biochemical and Structural Characterization of Cross-Linked Enzyme Aggregates (CLEAs) of Organic Solvent Tolerant Protease. Catalysts. 2020; 10(1):55. https://doi.org/10.3390/catal10010055
Chicago/Turabian StyleRazib, Muhammad Syafiq Mohd, Raja Noor Zaliha Raja Abd Rahman, Fairolniza Mohd Shariff, and Mohd Shukuri Mohamad Ali. 2020. "Biochemical and Structural Characterization of Cross-Linked Enzyme Aggregates (CLEAs) of Organic Solvent Tolerant Protease" Catalysts 10, no. 1: 55. https://doi.org/10.3390/catal10010055
APA StyleRazib, M. S. M., Rahman, R. N. Z. R. A., Shariff, F. M., & Ali, M. S. M. (2020). Biochemical and Structural Characterization of Cross-Linked Enzyme Aggregates (CLEAs) of Organic Solvent Tolerant Protease. Catalysts, 10(1), 55. https://doi.org/10.3390/catal10010055






