Exploring Metagenomic Enzymes: A Novel Esterase Useful for Short-Chain Ester Synthesis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Analysis of EST5
2.2. Obtaining Protein EST5
2.3. Enzyme Activity and Its Affecting Factors
2.3.1. Effect of pH on Activity and Structure of EST5
2.3.2. Effect of Temperature on the Activity and Stability of EST5
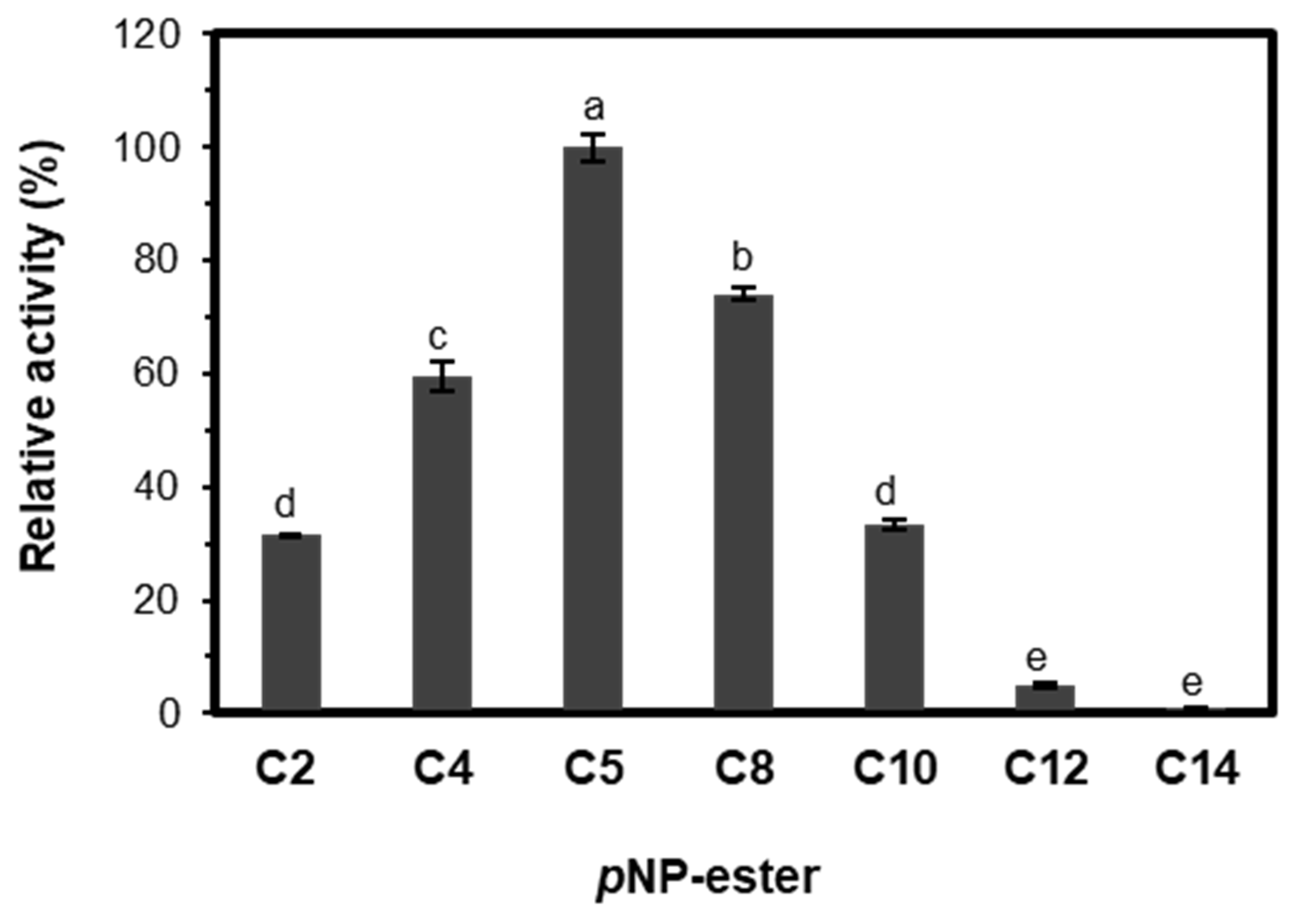
2.3.3. Substrate Specificity and Kinetic Parameters of EST5
2.3.4. Effect of Additives on EST5 Activity
2.4. EST5 Showed Esterification Activity
3. Materials and Methods
3.1. Sequence and Phylogenetic Analysis
3.2. Expression and Purification of the Recombinant Enzyme
3.3. Biophysical Analysis
3.4. Enzyme Characterization
3.4.1. Substrate Selectivity
3.4.2. Effect of pH and Temperature on Enzyme Activity
3.4.3. Effect of Additives on Enzyme Activity
3.5. Esterification Assays
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezborodov, A.M.; Zagustina, N.A. Lipases in catalytic reactions of organic chemistry. Appl. Biochem. Microbiol. 2014, 50, 313–337. [Google Scholar] [CrossRef]
- De Miranda, A.S.; Miranda, L.S.M.; de Souza, R.O.M.A. Lipases: Valuable catalysts for dynamic kinetic resolutions. Biotechnol. Adv. 2015, 33, 372–393. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef] [Green Version]
- Littlechild, J.A. Improving the ‘tool box’ for robust industrial enzymes. J. Ind. Microbiol. Biotechnol. 2017, 44, 711–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornscheuer, U. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef]
- Ferrer, M.; Bargiela, R.; Martínez-Martínez, M.; Mir, J.; Koch, R.; Golyshina, O.V.; Golyshin, P.N. Biodiversity for biocatalysis: A review of the α/β-hydrolase fold superfamily of esterases-lipases discovered in metagenomes. Biocatal. Biotransform. 2016, 2422, 1–15. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Kanmani, P.; Aravind, J.; Kumaresan, K. An insight into microbial lipases and their environmental facet. Int. J. Environ. Sci. Technol. 2015, 12, 1147–1162. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, A.; Fonseca, T.; Mattos, M.; Oliveira, M.; Lemos, T.; Molinari, F.; Romano, D.; Serra, I. Recent Advances in Lipase-Mediated Preparation of Pharmaceuticals and Their Intermediates. Int. J. Mol. Sci. 2015, 16, 29682–29716. [Google Scholar] [CrossRef] [Green Version]
- Trono, D. Recombinant Enzymes in the Food and Pharmaceutical Industries. In Advances in Enzyme Technology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 349–387. [Google Scholar]
- Samoylova, Y.V.; Sorokina, K.N.; Piligaev, A.V.; Parmon, V.N. Application of Bacterial Thermostable Lipolytic Enzymes in the Modern Biotechnological Processes: A Review. Catal. Ind. 2019, 11, 168–178. [Google Scholar] [CrossRef]
- Khambhaty, Y. Applications of enzymes in leather processing. Environ. Chem. Lett. 2020, 18, 747–769. [Google Scholar] [CrossRef]
- Zhang, Z.; Lan, D.; Zhou, P.; Li, J.; Yang, B.; Wang, Y. Control of sticky deposits in wastepaper recycling with thermophilic esterase. Cellulose 2017, 24, 311–321. [Google Scholar] [CrossRef]
- Sahay, S.; Chouhan, D. Study on the potential of cold-active lipases from psychrotrophic fungi for detergent formulation. J. Genet. Eng. Biotechnol. 2018, 16, 319–325. [Google Scholar] [CrossRef]
- Kumar, A.; Gudiukaite, R.; Gricajeva, A.; Sadauskas, M.; Malunavicius, V.; Kamyab, H.; Sharma, S.; Sharma, T.; Pant, D. Microbial lipolytic enzymes—Promising energy-efficient biocatalysts in bioremediation. Energy 2020, 192, 116674. [Google Scholar] [CrossRef]
- López-López, O.; Cerdán, M.E.; Gonzalez-Siso, M.I. New Extremophilic Lipases and Esterases from Metagenomics. Curr. Protein Pept. Sci. 2014, 15, 445–455. [Google Scholar] [CrossRef]
- Markets and Markets. Lipase Market by Source (Microbial Lipases, Animal Lipases), Application (Animal Feed, Dairy, Bakery, Confectionery, Others), & by Geography (North America, Europe, Asia-Pacific, Latin America, RoW)—Global Forecast to 2020; Markets and Markets: Pune, India, 2015. [Google Scholar]
- Fojan, P.; Jonson, P.H.; Petersen, M.T.N.; Petersen, S.B. What distinguishes an esterase from a lipase: A novel structural approach. Biochimie 2000, 82, 1033–1041. [Google Scholar] [CrossRef]
- Arpigny, J.L.; Jaeger, K.-E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 1999, 343, 177–183. [Google Scholar] [CrossRef]
- Salgado, V.; Fonseca, C.; Lopes da Silva, T.; Roseiro, J.C.; Eusébio, A. Isolation and Identification of Magnusiomyces capitatus as a Lipase-Producing Yeast from Olive Mill Wastewater. Waste Biomass Valor. 2020, 11, 3207–3221. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.C.; Chi, Z.; Liu, G.L.; Jiang, H.; Hu, Z.; Chi, Z.M. Production, purification, characterization and gene cloning of an esterase produced by Aureobasidium melanogenum HN6.2. Process Biochem. 2017, 53, 69–79. [Google Scholar] [CrossRef]
- Rade, L.L.; da Silva, M.N.P.; Vieira, P.S.; Milan, N.; de Souza, C.M.; de Melo, R.R.; Klein, B.C.; Bonomi, A.; de Castro, H.F.; Murakami, M.T.; et al. A Novel Fungal Lipase With Methanol Tolerance and Preference for Macaw Palm Oil. Front. Bioeng. Biotechnol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Turati, D.F.M.; Almeida, A.F.; Terrone, C.C.; Nascimento, J.M.F.; Terrasan, C.R.F.; Fernandez-Lorente, G.; Pessela, B.C.; Guisan, J.M.; Carmona, E.C. Thermotolerant lipase from Penicillium sp. section Gracilenta CBMAI 1583: Effect of carbon sources on enzyme production, biochemical properties of crude and purified enzyme and substrate specificity. Biocatal. Agric. Biotechnol. 2019, 17, 15–24. [Google Scholar] [CrossRef]
- Lin, X.-S.; Zhao, K.-H.; Zhou, Q.-L.; Xie, K.-Q.; Halling, P.J.; Yang, Z. Aspergillus oryzae lipase-catalyzed synthesis of glucose laurate with excellent productivity. Bioresour. Bioprocess. 2016, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, K.K.; Dogra, A.; Kapoor, S.; Mehta, A.; Gupta, R. Purification and Properties of an Esterase from Bacillus licheniformis and it’s Application in Synthesis of Octyl Acetate. Open Microbiol. J. 2020, 14, 113–121. [Google Scholar] [CrossRef]
- Noor, H.; Satti, S.M.; Din, S.U.; Farman, M.; Hasan, F.; Khan, S.; Badshah, M.; Shah, A.A. Insight on esterase from Pseudomonas aeruginosa strain S3 that depolymerize poly(lactic acid) (PLA) at ambient temperature. Polym. Degrad. Stab. 2020, 174, 109096. [Google Scholar] [CrossRef]
- Bharathi, D.; Rajalakshmi, G.; Komathi, S. Optimization and production of lipase enzyme from bacterial strains isolated from petrol spilled soil. J. King Saud Univ. Sci. 2019, 31, 898–901. [Google Scholar] [CrossRef]
- Boran, R.; Ugur, A.; Sarac, N.; Ceylan, O. Characterisation of Streptomyces violascens OC125-8 lipase for oily wastewater treatment. 3 Biotech 2019, 9, 5. [Google Scholar] [CrossRef]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 2. [Google Scholar] [CrossRef] [Green Version]
- Lai, O.-M.; Lee, Y.-Y.; Phuah, E.-T.; Akoh, C.C. Lipase/Esterase: Properties and Industrial Applications. In Encyclopedia of Food Chemistry; Elsevier: Amsterdam, The Netherlands, 2019; pp. 158–167. [Google Scholar]
- Gao, W.; Wu, K.; Chen, L.; Fan, H.; Zhao, Z.; Gao, B.; Wang, H.; Wei, D. A novel esterase from a marine mud metagenomic library for biocatalytic synthesis of short-chain flavor esters. Microb. Cell Fact. 2016, 15, 41. [Google Scholar] [CrossRef] [Green Version]
- Brault, G.; Shareck, F.; Hurtubise, Y.; Lépine, F.; Doucet, N. Short-chain flavor ester synthesis in organic media by an E. coli whole-cell biocatalyst expressing a newly characterized heterologous lipase. PLoS ONE 2014, 9, e91872. [Google Scholar] [CrossRef] [Green Version]
- SÁ, A.G.A.; Meneses, A.C.D.; Araújo, P.H.H.D.; Oliveira, D.D. A review on enzymatic synthesis of aromatic esters used as flavor ingredients for food, cosmetics and pharmaceuticals industries. Trends Food Sci. Technol. 2017, 69, 95–105. [Google Scholar] [CrossRef]
- Zhong, X.L.; Tian, Y.Z.; Jia, M.L.; Liu, Y.D.; Cheng, D.; Li, G. Characterization and purification via nucleic acid aptamers of a novel esterase from the metagenome of paper mill wastewater sediments. Int. J. Biol. Macromol. 2020, 153, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.M.; Martini, V.P.; Iulek, J.; Alnoch, R.C.; Moure, V.R.; Müller-Santos, M.; Souza, E.M.; Mitchell, D.A.; Krieger, N. Biochemical characterization and application of a new lipase and its cognate foldase obtained from a metagenomic library derived from fat-contaminated soil. Int. J. Biol. Macromol. 2019, 137, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.A.M.; Pereira, M.R.; Maester, T.C.; de Macedo Lemos, E.G. Investigation, Expression, and Molecular Modeling of ORF2, a Metagenomic Lipolytic Enzyme. Appl. Biochem. Biotechnol. 2015, 175, 3875–3887. [Google Scholar] [CrossRef]
- Pereira, M.R.; Mercaldi, G.F.; Maester, T.C.; Balan, A.; Lemos, E.G.M. Est16, a new esterase isolated from a metagenomic library of a microbial consortium specializing in diesel oil degradation. PLoS ONE 2015, 10, e0133723. [Google Scholar] [CrossRef]
- Maester, T.C.; Pereira, M.R.; Machado Sierra, E.G.; Balan, A.; de Macedo Lemos, E.G. Characterization of EST3: A metagenome-derived esterase with suitable properties for biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 5815–5827. [Google Scholar] [CrossRef] [Green Version]
- Greenfield, N.J. Using circular dichroism collected as a funcion of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2009, 1, 2527–2535. [Google Scholar] [CrossRef]
- Greenfield, N.J. Biomacromolecular Applications of Circular Dichroism and ORD BT—Encyclopedia of Spectroscopy and Spectrometry. In Encyclopedia of Spectroscopy and Spectrometry, 2nd ed.; Lindon, J.C., Tranter, G.E., Holmes, J.L., Eds.; Academic Press: San Diego, CA, USA, 1999; Volume 1, pp. 153–165. [Google Scholar]
- Sharma, P.K.; Singh, K.; Singh, R.; Capalash, N.; Ali, A.; Mohammad, O.; Kaur, J. Characterization of a thermostable lipase showing loss of secondary structure at ambient temperature. Mol. Biol. Rep. 2012, 39, 2795–2804. [Google Scholar] [CrossRef]
- Fu, J.; Leiros, H.K.S.; De Pascale, D.; Johnson, K.A.; Blencke, H.M.; Landfald, B. Functional and structural studies of a novel cold-adapted esterase from an Arctic intertidal metagenomic library. Appl. Microbiol. Biotechnol. 2013, 97, 3965–3978. [Google Scholar] [CrossRef]
- Dong, J.; Gasmalla, M.A.A.; Zhao, W.; Sun, J.; Liu, W.; Wang, M.; Han, L.; Yang, R. Characterisation of a cold adapted esterase and mutants from a psychotolerant Pseudomonas sp. strain. Biotechnol. Appl. Biochem. 2016, 64, 686–699. [Google Scholar] [CrossRef]
- Tchigvintsev, A.; Tran, H.; Popovic, A.; Kovacic, F.; Brown, G.; Flick, R.; Hajighasemi, M.; Egorova, O.; Somody, J.C.; Tchigvintsev, D.; et al. The environment shapes microbial enzymes: Five cold-active and salt-resistant carboxylesterases from marine metagenomes. Appl. Microbiol. Biotechnol. 2015, 99, 2165–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Zhang, S.; Gao, H.; Hu, N. Characterization of a cold-active esterase from Serratia sp. and improvement of thermostability by directed evolution. BMC Biotechnol. 2016, 16, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borchert, E.; Selvin, J.; Kiran, S.G.; Jackson, S.A.; O’Gara, F.; Dobson, A.D.W. A novel cold active esterase from a deep sea sponge stelletta normani metagenomic library. Front. Mar. Sci. 2017, 4, 287. [Google Scholar] [CrossRef] [Green Version]
- De Santi, C.; Altermark, B.; Pierechod, M.M.; Ambrosino, L.; De Pascale, D.; Willassen, N.P. Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries. BMC Biochem. 2016, 17, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaeger, K.-E.; Dijkstra, B.W.; Reetz, M.T. Bacterial Biocatalysts: Molecular Biology, Three-Dimensional Structures, and Biotechnological Applications of Lipases. Annu. Rev. Microbiol. 1999, 53, 315–351. [Google Scholar] [CrossRef] [Green Version]
- Tirawongsaroj, P.; Sriprang, R.; Harnpicharnchai, P.; Thongaram, T.; Champreda, V.; Tanapongpipat, S.; Pootanakit, K.; Eurwilaichitr, L. Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J. Biotechnol. 2008, 133, 42–49. [Google Scholar] [CrossRef]
- Okano, H.; Hong, X.; Kanaya, E.; Angkawidjaja, C.; Kanaya, S. Structural and biochemical characterization of a metagenome-derived esterase with a long N-terminal extension. Protein Sci. 2015, 24, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Zarafeta, D.; Szabo, Z.; Moschidi, D.; Phan, H.; Chrysina, E.D.; Peng, X.; Ingham, C.J.; Kolisis, F.N.; Skretas, G. EstDZ3: A new esterolytic enzyme exhibiting remarkable thermostability. Front. Microbiol. 2016, 7, 1779. [Google Scholar] [CrossRef] [Green Version]
- Sukul, P.; Lupilov, N.; Leichert, L.I. Characterization of ML-005, a novel metaproteomics-derived esterase. Front. Microbiol. 2018, 9, 2716. [Google Scholar] [CrossRef]
- Bofill, C.; Prim, N.; Mormeneo, M.; Manresa, A.; Javier Pastor, F.I.; Diaz, P. Differential behaviour of Pseudomonas sp. 42A2 LipC, a lipase showing greater versatility than its counterpart LipA. Biochimie 2010, 92, 307–316. [Google Scholar] [CrossRef]
- Kumar, S.; Mathur, A.; Singh, V.; Nandy, S.; Khare, S.K.; Negi, S. Bioremediation of waste cooking oil using a novel lipase produced by Penicillium chrysogenum SNP5 grown in solid medium containing waste grease. Bioresour. Technol. 2012, 120, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Maqbool, Q.U.A.; Johri, S.; Rasool, S.; Riyaz-ul-Hassan, S.; Verma, V.; Nargotra, A.; Koul, S.; Qazi, G.N. Molecular cloning of carboxylesterase gene and biochemical characterization of encoded protein from Bacillus subtilis (RRL BB1). J. Biotechnol. 2006, 125, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Sun, B. Identification of novel esterase from metagenomic library of Yangtze River. J. Microbiol. Biotechnol. 2009, 19, 187–193. [Google Scholar] [PubMed] [Green Version]
- Jeon, J.H.; Kim, J.T.; Lee, H.S.; Kim, S.J.; Kang, S.G.; Choi, S.H.; Lee, J.H. Novel lipolytic enzymes identified from metagenomic library of deep-sea sediment. Evid. Based Complement. Altern. Med. 2011, 2011, 271419. [Google Scholar] [CrossRef]
- Esteban-Torres, M.; Mancheño, J.M.; de las Rivas, B.; Muñoz, R. Characterization of a halotolerant lipase from the lactic acid bacteria Lactobacillus plantarum useful in food fermentations. LWT Food Sci. Technol. 2015, 60, 246–252. [Google Scholar] [CrossRef] [Green Version]
- Glogauer, A.; Martini, V.P.; Faoro, H.; Couto, G.H.; Müller-Santos, M.; Monteiro, R.A.; Mitchell, D.A.; de Souza, E.M.; Pedrosa, F.O.; Krieger, N. Identification and characterization of a new true lipase isolated through metagenomic approach. Microb. Cell Fact. 2011, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Nordblad, M.; Nielsen, P.M.; Brask, J.; Woodley, J.M. In situ visualization and effect of glycerol in lipase-catalyzed ethanolysis of rapeseed oil. J. Mol. Catal. B Enzym. 2011, 72, 213–219. [Google Scholar] [CrossRef]
- Rao, L.; Xue, Y.; Zheng, Y.; Lu, J.R.; Ma, Y. A Novel Alkaliphilic Bacillus Esterase Belongs to the 13th Bacterial Lipolytic Enzyme Family. PLoS ONE 2013, 8, e60645. [Google Scholar] [CrossRef]
- Cavalcanti, E.D.C.; Aguieiras, É.C.G.; da Silva, P.R.; Duarte, J.G.; Cipolatti, E.P.; Fernandez-Lafuente, R.; da Silva, J.A.C.; Freire, D.M.G. Improved production of biolubricants from soybean oil and different polyols via esterification reaction catalyzed by immobilized lipase from Candida rugosa. Fuel 2018, 215, 705–713. [Google Scholar] [CrossRef]
- Tang, S.J.; Sun, K.H.; Sun, G.H.; Chang, T.Y.; Lee, G.C. Recombinant expression of the Candida rugosa lip4 lipase in Escherichia coli. Protein Expr. Purif. 2000, 20, 308–313. [Google Scholar] [CrossRef]
- Lee, G.-C.; Lee, L.-C.; Sava, V.; Shaw, J.-F. Multiple mutagenesis of non-universal serine codons of the Candida rugosa LIP2 gene and biochemical characterization of purified recombinant LIP2 lipase overexpressed in Pichia pastoris. Biochem. J. 2002, 366, 603–611. [Google Scholar] [CrossRef]
- Liu, T.; Vora, H.; Khosla, C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab. Eng. 2010, 12, 378–386. [Google Scholar] [CrossRef]
- De Oliveira, D.; Feihrmann, A.C.; Dariva, C.; Cunha, A.G.; Bevilaqua, J.V.; Destain, J.; Oliveira, J.V.; Freire, D.M.G. Influence of compressed fluids treatment on the activity of Yarrowia lipolytica lipase. J. Mol. Catal. B Enzym. 2006, 39, 117–123. [Google Scholar] [CrossRef]
- Smaniotto, A.; Skovronski, A.; Rigo, E.; Tsai, S.M.; Durrer, A.; Foltran, L.L.; Paroul, N.; Di Luccio, M.; Vladimir Oliveira, J.; de Oliveira, D.; et al. Concentration, characterization and application of lipases from Sporidiobolus pararoseus strain. Braz. J. Microbiol. 2014, 45, 294–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Xu, H.; Yan, Q.; Yang, S.; Duan, X.; Jiang, Z. Biochemical characterization of a first fungal esterase from Rhizomucor miehei showing high efficiency of ester synthesis. PLoS ONE 2013, 8, e77856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radzi, S.M.; Mustafa, W.A.F.; Othman, S.S.; Noor, H.M. Green Synthesis of Butyl Acetate, A Pineapple Flavour via Lipase-Catalyzed Reaction. World Acad. Sci. Eng. Technol. 2011, 132, 7038–7042. [Google Scholar]
- Pires-Cabral, P.; da Fonseca, M.M.R.; Ferreira-Dias, S. Esterification activity and operational stability of Candida rugosa lipase immobilized in polyurethane foams in the production of ethyl butyrate. Biochem. Eng. J. 2010, 48, 246–252. [Google Scholar] [CrossRef]
- Kaur, M.; Mehta, A.; Gupta, R. Synthesis of methyl butyrate catalyzed by lipase from Aspergillus fumigatus. J. Oleo Sci. 2019, 68, 989–993. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.-L.; He, L.; Tao, G.-H.; Hu, C. High yield of ethyl valerate from the esterification of renewable valeric acid catalyzed by amino acid ionic liquids. RSC Adv. 2013, 3, 4806. [Google Scholar] [CrossRef]
- Paes, F.C.; Kanda, L.R.S.; Voll, F.A.P.; Corazza, M.L. Liquid-liquid equilibrium of ternary systems comprising ethyl valerate(1), water(2), ethanol(3) and valeric acid(4). J. Chem. Thermodyn. 2017, 111, 185–190. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Wang, X.; Shang, M.; Huang, J.; Guan, G.; Li, Y.; Shi, B. Isolation of a novel alkaline-stable lipase from a metagenomic library and its specific application for milkfat flavor production. Microb. Cell Fact. 2014, 13, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakiba, M.H.; Ali, M.S.M.; Rahman, R.N.Z.R.A.; Salleh, A.B.; Leow, T.C. Cloning, expression and characterization of a novel cold-adapted GDSL family esterase from Photobacterium sp. strain J15. Extremophiles 2016, 20, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Jung, W.K.; Kim, Y.H.; Ryu, B.H.; Doohun Kim, T.; Kim, J.; Kim, H. Characterization of a novel alkaline family viii esterase with S-enantiomer preference from a compost metagenomic library. J. Microbiol. Biotechnol. 2016, 26, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]





| Substrate | Vmax (µM·s−1) | Km (mM) | Kcat (s−1) | Kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| pNP-acetate (C2) | 0.5 ± 0.03 | 1.1 ± 0.13 | 18.97 ± 1.18 | 18.1 |
| pNP-butyrate (C4) | 0.78 ± 0.04 | 0.4 ± 0.05 | 29.0 ± 1.46 | 67.7 |
| pNP-valerate (C5) | 2.4 ± 0.04 | 0.1 ± 0.01 | 88.9 ± 1.02 | 1533.27 |
| pNP-octanoate (C8) | 1.4 ± 0.03 | 0.3 ± 0.04 | 52.2 ± 1.28 | 158.24 |
| pNP-decanoate (C10) | 0.3 ± 0.02 | 0.73 ± 0.1 | 12.79 ± 0.74 | 17.5 |
| Enzyme | Substrate | Esterification (U·mg−1) | Reference |
|---|---|---|---|
| EST5 | Butyric acid/Methanol | 127.04 | This study |
| LIP4 | Butyric acid/Propanol | 86.5 | [64,65] |
| CL | Butyric acid/Propanol | 15.83 | [65] |
| LIP2 | Butyric acid/Propanol | 166 | [65] |
| Immobilized Lipase PS | Lauric acid/Dodecanol | 8.87 | [66] |
| Novozyme 435 | Lauric acid/Dodecanol | 9.89 | [66] |
| Lipozyme TLIM | Lauric acid/Dodecanol | 3.54 | [66] |
| YLL | Lauric acid/Propanol | 0.35 | [67] |
| Sporidiobolus pararoseus lipase | Oleic Acid/Ethanol | 489.65 | [68] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maester, T.C.; Pereira, M.R.; Malaman, A.M.G.; Borges, J.P.; Pereira, P.A.M.; Lemos, E.G.M. Exploring Metagenomic Enzymes: A Novel Esterase Useful for Short-Chain Ester Synthesis. Catalysts 2020, 10, 1100. https://doi.org/10.3390/catal10101100
Maester TC, Pereira MR, Malaman AMG, Borges JP, Pereira PAM, Lemos EGM. Exploring Metagenomic Enzymes: A Novel Esterase Useful for Short-Chain Ester Synthesis. Catalysts. 2020; 10(10):1100. https://doi.org/10.3390/catal10101100
Chicago/Turabian StyleMaester, Thaís Carvalho, Mariana Rangel Pereira, Aliandra M. Gibertoni Malaman, Janaina Pires Borges, Pâmela Aparecida Maldaner Pereira, and Eliana G. M. Lemos. 2020. "Exploring Metagenomic Enzymes: A Novel Esterase Useful for Short-Chain Ester Synthesis" Catalysts 10, no. 10: 1100. https://doi.org/10.3390/catal10101100
APA StyleMaester, T. C., Pereira, M. R., Malaman, A. M. G., Borges, J. P., Pereira, P. A. M., & Lemos, E. G. M. (2020). Exploring Metagenomic Enzymes: A Novel Esterase Useful for Short-Chain Ester Synthesis. Catalysts, 10(10), 1100. https://doi.org/10.3390/catal10101100






