Rhizopus oryzae Lipase, a Promising Industrial Enzyme: Biochemical Characteristics, Production and Biocatalytic Applications
Abstract
:1. Introduction
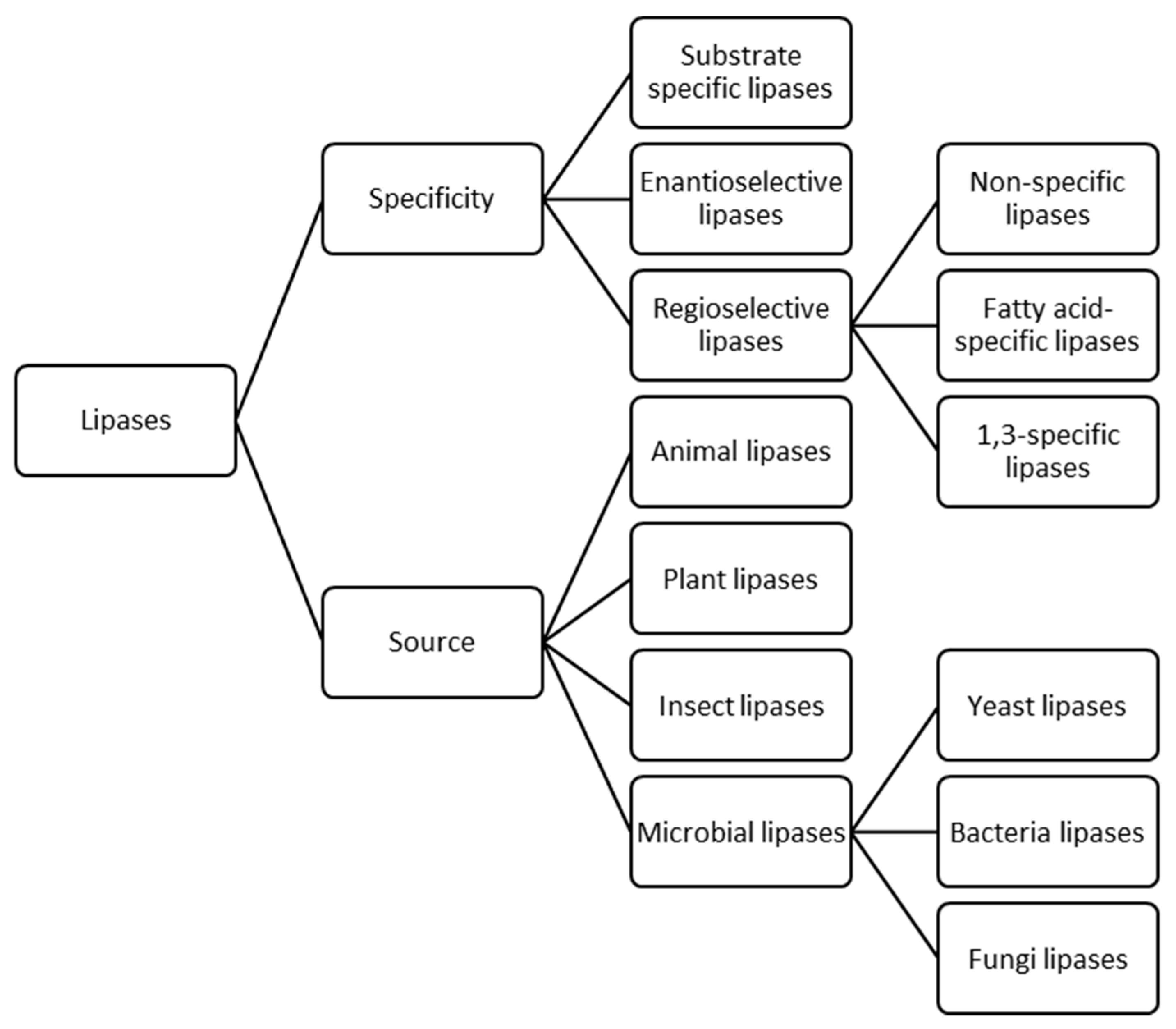
2. Biochemical Properties
3. Rhizopus oryzae Lipase Production and Bioprocess Engineering
3.1. Komogatella Phaffii Cell Factory
3.1.1. rROL PAOX
3.1.2. rROL PFLD1
3.1.3. proROL PAOX
3.1.4. rROL and proROL PGAP
3.1.5. 28proROL PAOX
3.1.6. Whole Cells
4. Industrial Applications of Rhizopus oryzae Lipase
4.1. Biodiesel Production
4.2. Structured Lipids Production
4.3. Flavour Esters Production
4.4. Resolution of Racemic Mixtures
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 28proROL-gene | Gene encoding a truncated prosequence of Rhizopus oryzae lipase 28 C-terminal amino acids fused to the N-terminal of the mature lipase region |
| 2-MAG | 2-monoacylglycerol |
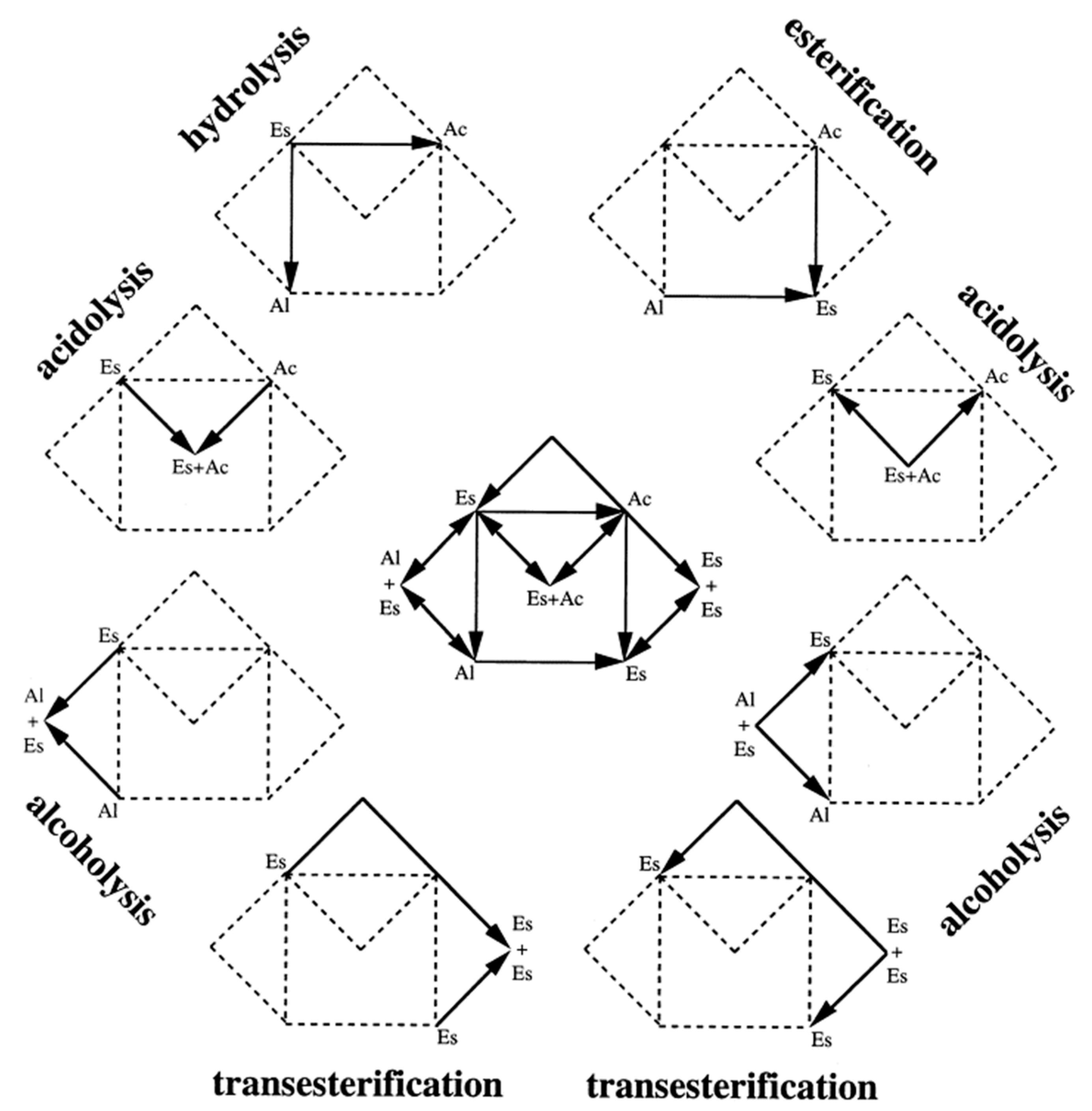
| Ac | Acids |
| Al | Alcohols |
| ALO | Alperujo oil |
| AOX | Alcohol oxidase |
| BiP | Binding proteins |
| Bmh2 | 14-3-3 protein |
| BR | Batch Reactor |
| C | TAG or FFA conversion (%) |
| CA | Capric acid |
| CBE | Cocoa butter equivalents |
| CI | Covalently immobilised or stabilised biocatalyst through crosslinking |
| CO | Canola oil |
| CRA | Caprylic acid |
| CRL | Candida rugosa lipase |
| DAG | Diacylglycerol |
| DCW | Dry cell weight |
| DO | Dissolved oxygen |
| DoE | Design of experiments |
| EDTA | Ethylenediaminetetraacetic acid |
| ee | Enantiomeric excess |
| EF-1α | Translation elongation factor 1α |
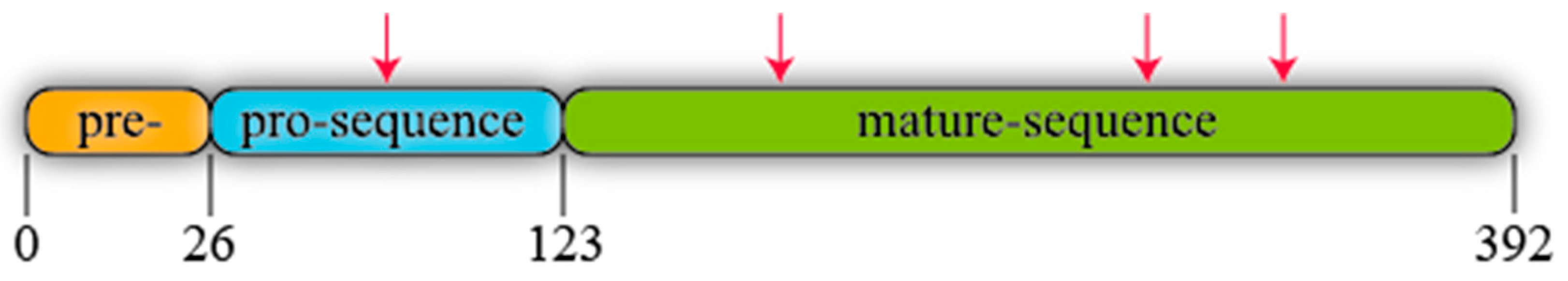
| entire-proROL | Rhizopus oryzae lipase including the whole prosequence and mature sequence |
| EPAX 1050TG | TAG rich in omega-3 PUFAs |
| ERAD | Endoplasmatic-reticulum associated protein degradation |
| ERO1 | Endoplasmatic-reticulum oxidoreductin |
| Es | Esters |
| EtOH | Ethanol |
| FAME | Fatty acid methyl esters |
| FFA | Free fatty acid |
| FLD | Formaldehyde dehydrogenase |
| GAP | Glyceraldehyde-3-phosphate dehydrogenase |
| GAS | β 1-3-glucanosytransglycosylase |
| HAC1 | UPR transcriptional factor |
| His | Histidine |
| HMFS | Human milk fat substitutes |
| HRD1 | Polytopic E3 ubiquitin ligase |
| IA | Immobilisation through adsorption |
| ICL | Isocitrate lyase |
| ID | Incorporation degree (%) |
| IE | Immobilisation through physical entrapment |
| JO | Jatropha oil |
| KO | Karanja oil |
| L | Long-chain fatty acid |
| M | Medium-chain fatty acid |
| MAG | Monoacylglycerol |
| MC | Multicopy |
| MeOH | Methanol |
| MLFB | Methanol limited fed-batch |
| MNLFB | Methanol non limited fed-batch |
| MSFBR | Magnetically-stabilised fluidised bed reactor |
| Mut+ | Methanol utilisation plus phenotype |
| Muts | Methanol utilisation slow phenotype |
| MW | Molecular weight (kDa) |
| NBS | N-Bromosuccinimide |
| OA | Oleic acid |
| OO | Olive oil |
| OP | Olive pomace |
| OPO | TAG with oleic acid in sn-1,3 positions and palmitic acid in sn-2 position. |
| OS | Operational stability |
| PA | Palmitic acid |
| PAOX | Inducible Alcohol oxidase promoter |
| PBR | Packed bed reactor |
| PDI | Protein disulphide isomerase |
| PFL | Pseudomonas fluorescens lipase |
| PFLD1 | Inducible formaldehyde dehydrogenase 1 promoter |
| PGAP | Constitutive glyceraldehyde-3-phosphate dehydrogenase promoter |
| Pmax | Maximum production |
| PMSF | Phenylmethylsulfonyl fluoride |
| proROL | R. oryzae lipase containing the N-terminal of mature sequence attached to 28 C-terminal amino acids of the prosequence |
| proROL-gene | Gene encoding the prosequence of 97 amino acids fused to the N-terminal of the mature lipase region of 269 amino acids |
| PUFA | Polyunsaturated fatty acids |
| PVA | Polyvinylalcohol |
| qp | Specific production rate (AU gX−1 h−1) |
| RO | Rapeseed oil |
| ROL | Rhizopus oryzae lipase |
| rROL | Rhizopus oryzae lipase containing mature sequence of R. oryzae lipase |
| rROL-gene | Gene encoding the mature lipase |
| S | Short-chain fatty acid |
| SA | Stearic acid |
| SC | Single copy |
| SCG | Spent coffee ground |
| SGLB | Solid gas liquid bioreactor |
| SL | structured lipid |
| SLLB | Solid liquid liquid bioreactor |
| SNLFB | Sorbitol non limited fed-batch |
| SO | Sunflower oil |
| Ssa4 | Cytosolic chaperone |
| Sso2 | Secretion helper factor |
| STR | Stirred tank reactor |
| SYO | Soybean oil |
| TAGs | Triacylglycerols |
| TGA40 | commercial oil |
| TGA55E | Hydrolysed TGA40 oil |
| TGA58F | Mortierella alpina single-cell oil |
| TPB | Three phase bioreactor |
| UBC1 | Ubiquitin-conjugating enzyme |
| UPR | Unfolding protein response |
| Vhb | Vitreoscilla haemoglobin |
| WCB | Whole cells biocatalyst |
| WCO | Waste cooking oil |
| Y | Yield (%) |
| YP/X | Product-biomass yield (AU gX−1) |
| μ | Specific growth rate (h-1) |
References
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial enzymes: Industrial progress in 21st century. 3 Biotech 2016, 6, 174–189. [Google Scholar] [CrossRef] [Green Version]
- Bilal, M.; Cui, J.; Iqbal, H.M.N. Tailoring enzyme microenvironment: State-of-the-art strategy to fulfill the quest for efficient bio-catalysis. Int. J. Biol. Macromol. 2019, 130, 186–196. [Google Scholar] [CrossRef]
- Girelli, A.M.; Astolfi, M.L.; Scuto, F.R. Agro-industrial wastes as potential carriers for enzyme immobilization: A review. Chemosphere 2020, 244, 1–12. [Google Scholar] [CrossRef]
- Raveendran, S.; Parameswaran, B.; Ummalyma, S.B.; Abraham, A.; Mathew, A.K.; Madhavan, A.; Rebello, S.; Pandey, A. Applications of microbial enzymes in food industry. Food Technol. Biotechnol. 2018, 56, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes—A review. Mol. Catal. 2019, 479, 1–20. [Google Scholar] [CrossRef]
- Ravindran, R.; Hassan, S.S.; Williams, G.A.; Jaiswal, A.K. A review on bioconversion of agro-industrial wastes to industrially important enzymes. Bioengineering 2018, 5, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodley, J.M. Accelerating the implementation of biocatalysis in industry. Appl. Microbiol. Biotechnol. 2019, 103, 4733–4739. [Google Scholar] [CrossRef]
- Singh, G.; Arya, S.K. Utility of laccase in pulp and paper industry: A progressive step towards the green technology. Int. J. Biol. Macromol. 2019, 134, 1071–1084. [Google Scholar] [CrossRef]
- Publishing, B. Global Markets for Enzymes in Industrial Applications; BCC Publishing: Wellesley, MA, USA, 2018. [Google Scholar]
- Li, S.; Yang, X.; Yang, S.; Zhu, M.; Wang, X. Technology prospecting on enzymes: Application, marketing and engineering. Comput. Struct. Biotechnol. J. 2012, 2, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Haki, G.D.; Rakshit, S.K. Developments in industrially important thermostable enzymes: A review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef]
- Dalla-Vecchia, R.; Sebrão, D.; Nascimento, M.D.G.; Soldi, V. Carboxymethylcellulose and poly(vinyl alcohol) used as a film support for lipases immobilization. Process Biochem. 2005, 40, 2677–2682. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Sarmah, N.; Revathi, D.; Sheelu, G.; Yamuna Rani, K.; Sridhar, S.; Mehtab, V.; Sumana, C. Recent advances on sources and industrial applications of lipases. Biotechnol. Prog. 2018, 34, 5–28. [Google Scholar] [CrossRef]
- Yu, X.W.; Xu, Y.; Xiao, R. Lipases from the genus Rhizopus: Characteristics, expression, protein engineering and application. Prog. Lipid Res. 2016, 64, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Melani, N.B.; Tambourgi, E.B.; Silveira, E. Lipases: From Production to Applications. Sep. Purif. Rev. 2020, 49, 143–158. [Google Scholar] [CrossRef]
- Lima, R.N.; Porto, A.L.M. Biocatalytic aminolysis of ethyl (S)-mandelate by lipase from Candida antarctica. Catal. Commun. 2017, 100, 157–163. [Google Scholar] [CrossRef]
- Paiva, A.L.; Balcão, V.M.; Malcata, F.X. Kinetics and mechanisms of reactions catalyzed by immobilized lipases. Enzyme Microb. Technol. 2000, 27, 187–204. [Google Scholar] [CrossRef]
- Zeng, S.; Liu, J.; Anankanbil, S.; Chen, M.; Guo, Z.; Adams, J.P.; Snajdrova, R.; Li, Z. Amide Synthesis via Aminolysis of Ester or Acid with an Intracellular Lipase. ACS Catal. 2018, 8, 8856–8865. [Google Scholar] [CrossRef]
- Ken Ugo, A.; Vivian Amara, A.; CN, I.; Kenechuwku, U. Microbial lipases: A prospect for biotechnological industrial catalysis for green products: A review. Ferment Technol. 2017, 6, 144–156. [Google Scholar] [CrossRef] [Green Version]
- Bharathi, D.; Rajalakshmi, G. Microbial lipases: An overview of screening, production and purification. Biocatal. Agric. Biotechnol. 2019, 22, 101368–101375. [Google Scholar] [CrossRef]
- Borza, P.; Peter, F.; Paul, C. Improved enantioselectivity of Candida antarctica A lipase through sol-gel entrapment. Chem. Bull. Politehnica Univ. 2015, 60, 49–54. [Google Scholar]
- Fernandez-Lafuente, R. Lipase from Thermomyces lanuginosus: Uses and prospects as an industrial biocatalyst. J. Mol. Catal. B Enzym. 2010, 62, 197–212. [Google Scholar] [CrossRef]
- Guncheva, M.; Zhiryakova, D. Catalytic properties and potential applications of Bacillus lipases. J. Mol. Catal. B Enzym. 2011, 68, 1–21. [Google Scholar] [CrossRef]
- Jachmanián, I.; Schulte, E.; Mukherjee, K.D. Substrate selectivity in esterification of less common fatty acids catalysed by lipases from different sources. Appl. Microbiol. Biotechnol. 1996, 44, 563–567. [Google Scholar] [CrossRef]
- Barros, M.; Fleuri, L.F.; MacEdo, G.A. Seed lipases: Sources, applications and properties—A review. Brazilian J. Chem. Eng. 2010, 27, 15–29. [Google Scholar] [CrossRef] [Green Version]
- Dhake, K.P.; Thakare, D.D.; Bhanage, B.M. Lipase: A potential biocatalyst for the synthesis of valuable flavour and fragrance ester compounds. Flavour Fragr. J. 2013, 28, 71–83. [Google Scholar] [CrossRef]
- Su, E.; Xu, J.; You, P. Functional expression of Serratia marcescens H30 lipase in Escherichia coli for efficient kinetic resolution of racemic alcohols in organic solvents. J. Mol. Catal. B Enzym. 2014, 106, 11–16. [Google Scholar] [CrossRef]
- Pomeisl, K.; Lamatová, N.; Šolínová, V.; Pohl, R.; Brabcová, J.; Kašička, V.; Krečmerová, M. Enantioselective resolution of side-chain modified gem-difluorinated alcohols catalysed by Candida antarctica lipase B and monitored by capillary electrophoresis. Bioorg. Med. Chem. 2019, 27, 1246–1253. [Google Scholar] [CrossRef]
- Javed, S.; Azeem, F.; Hussain, S.; Rasul, I.; Siddique, M.H.; Riaz, M.; Afzal, M.; Kouser, A.; Nadeem, H. Bacterial lipases: A review on purification and characterization. Prog. Biophys. Mol. Biol. 2018, 132, 23–34. [Google Scholar] [CrossRef]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Process. Biochem. 2012, 57, 555–569. [Google Scholar] [CrossRef]
- Ribeiro, B.D.; De Castro, A.M.; Coelho, M.A.Z.; Freire, D.M.G. Production and use of lipases in bioenergy: A review from the feedstocks to biodiesel production. Enzyme Res. 2011, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savaghebi, D.; Safari, M.; Rezaei, K.; Ashtari, P.; Farmani, J. Structured lipids produced through lipase-catalyzed acidolysis of canola oil. J. Agric. Sci. Technol. 2012, 14, 1297–1310. [Google Scholar]
- Functional Foods from Soybean Oil Deodorizer Distillate using Candida rugosa and Candida Antarctica lipases. Chem. Sci. Trans. 2019, 8, 268–272.
- Jensen, R.G. Characteristics of the lipase from the mold, Geotrichum candidum: A review. Lipids 1974, 9, 149–157. [Google Scholar] [CrossRef]
- Miettinen, H.; Nyyssölä, A.; Rokka, S.; Kontkanen, H.; Kruus, K. Screening of microbes for lipases specific for saturated medium and long-chain fatty acids of milk fat. Int. Dairy J. 2013, 32, 61–67. [Google Scholar] [CrossRef]
- Zhao, J.-f.; Lin, J.-p.; Yang, L.-r.; Wu, M.-b. Enhanced performance of Rhizopus oryzae lipase by reasonable immobilization on magnetic nanoparticles and its application in synthesis 1,3-diacyglycerol. Appl. Biochem. Biotechnol. 2019, 188, 677–689. [Google Scholar] [CrossRef]
- López-Fernández, J.; Barrero, J.J.; Benaiges, M.D.; Valero, F. Truncated prosequence of Rhizopus oryzae lipase: Key factor for production improvement and biocatalyst stability. Catalysts 2019, 9, 961. [Google Scholar] [CrossRef] [Green Version]
- da S. Pereira, A.; Fontes-Sant’Ana, G.C.; Amaral, P.F.F. Mango agro-industrial wastes for lipase production from Yarrowia lipolytica and the potential of the fermented solid as a biocatalyst. Food Bioprod. Process. 2019, 115, 68–77. [Google Scholar]
- Filho, D.G.; Silva, A.G.; Guidini, C.Z. Lipases: Sources, immobilization methods, and industrial applications. Appl. Microbiol. Biotechnol. 2019, 103, 7399–7423. [Google Scholar] [CrossRef]
- Thapa, S.; Li, H.; OHair, J.; Bhatti, S.; Chen, F.C.; Nasr, K.A.; Johnson, T.; Zhou, S. Biochemical characteristics of microbial enzymes and their significance from industrial perspectives. Mol. Biotechnol. 2019, 61, 579–601. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Pandey, N.; Dhakar, K.; Jain, R.; Pandey, A. Temperature dependent lipase production from cold and pH tolerant species of Penicillium. Mycosphere 2016, 7, 1533–1545. [Google Scholar] [CrossRef]
- Mehta, A.; Bodh, U.; Gupta, R. Fungal lipases: A review. J. Biotech. Res. 2017, 8, 58–77. [Google Scholar]
- Gryganskyi, A.P.; Golan, J.; Dolatabadi, S.; Mondo, S.; Robb, S.; Idnurm, A.; Muszewska, A.; Steczkiewicz, K.; Masonjones, S.; Liao, H.L.; et al. Phylogenetic and phylogenomic definition of Rhizopus species. G3 Genes Genomes Genet. 2018, 8, 2007–2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MAA Schipper A revision of the genus Rhizopus. I. The Rhizopus stolonifer-group and Rhizopus oryzae. Stud. Mycol. 1984, 25, 1–19. [Google Scholar]
- Abe, A.; Oda, Y.; Asano, K.; Sone, T. The molecular phylogeny of the genus Rhizopus based on rDNA sequences. Biosci. Biotechnol. Biochem. 2006, 70, 2387–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.-Y.; Huang, H.; Zheng, R.-Y. Molecular phylogenetic relationships within Rhizopus based on combined analyses of ITS rDNA and pyrG gene sequences. Sydowia 2007, 59, 235–253. [Google Scholar]
- Zheng, R.Y.; Chen, G.Q.; Huang, H.; Liu, X.Y. A monograph of Rhizopus. Sydowia 2007, 59, 273–372. [Google Scholar]
- Abe, A.; Asano, K.; Sone, T. A molecular phylogeny-based taxonomy of the genus Rhizopus. Biosci. Biotechnol. Biochem. 2010, 74, 1325–1331. [Google Scholar] [CrossRef] [Green Version]
- Londoño-Hernández, L.; Ramírez-Toro, C.; Ruiz, H.A.; Ascacio-Valdés, J.A.; Aguilar-Gonzalez, M.A.; Rodríguez-Herrera, R.; Aguilar, C.N. Rhizopus oryzae—Ancient microbial resource with importance in modern food industry. Int. J. Food Microbiol. 2017, 257, 110–127. [Google Scholar] [CrossRef]
- Sebastian, J.; Hegde, K.; Kumar, P.; Rouissi, T.; Brar, S.K. Bioproduction of fumaric acid: An insight into microbial strain improvement strategies. Crit. Rev. Biotechnol. 2019, 39, 817–834. [Google Scholar] [CrossRef]
- Benabda, O.; M’Hir, S.; Kasmi, M.; Mnif, W.; Hamdi, M. Optimization of protease and amylase production by Rhizopus oryzae cultivated on bread waste using solid-state fermentation. J. Chem. 2019, 2019, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, B.; Ray, R.R. Current commercial perspective of Rhizopus oryzae: A review. J. Appl. Sci. 2011, 11, 2470–2486. [Google Scholar] [CrossRef] [Green Version]
- Beer, H.D.; McCarthy, J.E.G.; Bornscheuer, U.T.; Schmid, R.D. Cloning, expression, characterization and role of the leader sequence of a lipase from Rhizopus oryzae. Biochim. Biophys. Acta Gene Struct. Expr. 1998, 1399, 173–180. [Google Scholar] [CrossRef]
- Salah, R.B.; Mosbah, H.; Fendri, A.; Gargouri, A.; Gargouri, Y.; Mejdoub, H. Biochemical and molecular characterization of a lipase produced by Rhizopus oryzae. FEMS Microbiol. Lett. 2006, 260, 241–248. [Google Scholar] [CrossRef] [Green Version]
- Sayari, A.; Frikha, F.; Miled, N.; Mtibaa, H.; Ali, Y.B.; Verger, R.; Gargouri, Y. N-terminal peptide of Rhizopus oryzae lipase is important for its catalytic properties. FEBS Lett. 2005, 579, 976–982. [Google Scholar] [CrossRef] [Green Version]
- Derewenda, U.; Swenson, L.; Wei, Y.; Green, R.; Kobos, P.M.; Joerger, R.; Haas, M.J.; Derewenda, Z.S. Conformational lability of lipases observed in the absence of an oil- water interface: Crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar. J. Lipid Res. 1994, 35, 524–534. [Google Scholar]
- Kohno, M.; Kugimiya, W.; Hashimoto, Y.; Morita, Y. Purification, characterization, and crystallization of two types of lipase from Rhizopus niveus. Biosci. Biotechnol. Biochem. 1994, 58, 1007–1012. [Google Scholar] [CrossRef]
- Yang, M.; Yu, X.W.; Zheng, H.; Sha, C.; Zhao, C.; Qian, M.; Xu, Y. Role of N-linked glycosylation in the secretion and enzymatic properties of Rhizopus chinensis lipase expressed in Pichia pastoris. Microb. Cell Fact. 2015, 14, 40–54. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.W.; Yang, M.; Jiang, C.; Zhang, X.; Xu, Y. N-Glycosylation engineering to improve the constitutive expression of Rhizopus oryzae lipase in Komagataella phaffii. J. Agric. Food Chem. 2017, 65, 6009–6015. [Google Scholar] [CrossRef]
- Beer, H.D.; Wohlfahrt, G.; Schmid, R.D.; Mccarthy, J.E.G. The folding and activity of the extracellular lipase of Rhizopus oryzae are modulated by a prosequence. Biochem. J. 1996, 319, 351–359. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.J.; Inouye, M. The intramolecular chaperone-mediated protein folding. Curr. Opin. Struct. Biol. 2008, 18, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Ueda, M.; Atomi, H.; Beer, H.D.; Bornscheuer, U.T.; Schmid, R.D.; Tanaka, A. Extracellular production of active Rhizopus oryzae lipase by Saccharomyces cerevisiae. J. Ferment. Bioeng. 1998, 86, 164–168. [Google Scholar] [CrossRef]
- Ueda, M.; Takahashi, S.; Washida, M.; Shiraga, S.; Tanaka, A. Expression of Rhizopus oryzae lipase gene in Saccharomyces cerevisiae. J. Mol. Catal. B Enzym. 2002, 17, 113–124. [Google Scholar] [CrossRef]
- Niu, W.; Li, Z.; Tan, T. Secretion of pro- and mature Rhizopus arrhizus lipases by Pichia pastoris and properties of the proteins. Mol. Biotechnol. 2006, 32, 73–81. [Google Scholar] [CrossRef]
- Wang, J.R.; Li, Y.Y.; De Xu, S.; Li, P.; Liu, J.S.; Liu, D.N. High-level expression of pro-form lipase from Rhizopus oryzae in Pichia pastoris and its purification and characterization. Int. J. Mol. Sci. 2014, 15, 203–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, S.; Ueda, M.; Tanaka, A. Function of the prosequence for in vivo folding and secretion of active Rhizopus oryzae lipase in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2001, 55, 454–462. [Google Scholar] [CrossRef]
- Ben Salah, A.; Sayari, A.; Verger, R.; Gargouri, Y. Kinetic studies of Rhizopus oryzae lipase using monomolecular film technique. Biochimie 2001, 83, 463–469. [Google Scholar] [CrossRef]
- Takahashi, S.; Ueda, M.; Tanaka, A. Independent production of two molecular forms of a recombinant Rhizopus oryzae lipase by KEX2-engineered strains of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1999, 52, 534–540. [Google Scholar] [CrossRef]
- Hama, S.; Tamalampudi, S.; Shindo, N.; Numata, T.; Yamaji, H.; Fukuda, H.; Kondo, A. Role of N-terminal 28-amino-acid region of Rhizopus oryzae lipase in directing proteins to secretory pathway of Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2008, 79, 1009–1018. [Google Scholar] [CrossRef]
- Minning, S.; Schmidt-Dannert, C.; Schmid, R.D. Functional expression of Rhizopus oryzae lipase in Pichia pastoris: High-level production and some properties. J. Biotechnol. 1998, 66, 147–156. [Google Scholar] [CrossRef]
- Takó, M.; Kotogán, A.; Papp, T.; Kadaikunnan, S.; Alharbi, N.S.; Vágvölgyi, C. Purification and properties of extracellular lipases with transesterification activity and 1,3-regioselectivity from Rhizomucor miehei and Rhizopus oryzae. J. Microbiol. Biotechnol. 2017, 27, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Hiol, A.; Jonzo, M.D.; Rugani, N.; Druet, D.; Sarda, L.; Comeau, L.C. Purification and characterization of an extracellular lipase from a thermophilic Rhizopus oryzae strain isolated from palm fruit. Enzyme Microb. Technol. 2000, 26, 421–430. [Google Scholar] [CrossRef]
- Shimada, Y.; Iwai, M.; Tsujisaka, Y. Reversibility of the modification of Rhizopus delemar lipase by phosphatidylcholine. J. Biochem. 1981, 89, 937–942. [Google Scholar] [CrossRef]
- Schrag, J.D.; Cygler, M. 1.8 A refined structure of the lipase from Geotrichum candidum. J. Mol. Biol. 1993, 230, 575–591. [Google Scholar] [CrossRef]
- Grochulski, P.; Li, Y.; Schrag, J.D.; Bouthillier, F.; Smith, P.; Harrison, D.; Rubin, B.; Cygler, M. Insights into interfacial activation from an open structure of Candida rugosa lipase. J. Biol. Chem. 1993, 268, 12843–12847. [Google Scholar]
- Noble, M.E.M.; Cleasby, A.; Johnson, L.N.; Egmond, M.R.; Frenken, L.G.J. The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Lett. 1993, 331, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Derewenda, U.; Swenson, L.; Green, R.; Wei, Y.; Dodson, G.G.; Yamaguchi, S.; Haas, M.J.; Derewenda, Z.S. An unusual buried polar cluster in a family of fungal lipases. Nat. Struct. Biol. 1994, 1, 36–47. [Google Scholar] [CrossRef]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 2017, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Satomura, A.; Kuroda, K.; Ueda, M. Generation of a functionally distinct Rhizopus oryzae lipase through protein folding memory. PLoS ONE 2015, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiraga, S.; Ueda, M.; Takahashi, S.; Tanaka, A. Construction of the combinatorial library of Rhizopus oryzae lipase mutated in the lid domain by displaying on yeast cell surface. J. Mol. Catal. B Enzym. 2002, 17, 167–173. [Google Scholar] [CrossRef]
- Adlercreutz, P. Immobilisation and application of lipases in organic media. Chem. Soc. Rev. 2013, 7, 6406–6436. [Google Scholar] [CrossRef] [Green Version]
- Verger, R. “Interfacial activation” of lipases: Facts and artifacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar] [CrossRef]
- Reis, P.; Holmberg, K.; Watzke, H.; Leser, M.E.; Miller, R. Lipases at interfaces: A review. Adv. Colloid Interface Sci. 2009, 147–148, 237–250. [Google Scholar] [CrossRef]
- Kourist, R.; Brundiek, H.; Bornscheuer, U.T. Protein engineering and discovery of Lipases. Eur. J. Lipid Sci. Technol. 2010, 112, 64–74. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Gao, X.; Jiang, W.; Li, Z.; Yao, Q.; Yang, F.; Wang, F.; Liu, J. Kinetic model of the enzymatic Michael addition for synthesis of mitomycin analogs catalyzed by immobilized lipase from T. laibacchii. Mol. Catal. 2019, 466, 146–156. [Google Scholar] [CrossRef]
- Shiraga, S.; Ishiguro, M.; Fukami, H.; Nakao, M.; Ueda, M. Creation of Rhizopus oryzae lipase having a unique oxyanion hole by combinatorial mutagenesis in the lid domain. Appl. Microbiol. Biotechnol. 2005, 68, 779–785. [Google Scholar] [CrossRef]
- Guillén, M.; Benaiges, M.D.; Valero, F. Comparison of the biochemical properties of a recombinant lipase extract from Rhizopus oryzae expressed in Pichia pastoris with a native extract. Biochem. Eng. J. 2011, 54, 117–123. [Google Scholar] [CrossRef]
- Hermanová, S.; Zarevúcká, M.; Bouša, D.; Pumera, M.; Sofer, Z. Graphene oxide immobilized enzymes show high thermal and solvent stability. Nanoscale 2015, 7, 5852–5858. [Google Scholar] [CrossRef] [Green Version]
- Pashangeh, K.; Akhond, M.; Karbalaei-Heidari, H.R.; Absalan, G. Biochemical characterization and stability assessment of Rhizopus oryzae lipase covalently immobilized on amino-functionalized magnetic nanoparticles. Int. J. Biol. Macromol. 2017, 105, 300–307. [Google Scholar] [CrossRef]
- Haas, M.J.; Cichowicz, D.J.; Bailey, D.G. Purification and characterization of an extracellular lipase from the fungus Rhizopus delemar. Lipids 1992, 27, 571–576. [Google Scholar] [CrossRef]
- Essamri, M.; Deyris, V.; Comeau, L. Optimization of lipase production by Rhizopus oryzae and study on the stability of lipase activity in organic solvents. J. Biotechnol. 1998, 60, 97–103. [Google Scholar] [CrossRef]
- Yu, X.W.; Sha, C.; Guo, Y.L.; Xiao, R.; Xu, Y. High-level expression and characterization of a chimeric lipase from Rhizopus oryzae for biodiesel production. Biotechnol. Biofuels 2013, 6, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Ben Salah, R.; Gargouri, A.; Verger, R.; Gargouri, Y.; Mejdoub, H. Expression in Pichia pastoris X33 of his-tagged lipase from a novel strain of Rhizopus oryzae and its mutant Asn 134 his: Purification and characterization. World J. Microbiol. Biotechnol. 2009, 25, 1375–1384. [Google Scholar] [CrossRef]
- Kantak, J.B.; Prabhune, A.A. Characterization of smallest active monomeric lipase from novel Rhizopus strain: Application in transesterification. Appl. Biochem. Biotechnol. 2012, 166, 1769–1780. [Google Scholar] [CrossRef]
- Li, C.; Zhang, G.; Liu, N.; Liu, L. Preparation and properties of Rhizopus oryzae lipase immobilized using an adsorption-crosslinking method. Int. J. Food Prop. 2015, 19, 1776–1785. [Google Scholar] [CrossRef] [Green Version]
- Razak, C.N.A.; Salleh, A.B.; Musani, R.; Samad, M.Y.; Basri, M. Some characteristics of lipases from thermophilic fungi isolated from palm oil mill effluent. J. Mol. Catal. B Enzym. 1997, 3, 153–159. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Wang, Y.; Wang, Y.; Wang, F.; Jiang, J. Expression and characterization of recombinant Rhizopus oryzae lipase for enzymatic biodiesel production. Bioresour. Technol. 2011, 10, 9810–9813. [Google Scholar] [CrossRef]
- Song, X.; Qi, X.; Hao, B.; Qu, Y. Studies of substrate specificities of lipases from different sources. Eur. J. Lipid Sci. Technol. 2008, 110, 1095–1101. [Google Scholar] [CrossRef]
- Pereira, R.M.; Andrade, G.S.S.; De Castro, H.F.; Campos, M.G.N. Performance of chitosan/glycerol phosphate hydrogel as a support for lipase immobilization. Mater. Res. 2017, 20, 190–201. [Google Scholar] [CrossRef] [Green Version]
- Yuzbashev, T.V.; Yuzbasheva, E.Y.; Vibornaya, T.V.; Sobolevskaya, T.I.; Laptev, I.A.; Gavrikov, A.V.; Sineoky, S.P. Production of recombinant Rhizopus oryzae lipase by the yeast Yarrowia lipolytica results in increased enzymatic thermostability. Protein Expr. Purif. 2012, 82, 83–89. [Google Scholar] [CrossRef]
- Adak, S.; Banerjee, R. Biochemical characterisation of a newly isolated low molecular eeight lipase from Rhizopus oryzae NRRL. Enzym Eng. 2013, 2, 118–125. [Google Scholar]
- Karra-Châabouni, M.; Bouaziz, I.; Boufi, S.; Botelho do Rego, A.M.; Gargouri, Y. Physical immobilization of Rhizopus oryzae lipase onto cellulose substrate: Activity and stability studies. Colloids Surfaces B Biointerfaces 2008, 66, 168–177. [Google Scholar] [CrossRef]
- Ebrahimpour, A.; Rahman, R.N.Z.R.A.; Basri, M.; Salleh, A.B. High level expression and characterization of a novel thermostable, organic solvent tolerant, 1,3-regioselective lipase from Geobacillus sp. strain ARM. Bioresour. Technol. 2011, 102, 6972–6981. [Google Scholar] [CrossRef]
- Lin, S.F. Production and stabilization of a solvent-tolerant alkaline lipase from Pseudomonas pseudoalcaligenes F-111. J. Ferment. Bioeng. 1996, 82, 448–451. [Google Scholar] [CrossRef]
- Lesuisse, E.; Schanck, K.; Colson, C. Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur. J. Biochem. 1993, 216, 155–160. [Google Scholar] [CrossRef]
- Bose, A.; Keharia, H. Production, characterization and applications of organic solvent tolerant lipase by Pseudomonas aeruginosa AAU2. Biocatal. Agric. Biotechnol. 2013, 2, 255–266. [Google Scholar] [CrossRef]
- Yan, J.; Yang, J.; Xu, L.; Yan, Y. Gene cloning, overexpression and characterization of a novel organic solvent tolerant and thermostable lipase from Galactomyces geotrichum Y05. J. Mol. Catal. B Enzym. 2007, 49, 28–35. [Google Scholar] [CrossRef]
- Zhao, H.; Zheng, L.; Wang, X.; Liu, Y.; Xu, L.; Yan, Y. Cloning, expression and characterization of a new lipase from Yarrowia lipolytica. Biotechnol. Lett. 2011, 33, 2445–2452. [Google Scholar] [CrossRef]
- Katiyar, M.; Ali, A. Effect of metal ions on the hydrolytic and transesterification activities of Candida rugosa Lipase. J. Oleo Sci. 2013, 62, 912–924. [Google Scholar] [CrossRef] [Green Version]
- Ateşlier, Z.B.B.; Metin, K. Production and partial characterization of a novel thermostable esterase from a thermophilic Bacillus sp. Enzyme Microb. Technol. 2006, 38, 628–635. [Google Scholar] [CrossRef]
- Li, W.; Li, R.W.; Li, Q.; Du, W.; Liu, D. Acyl migration and kinetics study of 1(3)-positional specific lipase of Rhizopus oryzae-catalyzed methanolysis of triglyceride for biodiesel production. Process. Biochem. 2010, 45, 1888–1893. [Google Scholar] [CrossRef]
- Šinkuniene, D.; Adlercreutz, P. Effects of regioselectivity and lipid class specificity of lipases on transesterification, exemplified by biodiesel production. J. Am. Oil Chem. Soc. 2014, 91, 1283–1290. [Google Scholar] [CrossRef] [Green Version]
- Okumura, S.; Iwai, M.; Tsujisaka, Y. Positional specificities of four kinds of microbial lipases. Agric. Biol. Chem. 1976, 40, 655–660. [Google Scholar] [CrossRef]
- Canet, A.; Benaiges, M.D.; Valero, F.; Adlercreutz, P. Exploring substrate specificities of a recombinant Rhizopus oryzae lipase in biodiesel synthesis. N. Biotechnol. 2017, 39, 59–67. [Google Scholar] [CrossRef]
- Cao, X.; Mangas-Sánchez, J.; Feng, F.; Adlercreutz, P. Acyl migration in enzymatic interesterification of triacylglycerols: Effects of lipases from Thermomyces lanuginosus and Rhizopus oryzae, support material, and water activity. Eur. J. Lipid Sci. Technol. 2016, 118, 1579–1587. [Google Scholar] [CrossRef]
- Zaks, A.; Klibanov, A.M. Enzymatic catalysis in nonaqueous solvents. J. Biol. Chem. 1988, 263, 3194–3201. [Google Scholar]
- Ben Salah, A.; Fendri, K.; Gargoury, Y. The Enzyme of Rhizopus oryzae—Production, Purification and Biochemical Characteristics. Rev. Fr. Des Corps Gras 1994. [Google Scholar]
- Di Lorenzo, M.; Hidalgo, A.; Haas, M.; Bornscheuer, U.T. Heterologous production of functional forms of Rhizopus oryzae lipase in Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 8974–8977. [Google Scholar] [CrossRef] [Green Version]
- Çelik, E.; Çalik, P. Production of recombinant proteins by yeast cells. Biotechnol. Adv. 2012, 30, 1108–1118. [Google Scholar] [CrossRef]
- Juturu, V.; Wu, J.C. Heterologous protein expression in Pichia pastoris: Latest research progress and applications. ChemBioChem 2018, 19, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Hirz, M.; Pichler, H.; Schwab, H. Protein expression in Pichia pastoris: Recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biotechnol. 2014, 98, 5301–5317. [Google Scholar] [CrossRef] [Green Version]
- García-Ortega, X.; Cámara, E.; Ferrer, P.; Albiol, J.; Montesinos-Seguí, J.L.; Valero, F. Rational development of bioprocess engineering strategies for recombinant protein production in Pichia pastoris (Komagataella phaffii) using the methanol-free GAP promoter. Where do we stand? N. Biotechnol. 2019, 53, 24–34. [Google Scholar] [CrossRef]
- Mattanovich, D.; Graf, A.; Stadlmann, J.; Dragosits, M.; Redl, A.; Maurer, M.; Kleinheinz, M.; Sauer, M.; Altmann, F.; Gasser, B. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microb. Cell Fact. 2009, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemmerich, J.; Adelantado, N.; Barrigón, J.M.; Ponte, X.; Hörmann, A.; Ferrer, P.; Kensy, F.; Valero, F. Comprehensive clone screening and evaluation of fed-batch strategies in a microbioreactor and lab scale stirred tank bioreactor system: Application on Pichia pastoris producing Rhizopus oryzae lipase. Microb. Cell Fact. 2014, 13, 36–52. [Google Scholar] [CrossRef] [Green Version]
- Minning, S.; Serrano, A.; Ferrer, P.; Solá, C.; Schmid, R.D.; Valero, F. Optimization of the high-level production of Rhizopus oryzae lipase in Pichia pastoris. J. Biotechnol. 2001, 86, 59–70. [Google Scholar] [CrossRef]
- Cos, O.; Serrano, A.; Montesinos, J.L.; Ferrer, P.; Cregg, J.M.; Valero, F. Combined effect of the methanol utilization (Mut) phenotype and gene dosage on recombinant protein production in Pichia pastoris fed-batch cultures. J. Biotechnol. 2005, 116, 321–335. [Google Scholar] [CrossRef]
- Surribas, A.; Stahn, R.; Montesinos, J.L.; Enfors, S.O.; Valero, F.; Jahic, M. Production of a Rhizopus oryzae lipase from Pichia pastoris using alternative operational strategies. J. Biotechnol. 2007, 130, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Barrigón, J.M.; Montesinos, J.L.; Valero, F. Searching the best operational strategies for Rhizopus oryzae lipase production in Pichia pastoris Mut+phenotype: Methanol limited or methanol non-limited fed-batch cultures? Biochem. Eng. J. 2013, 75, 47–54. [Google Scholar] [CrossRef]
- Ponte, X.; Montesinos-Seguí, J.L.; Valero, F. Bioprocess efficiency in Rhizopus oryzae lipase production by Pichia pastoris under the control of PAOX1 is oxygen tension dependent. Process. Biochem. 2016, 51, 1954–1963. [Google Scholar] [CrossRef]
- Canales, C.; Altamirano, C.; Berrios, J. Effect of dilution rate and methanol-glycerol mixed feeding on heterologous Rhizopus oryzae lipase production with Pichia pastoris Mut+ phenotype in continuous culture. Biotechnol. Prog. 2015, 31, 707–714. [Google Scholar] [CrossRef]
- Cos, O.; Ramon, R.; Montesinos, J.L.; Valero, F. A simple model-based control for Pichia pastoris allows a more efficient heterologous protein production bioprocess. Biotechnol. Bioeng. 2006, 95, 145–154. [Google Scholar] [CrossRef]
- Arnau, C.; Ramon, R.; Casas, C.; Valero, F. Optimization of the heterologous production of a Rhizopus oryzae lipase in Pichia pastoris system using mixed substrates on controlled fed-batch bioprocess. Enzyme Microb. Technol. 2010, 46, 494–500. [Google Scholar] [CrossRef]
- Arnau, C.; Casas, C.; Valero, F. The effect of glycerol mixed substrate on the heterologous production of a Rhizopus oryzae lipase in Pichia pastoris system. Biochem. Eng. J. 2011, 57, 30–37. [Google Scholar] [CrossRef]
- Resina, D.; Cos, O.; Ferrer, P.; Valero, F. Developing high cell density fed-batch cultivation strategies for heterologous protein production in Pichia pastoris using the nitrogen source-regulated FLD1 promoter. Biotechnol. Bioeng. 2005, 91, 760–767. [Google Scholar] [CrossRef]
- Resina, D.; Maurer, M.; Cos, O.; Arnau, C.; Carnicer, M.; Marx, H.; Gasser, B.; Valero, F.; Mattanovich, D.; Ferrer, P. Engineering of bottlenecks in Rhizopus oryzae lipase production in Pichia pastoris using the nitrogen source-regulated FLD1 promoter. N. Biotechnol. 2009, 25, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.W.; Lu, X.; Zhao, L.S.; Xu, Y. Impact of NH4+ nitrogen source on the production of Rhizopus oryzae lipase in Pichia pastoris. Process. Biochem. 2013, 48, 1462–1468. [Google Scholar] [CrossRef]
- Jiao, L.; Zhou, Q.; Su, Z.; Xu, L.; Yan, Y. High-level extracellular production of Rhizopus oryzae lipase in Pichia pastoris via a strategy combining optimization of gene-copy number with co-expression of ERAD-related proteins. Protein Expr. Purif. 2018, 147, 1–12. [Google Scholar] [CrossRef]
- Jiao, L.; Zhou, Q.; Su, Z.; Yan, Y. Efficient heterologous production of Rhizopus oryzae lipase via optimization of multiple expression-related helper proteins. Int. J. Mol. Sci. 2018, 19, 3372. [Google Scholar] [CrossRef] [Green Version]
- Ponte, X.; Barrigón, J.M.; Maurer, M.; Mattanovich, D.; Valero, F.; Montesinos-Seguí, J.L. Towards optimal substrate feeding for heterologous protein production in Pichia pastoris (Komagataella spp) fed-batch processes under PAOX1 control: A modeling aided approach. J. Chem. Technol. Biotechnol. 2018, 93, 3208–3218. [Google Scholar] [CrossRef]
- Li, X.; He, X.; Li, Z.; Wang, F. Combined strategies for improving the production of recombinant Rhizopus oryzae lipase in Pichia pastoris. BioResources 2013, 8, 2867–2880. [Google Scholar] [CrossRef] [Green Version]
- Ramón, R.; Ferrer, P.; Valero, F. Sorbitol co-feeding reduces metabolic burden caused by the overexpression of a Rhizopus oryzae lipase in Pichia pastoris. J. Biotechnol. 2007, 130, 39–46. [Google Scholar] [CrossRef]
- Resina, D.; Serrano, A.; Valero, F.; Ferrer, P. Expression of a Rhizopus oryzae lipase in Pichia pastoris under control of the nitrogen source-regulated formaldehyde dehydrogenase promoter. J. Biotechnol. 2004, 109, 103–113. [Google Scholar] [CrossRef]
- Cos, O.; Resina, D.; Ferrer, P.; Montesinos, J.L.; Valero, F. Heterologous production of Rhizopus oryzae lipase in Pichia pastoris using the alcohol oxidase and formaldehyde dehydrogenase promoters in batch and fed-batch cultures. Biochem. Eng. J. 2005, 26, 86–94. [Google Scholar] [CrossRef]
- Alhadeff, E.M.; Salgado, A.M.; Cos, O.; Pereira, N.; Valdman, B.; Valero, F. Enzymatic microreactors for the determination of ethanol by an automatic sequential injection analysis system. Appl. Biochem. Biotechnol. 2007, 137, 17–25. [Google Scholar]
- Marx, H.; Sauer, M.; Resina, D.; Vai, M.; Porro, D.; Valero, F.; Ferrer, P.; Mattanovich, D. Cloning, disruption and protein secretory phenotype of the GAS1 homologue of Pichia pastoris. FEMS Microbiol. Lett. 2006, 264, 40–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasser, B.; Sauer, M.; Maurer, M.; Stadlmayr, G.; Mattanovich, D. Transcriptomics-based identification of novel factors enhancing heterologous protein secretion in yeasts. Appl. Environ. Microbiol. 2007, 73, 6499–6507. [Google Scholar] [CrossRef] [Green Version]
- Sha, C.; Yu, X.W.; Zhang, M.; Xu, Y. Efficient secretion of lipase r27RCL in Pichia pastoris by enhancing the disulfide bond formation pathway in the endoplasmic reticulum. J. Ind. Microbiol. Biotechnol. 2013, 40, 1241–1249. [Google Scholar] [CrossRef]
- Li, W.; Du, W.; Liu, D. Rhizopus oryzae IFO 4697 whole cell catalyzed methanolysis of crude and acidified rapeseed oils for biodiesel production in tert-butanol system. Process. Biochem. 2007, 42, 1481–1485. [Google Scholar] [CrossRef]
- Li, W.; Du, W.; Liu, D. Optimization of whole cell-catalyzed methanolysis of soybean oil for biodiesel production using response surface methodology. J. Mol. Catal. B Enzym. 2007, 45, 122–127. [Google Scholar] [CrossRef]
- Hama, S.; Yamaji, H.; Kaieda, M.; Oda, M.; Kondo, A.; Fukuda, H. Effect of fatty acid membrane composition on whole-cell biocatalysts for biodiesel-fuel production. Biochem. Eng. J. 2004, 21, 155–160. [Google Scholar] [CrossRef]
- Matsumoto, T.; Takahashi, S.; Kaieda, M.; Ueda, M.; Tanaka, A.; Fukuda, H.; Kondo, A. Yeast whole-cell biocatalyst constructed by intracellular overproduction of Rhizopus oryzae lipase is applicable to biodiesel fuel production. Appl. Microbiol. Biotechnol. 2001, 57, 515–520. [Google Scholar]
- Matsumoto, T.; Takahashi, S.; Ueda, M.; Tanaka, A.; Fukuda, H.; Kondo, A. Preparation of high activity yeast whole cell bioctalysts by optimization of intracellular production of recombinant Rhizopus oryzae lipase. J. Mol. Catal. B Enzym. 2002, 17, 143–149. [Google Scholar] [CrossRef]
- Tanino, T.; Fukuda, H.; Kondo, A. Construction of a Pichia pastoris cell-surface display system using Flo1p anchor system. Biotechnol. Prog. 2006, 22, 983–999. [Google Scholar] [CrossRef]
- Li, W.; Shi, H.; Ding, H.; Wang, L.; Zhang, Y.; Li, X.; Wang, F. Cell surface display and characterization of Rhizopus oryzae lipase in Pichia pastoris using Sed1p as an anchor protein. Curr. Microbiol. 2015, 71, 150–155. [Google Scholar] [CrossRef]
- Thomas, S.M.; DiCosimo, R.; Nagarajan, V. Biocatalysis: Applications and potentials for the chemical industry. Trends Biotechnol. 2002, 20, 238–242. [Google Scholar] [CrossRef]
- Tenenbaum, D.J. Food vs. fuel diversion of crops could cause more hunger. Environ. Health Perspect. 2008, 116, A254–A257. [Google Scholar] [CrossRef] [Green Version]
- Amin, A. Review of diesel production from renewable resources: Catalysis, process kinetics and technologies. Ain Shams Eng. J. 2019, 201, 112155–112170. [Google Scholar] [CrossRef]
- Singh, D.; Sharma, D.; Soni, S.L.; Sharma, S.; Kumar Sharma, P.; Jhalani, A. A review on feedstocks, production processes, and yield for different generations of biodiesel. Fuel 2020, 262, 116553–116568. [Google Scholar] [CrossRef]
- Yew, G.Y.; Lee, S.Y.; Show, P.L.; Tao, Y.; Law, C.L.; Nguyen, T.T.C.; Chang, J.S. Recent advances in algae biodiesel production: From upstream cultivation to downstream processing. Bioresour. Technol. Reports 2019, 7. [Google Scholar] [CrossRef]
- Sitepu, I.R.; Garay, L.A.; Sestric, R.; Levin, D.; Block, D.E.; German, J.B.; Boundy-Mills, K.L. Oleaginous yeasts for biodiesel: Current and future trends in biology and production. Biotechnol. Adv. 2014, 32, 1336–1360. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.; Sharma, D.; Soni, S.L.; Sharma, S.; Kumari, D. Chemical compositions, properties, and standards for different generation biodiesels: A review. Fuel 2019, 253, 60–71. [Google Scholar] [CrossRef]
- Ambat, I.; Srivastava, V.; Sillanpää, M. Recent advancement in biodiesel production methodologies using various feedstock: A review. Renew. Sustain. Energy Rev. 2018, 90, 356–369. [Google Scholar] [CrossRef]
- Ranganathan, S.V.; Narasimhan, S.L.; Muthukumar, K. An overview of enzymatic production of biodiesel. Bioresour. Technol. 2008, 99, 3975–3981. [Google Scholar] [CrossRef]
- Guldhe, A.; Singh, B.; Mutanda, T.; Permaul, K.; Bux, F. Advances in synthesis of biodiesel via enzyme catalysis: Novel and sustainable approaches. Renew. Sustain. Energy Rev. 2015, 41, 1447–1464. [Google Scholar] [CrossRef]
- Christopher, L.P.; Kumar, H.; Zambare, V.P. Enzymatic biodiesel: Challenges and opportunities. Appl. Energy 2014, 119, 497–520. [Google Scholar] [CrossRef]
- Santos, S.; Puna, J.; Gomes, J. A review on bio-based catalysts (immobilized enzymes) used for biodiesel production. Energies 2020, 13, 3013. [Google Scholar] [CrossRef]
- Bonet-Ragel, K.; Canet, A.; Benaiges, M.D.; Valero, F. Synthesis of biodiesel from high FFA alperujo oil catalysed by immobilised lipase. Fuel 2015, 161, 12–17. [Google Scholar] [CrossRef]
- Lin, Y.H.; Luo, J.J.; John Hwang, S.C.; Liau, P.R.; Lu, W.J.; Lee, H.T. The influence of free fatty acid intermediate on biodiesel production from soybean oil by whole cell biocatalyst. Biomass Bioenergy 2011, 35, 2217–2223. [Google Scholar] [CrossRef]
- Aghababaie, M.; Beheshti, M.; Razmjou, A.; Bordbar, A.K. Enzymatic biodiesel production from crude Eruca sativa oil using Candida rugosa lipase in a solvent-free system using response surface methodology. Biofuels 2020, 11, 93–99. [Google Scholar] [CrossRef]
- Guldhe, A.; Singh, P.; Renuka, N.; Bux, F. Biodiesel synthesis from wastewater grown microalgal feedstock using enzymatic conversion: A greener approach. Fuel 2019, 237, 1112–1118. [Google Scholar] [CrossRef]
- Moreira, K.S.; Moura, L.S.; Monteiro, R.R.C.; de Oliveira, A.L.B.; Valle, C.P.; Freire, T.M.; Fechine, P.B.A.; de Souza, M.C.M.; Fernandez-Lorente, G.; Guisan, J.M.; et al. Optimization of the Production of Enzymatic Biodiesel from Residual Babassu Oil (Orbignya sp.) via RSM. Catalysts 2020, 10, 414. [Google Scholar] [CrossRef] [Green Version]
- Fedosov, S.N.; Brask, J.; Pedersen, A.K.; Nordblad, M.; Woodley, J.M.; Xu, X. Kinetic model of biodiesel production using immobilized lipase Candida antarctica lipase B. J. Mol. Catal. B Enzym. 2013, 85–86, 156–168. [Google Scholar] [CrossRef]
- Liu, L.H.; Shih, Y.H.; Liu, W.L.; Lin, C.H.; Huang, H.Y. Enzyme Immobilized on nanoporous carbon derived from metal–organic framework: A new support for biodiesel synthesis. ChemSusChem 2017, 10, 1364–1369. [Google Scholar] [CrossRef]
- Shah, S.; Gupta, M.N. The effect of ultrasonic pre-treatment on the catalytic activity of lipases in aqueous and non-aqueous media. Chem. Cent. J. 2008, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estevez, R.; Aguado-Deblas, L.; Bautista, F.M.; Luna, D.; Luna, C.; Calero, J.; Posadillo, A.; Romero, A.A. Biodiesel at the crossroads: A critical review. Catalysts 2019, 9, 1033. [Google Scholar] [CrossRef] [Green Version]
- Dunn, R.O. Effects of monoacylglycerols on the cold flow properties of biodiesel. J. Am. Oil Chem. Soc. 2012, 89, 1509–1520. [Google Scholar] [CrossRef]
- Calero, J.; Verdugo, C.; Luna, D.; Sancho, E.D.; Luna, C.; Posadillo, A.; Bautista, F.M.; Romero, A.A. Selective ethanolysis of sunflower oil with Lipozyme RM IM, an immobilized Rhizomucor miehei lipase, to obtain a biodiesel-like biofuel, which avoids glycerol production through the monoglyceride formation. N. Biotechnol. 2014, 31, 596–601. [Google Scholar] [CrossRef]
- Bonet-Ragel, K.; Canet, A.; Benaiges, M.D.; Valero, F. Effect of acyl-acceptor stepwise addition strategy using alperujo oil as a substrate in enzymatic biodiesel synthesis. J. Chem. Technol. Biotechnol. 2018, 93, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Fredrick, E.; Moens, K.; Heyman, B.; Fischer, S.; Van der Meeren, P.; Dewettinck, K. Monoacylglycerols in dairy recombined cream: I. The effect on milk fat crystallization. Food Res. Int. 2013, 51, 892–898. [Google Scholar] [CrossRef]
- Itabaiana, I.; Gonçalves, K.M.; Cordeiro, Y.M.L.; Zoumpanioti, M.; Leal, I.C.R.; Miranda, L.S.M.; De Souza, R.O.M.A.; Xenakis, A. Kinetics and mechanism of lipase catalyzed monoacylglycerols synthesis. J. Mol. Catal. B Enzym. 2013, 96, 34–39. [Google Scholar] [CrossRef]
- Feltes, M.M.C.; de Oliveira, D.; Block, J.M.; Ninow, J.L. The production, benefits, and applications of monoacylglycerols and diacylglycerols of nutritional interest. Food Bioprocess. Technol. 2013, 6, 17–35. [Google Scholar] [CrossRef]
- Canet, A.; Bonet-Ragel, K.í.; Benaiges, M.D.; Valero, F. Biodiesel synthesis in a solvent-free system by recombinant Rhizopus oryzae: Comparative study between a stirred tank and a packed-bed batch reactor. Biocatal. Biotransform. 2017, 35, 35–40. [Google Scholar] [CrossRef]
- Ciudad, G.; Reyes, I.; Jorquera, M.A.; Azócar, L.; Wick, L.Y.; Navia, R. Novel three-phase bioreactor concept for fatty acid alkyl ester production using R. oryzae as whole cell catalyst. World J. Microbiol. Biotechnol. 2011, 27, 2505–2512. [Google Scholar] [CrossRef]
- Jang, M.G.; Kim, D.K.; Park, S.C.; Lee, J.S.; Kim, S.W. Biodiesel production from crude canola oil by two-step enzymatic processes. Renew. Energy 2012, 42, 99–104. [Google Scholar] [CrossRef]
- He, Q.; Shi, H.; Gu, H.; Naka, G.; Ding, H.; Li, X.; Zhang, Y.; Hu, B.; Wang, F. Immobilization of Rhizopus oryzae ly6 onto loofah sponge as a whole-cell biocatalyst for biodiesel production. BioResources 2016, 11, 850–860. [Google Scholar] [CrossRef] [Green Version]
- Su, F.; Li, G.L.; Fan, Y.L.; Yan, Y.J. Enhancing biodiesel production via a synergic effect between immobilized Rhizopus oryzae lipase and Novozym Fuel Process. Technol. 2015, 137, 298–304. [Google Scholar]
- Zhou, G.-x.; Chen, G.-y.; Yan, B.-b. Biodiesel production in a magnetically-stabilized, fluidized bed reactor with an immobilized lipase in magnetic chitosan microspheres. Biotechnol. Lett. 2014, 36, 63–68. [Google Scholar] [CrossRef]
- Hama, S.; Yamaji, H.; Fukumizu, T.; Numata, T.; Tamalampudi, S.; Kondo, A.; Noda, H.; Fukuda, H. Biodiesel-fuel production in a packed-bed reactor using lipase-producing Rhizopus oryzae cells immobilized within biomass support particles. Biochem. Eng. J. 2007, 34, 273–278. [Google Scholar] [CrossRef]
- Luna, C.; Verdugo, C.; Sancho, E.D.; Luna, D.; Calero, J.; Posadillo, A.; Bautista, F.M.; Romero, A.A. Biocatalytic behaviour of immobilized Rhizopus oryzae lipase in the 1,3-selective ethanolysis of sunflower oil to obtain a biofuel similar to biodiesel. Molecules 2014, 19, 11419–11439. [Google Scholar] [CrossRef] [Green Version]
- Luna, C.; Verdugo, C.; Sancho, E.D.; Luna, D.; Calero, J.; Posadillo, A.; Bautista, F.M.; Romero, A.A. A biofuel similar to biodiesel obtained by using a lipase from Rhizopus oryzae, optimized by response surface methodology. Energies 2014, 7, 3383–3399. [Google Scholar] [CrossRef] [Green Version]
- Meher, L.C.; Churamani, C.P.; Ahmed, Z.; Naik, S.N. Jatropha curcas as a renewable source for bio-fuels—A review. Renew. Sustain. Energy Rev. 2013, 26, 397–407. [Google Scholar] [CrossRef]
- Rodrigues, J.; Perrier, V.; Lecomte, J.; Dubreucq, E.; Ferreira-Dias, S. Biodiesel production from crude jatropha oil catalyzed by immobilized lipase/acyltransferase from Candida parapsilosis in aqueous medium. Bioresour. Technol. 2016, 218, 1224–1229. [Google Scholar] [CrossRef]
- Li, X.; He, X.Y.; Li, Z.L.; Wang, Y.D.; Wang, C.Y.; Shi, H.; Wang, F. Enzymatic production of biodiesel from Pistacia chinensis bge seed oil using immobilized lipase. Fuel 2012, 92, 89–93. [Google Scholar] [CrossRef]
- Arumugam, A.; Ponnusami, V. Biodiesel production from Calophyllum inophyllum oil using lipase producing Rhizopus oryzae cells immobilized within reticulated foams. Renew. Energy 2014, 64, 276–282. [Google Scholar] [CrossRef]
- Navarro López, E.; Robles Medina, A.; González Moreno, P.A.; Esteban Cerdán, L.; Martín Valverde, L.; Molina Grima, E. Biodiesel production from Nannochloropsis gaditana lipids through transesterification catalyzed by Rhizopus oryzae lipase. Bioresour. Technol. 2016, 203, 233–244. [Google Scholar] [CrossRef]
- Navarro López, E.; Robles Medina, A.; González Moreno, P.A.; Esteban Cerdán, L.; Molina Grima, E. Extraction of microalgal lipids and the influence of polar lipids on biodiesel production by lipase-catalyzed transesterification. Bioresour. Technol. 2016, 216, 904–913. [Google Scholar] [CrossRef]
- Araya, K.; Ugarte, A.; Azócar, L.; Valerio, O.; Wick, L.Y.; Ciudad, G. Whole cell three phase bioreactors allow for effective production of fatty acid alkyl esters derived from microalgae lipids. Fuel 2015, 144, 25–32. [Google Scholar] [CrossRef]
- Nematian, T.; Salehi, Z.; Shakeri, A. Conversion of bio-oil extracted from Chlorella vulgaris micro algae to biodiesel via modified superparamagnetic nano-biocatalyst. Renew. Energy 2020, 146, 1796–1804. [Google Scholar] [CrossRef]
- Muanruksa, P.; Kaewkannetra, P. Combination of fatty acids extraction and enzymatic esterification for biodiesel production using sludge palm oil as a low-cost substrate. Renew. Energy 2020, 146, 901–906. [Google Scholar] [CrossRef]
- Karmee, S.K.; Swanepoel, W.; Marx, S. Biofuel production from spent coffee grounds via lipase catalysis. Energy Sour. Part A Recovery Util. Environ. Eff. 2018, 40, 294–300. [Google Scholar] [CrossRef]
- Bharathiraja, B.; Ranjithkumar, R.; Chakravarthy, M.; Yogendran, D.; Vivek, P.; Yuvaraj, D.; Kumar, R.P.; Palani, S. Kinetic analysis of fatty acid alkyl esters using whole cell biocatalyst and lipase catalyzed transesterification from waste cooking oil. Asian J. Microbiol. Biotechnol. Environ. Sci. 2014, 16, 745–752. [Google Scholar]
- Syed, M.B.; Ali, M.Y.; Ishaq, M.; Bakkiyaraj, S.; Devanesan, M.G.; Tangavelu, V. Response surface optimization of biodiesel production using immobilized Rhizopus oryzae cells. Biofuels 2016, 7, 1–8. [Google Scholar] [CrossRef]
- Sun, T.; Du, W.; Liu, D. Comparative study on stability of whole cells during biodiesel production in solvent-free system. Process. Biochem. 2011, 46, 661–664. [Google Scholar] [CrossRef]
- Ban, K.; Hama, S.; Nishizuka, K.; Kaieda, M.; Matsumoto, T.; Kondo, A.; Noda, H.; Fukuda, H. Repeated use of whole-cell biocatalysts immobilized within biomass support particles for biodiesel fuel production. J. Mol. Catal. B Enzym. 2002, 17, 157–165. [Google Scholar] [CrossRef]
- Bonet-Ragel, K.; López-Pou, L.; Tutusaus, G.; Benaiges, M.D.; Valero, F. Rice husk ash as a potential carrier for the immobilization of lipases applied in the enzymatic production of biodiesel. Biocatal. Biotransform. 2018, 36, 151–158. [Google Scholar] [CrossRef]
- Duarte, S.H.; del Peso Hernández, G.L.; Canet, A.; Benaiges, M.D.; Maugeri, F.; Valero, F. Enzymatic biodiesel synthesis from yeast oil using immobilized recombinant Rhizopus oryzae lipase. Bioresour. Technol. 2015, 183, 175–180. [Google Scholar] [CrossRef]
- Su, F.; Li, G.; Zhang, H.; Yan, Y. Enhanced performance of Rhizopus oryzae lipase immobilized on hydrophobic carriers and its application in biorefinery of rapeseed oil deodorizer distillate. Bioenergy Res. 2014, 7, 935–945. [Google Scholar] [CrossRef]
- Bakkiyaraj, S.; Syed, M.B.; Devanesan, M.G.; Thangavelu, V. Production and optimization of biodiesel using mixed immobilized biocatalysts in packed bed reactor. Environ. Sci. Pollut. Res. 2016, 23, 9276–9283. [Google Scholar] [CrossRef]
- Balasubramanian, B.; Ramanujam, P.K.; Ravi Kumar, R.; Muninathan, C.; Dhinakaran, Y. Optimization of biological transesterification of waste cooking oil in different solvents using response surface methodology. Manag. Environ. Qual. An. Int. J. 2016, 27, 537–550. [Google Scholar] [CrossRef]
- Nematian, T.; Shakeri, A.; Salehi, Z.; Saboury, A.A. Lipase immobilized on functionalized superparamagnetic few-layer graphene oxide as an efficient nanobiocatalyst for biodiesel production from Chlorella vulgaris bio-oil. Biotechnol. Biofuels 2020, 13. [Google Scholar] [CrossRef] [Green Version]
- Athalye, S.; Sharma-Shivappa, R.; Peretti, S.; Kolar, P.; Davis, J.P. Producing biodiesel from cottonseed oil using Rhizopus oryzae ATCC #34612 whole cell biocatalysts: Culture media and cultivation period optimization. Energy Sustain. Dev. 2013, 17, 331–336. [Google Scholar]
- Vipin, V.C.; Sebastian, J.; Muraleedharan, C.; Santhiagu, A. Enzymatic transesterification of rubber seed oil using Rhizopus oryzae Lipase. Procedia Technol. 2016, 25, 1014–1021. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Kim, S.B.; Kang, S.W.; Song, Y.S.; Park, C.; Han, S.O.; Kim, S.W. Biodiesel production by a mixture of Candida rugosa and Rhizopus oryzae lipases using a supercritical carbon dioxide process. Bioresour. Technol. 2011, 102, 2105–2108. [Google Scholar] [CrossRef]
- Zeng, L.; He, Y.; Jiao, L.; Li, K.; Yan, Y. Preparation of biodiesel with liquid synergetic lipases from rapeseed oil deodorizer distillate. Appl. Biochem. Biotechnol. 2017, 183, 778–791. [Google Scholar] [CrossRef]
- Rodrigues, J.; Canet, A.; Rivera, I.; Osório, N.M.; Sandoval, G.; Valero, F.; Ferreira-Dias, S. Biodiesel production from crude Jatropha oil catalyzed by non-commercial immobilized heterologous Rhizopus oryzae and Carica papaya lipases. Bioresour. Technol. 2016, 213, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Mukhtar, H.; Khursheed, S.; Ikram-ul-Haq; Mumtaz, M.W.; Rashid, U.; Al-Resayes, S.I. Optimization of lipase biosynthesis from Rhizopus oryzae for biodiesel production using multiple oils. Chem. Eng. Technol. 2016, 39, 1–10. [Google Scholar] [CrossRef]
- Zhou, G.-x.; Chen, G.-y.; Yan, B. bei Two-step biocatalytic process using lipase and whole cell catalysts for biodiesel production from unrefined jatropha oil. Biotechnol. Lett. 2015, 37, 1959–1963. [Google Scholar] [CrossRef]
- Sattari, S.; Vahabzadeh, F.; Aghtaei, H.K. Performance of loofa-immobilized Rhizopus oryzae in the enzymatic production of biodiesel with use of oleic acid in n-hexane medium. Brazilian J. Chem. Eng. 2015, 32, 367–376. [Google Scholar] [CrossRef]
- Ramachandran, P.; Narayanan, G.K.; Gandhi, S.; Sethuraman, S.; Krishnan, U.M. Rhizopus oryzae lipase immobilized on hierarchical mesoporous silica supports for transesterification of rice bran oil. Appl. Biochem. Biotechnol. 2015, 175, 2332–2346. [Google Scholar] [CrossRef]
- Zarei, A.; Amin, N.A.S.; Talebian-Kiakalaieh, A.; Zain, N.A.M. Immobilized lipase-catalyzed transesterification of Jatropha curcas oil: Optimization and modeling. J. Taiwan Inst. Chem. Eng. 2014, 45, 444–451. [Google Scholar] [CrossRef]
- Canet, A.; Dolors Benaiges, M.; Valero, F. Biodiesel synthesis in a solvent-free system by recombinant Rhizopus oryzae lipase. Study of the catalytic reaction progress. J. Am. Oil Chem. Soc. 2014, 91, 1499–1506. [Google Scholar] [CrossRef]
- Andrade, G.S.S.; Freitas, L.; Oliveira, P.C.; De Castro, H.F. Screening, immobilization and utilization of whole cell biocatalysts to mediate the ethanolysis of babassu oil. J. Mol. Catal. B Enzym. 2012, 84, 183–188. [Google Scholar] [CrossRef]
- Deshmukh, S.; Kumar, R.; Bala, K. Microalgae biodiesel: A review on oil extraction, fatty acid composition, properties and effect on engine performance and emissions. Fuel Process. Technol. 2019, 191, 232–247. [Google Scholar] [CrossRef]
- Yin, Z.; Zhu, L.; Li, S.; Hu, T.; Chu, R.; Mo, F.; Hu, D.; Liu, C.; Li, B. A comprehensive review on cultivation and harvesting of microalgae for biodiesel production: Environmental pollution control and future directions. Bioresour. Technol. 2020, 301, 122804–122823. [Google Scholar] [CrossRef]
- Lisboa, P.; Rodrigues, A.R.; Martín, J.L.; Simões, P.; Barreiros, S.; Paiva, A. Economic analysis of a plant for biodiesel production from waste cooking oil via enzymatic transesterification using supercritical carbon dioxide. J. Supercrit. Fluids 2014, 85, 31–40. [Google Scholar] [CrossRef]
- Tsoutsos, T.; Tournaki, S.; Gkouskos, Z.; Paraíba, O.; Giglio, F.; García, P.Q.; Braga, J.; Adrianos, H.; Filice, M. Quality Characteristics of biodiesel produced from used cooking oil in Southern Europe. ChemEngineering 2019, 3, 19. [Google Scholar] [CrossRef] [Green Version]
- Yaakob, Z.; Mohammad, M.; Alherbawi, M.; Alam, Z.; Sopian, K. Overview of the production of biodiesel from waste cooking oil. Renew. Sustain. Energy Rev. 2013, 18, 184–193. [Google Scholar] [CrossRef]
- Mandolesi De Araújo, C.D.; De Andrade, C.C.; De Souza E Silva, E.; Dupas, F.A. Biodiesel production from used cooking oil: A review. Renew. Sustain. Energy Rev. 2013, 27, 445–452. [Google Scholar] [CrossRef]
- Zhang, Y.; Dubé, M.A.; McLean, D.D.; Kates, M. Biodiesel production from waste cooking oil: Economic assessment and sensitivity analysis. Bioresour. Technol. 2003, 90, 229–240. [Google Scholar] [CrossRef]
- Wahab, R.A.; Elias, N.; Abdullah, F.; Ghoshal, S.K. On the taught new tricks of enzymes immobilization: An all-inclusive overview. React. Funct. Polym. 2020, 152, 104613. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Woodley, J.M. Role of Biocatalysis in Sustainable Chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H.; Hama, S.; Tamalampudi, S.; Noda, H. Whole-cell biocatalysts for biodiesel fuel production. Trends Biotechnol. 2008, 26, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lafuente, R.; Armisén, P.; Sabuquillo, P.; Fernández-Lorente, G.; Guisán, J.M. Immobilization of lipases by selective adsorption on hydrophobic supports. Chem. Phys. Lipids 1998, 93, 185–197. [Google Scholar] [CrossRef]
- Fernandez-Lorente, G.; Rocha-Martín, J.; Guisan, J.M. Immobilization of Lipases by Adsorption on Hydrophobic Supports: Modulation of Enzyme Properties in Biotransformations in Anhydrous Media. Methods Mol. Biol. 2020, 21000, 143–158. [Google Scholar]
- Wang, Y.D.; Shen, X.Y.; Li, Z.L.; Li, X.; Wang, F.; Nie, X.A.; Jiang, J.C. Immobilized recombinant Rhizopus oryzae lipase for the production of biodiesel in solvent free system. J. Mol. Catal. B Enzym. 2010, 67, 45–51. [Google Scholar] [CrossRef]
- Tan, T.; Lu, J.; Nie, K.; Deng, L.; Wang, F. Biodiesel production with immobilized lipase: A review. Biotechnol. Adv. 2010, 28, 628–634. [Google Scholar] [CrossRef]
- Balasubramaniam, B.; Sudalaiyadum Perumal, A.; Jayaraman, J.; Mani, J.; Ramanujam, P. Comparative analysis for the production of fatty acid alkyl esterase using whole cell biocatalyst and purified enzyme from Rhizopus oryzae on waste cooking oil (sunflower oil). Waste Manag. 2012, 32, 1539–1547. [Google Scholar] [CrossRef]
- Zhao, X.; Qi, F.; Yuan, C.; Du, W.; Liu, D. Lipase-catalyzed process for biodiesel production: Enzyme immobilization, process simulation and optimization. Renew. Sustain. Energy Rev. 2015, 44, 182–197. [Google Scholar] [CrossRef]
- Mendes, A.A.; Freitas, L.; De Carvalho, A.K.F.; De Oliveira, P.C.; De Castro, H.F. Immobilization of a commercial lipase from Penicillium camembertii (Lipase G) by different strategies. Enzyme Res. 2011, 2011. [Google Scholar] [CrossRef] [Green Version]
- Illanes, A. Enzyme biocatalysis: Principles and applications. In Enzyme Biocatalysis: Principles and Applications; Springer: Valparaíso, Chile, 2008; pp. 172–173. ISBN 9781402083600. [Google Scholar]
- Norjannah, B.; Ong, H.C.; Masjuki, H.H.; Juan, J.C.; Chong, W.T. Enzymatic transesterification for biodiesel production: A comprehensive review. RSC Adv. 2016, 6, 60034–60055. [Google Scholar] [CrossRef]
- Lotti, M.; Pleiss, J.; Valero, F.; Ferrer, P. Effects of methanol on lipases: Molecular, kinetic and process issues in the production of biodiesel. Biotechnol. J. 2015, 10, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Dias, S.; Sandoval, G.; Plou, F.; Valero, F. The potential use of lipases in the production of fatty acid derivatives for the food and nutraceutical industries. Electron. J. Biotechnol. 2013, 16, 1–38. [Google Scholar]
- Guo, Y.; Cai, Z.; Xie, Y.; Ma, A.; Zhang, H.; Rao, P.; Wang, Q. Synthesis, physicochemical properties, and health aspects of structured lipids: A review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 759–800. [Google Scholar] [CrossRef]
- Smith, R.E.; Finley, J.W.; Leveille, G.A. Overview of SALATRIM, a Family of Low-Calorie Fats. J. Agric. Food Chem. 1994, 42, 432–434. [Google Scholar] [CrossRef]
- Osborn, H.T.; Akoh, C.C. Structured lipids-novel fats with medical, nutraceutical, and food applications. Compr. Rev. Food Sci. Food Saf. 2002, 1, 110–120. [Google Scholar] [CrossRef]
- López-López, A.; Castellote-Bargalló, A.I.; Campoy-Folgoso, C.; Rivero-Urgel, M.; Tormo-Carnicé, R.; Infante-Pina, D.; López-Sabater, M.C. The influence of dietary palmitic acid triacylglyceride position on the fatty acid, calcium and magnesium contents of at term newborn faeces. Early Hum. Dev. 2001, 65, S83–S94. [Google Scholar] [CrossRef]
- Şahín, N.; Akoh, C.C.; Karaalí, A. Human milk fat substitutes containing omega-3 fatty acids. J. Agric. Food Chem. 2006, 54, 3717–3722. [Google Scholar] [CrossRef]
- Biswas, N.; Cheow, Y.L.; Tan, C.P.; Siow, L.F. Physicochemical Properties of Enzymatically Produced Palm-Oil-Based Cocoa Butter Substitute (CBS) With Cocoa Butter Mixture. Eur. J. Lipid Sci. Technol. 2018, 120, 1700205. [Google Scholar] [CrossRef]
- Yamoneka, J.; Malumba, P.; Lognay, G.; Béra, F.; Blecker, C.; Danthine, S. Enzymatic Inter-Esterification of Binary Blends Containing Irvingia gabonensis Seed Fat to Produce Cocoa Butter Substitute. Eur. J. Lipid Sci. Technol. 2018, 120, 1700423. [Google Scholar] [CrossRef]
- Lakum, R.; Sonwai, S. Production of trans-free margarine fat by enzymatic interesterification of soybean oil, palm stearin and coconut stearin blend. Int. J. Food Sci. Technol. 2018, 53, 2761–2769. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, J.; Xie, X.; Zhang, Z.; Zhang, N.; Wang, Y. A low trans margarine fat analog to beef tallow for healthier formulations: Optimization of enzymatic interesterification using soybean oil and fully hydrogenated palm oil. Food Chem. 2018, 255, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, A.; Shimada, Y.; Yamamoto, M.; Sugihara, A.; Nagao, T.; Komemushi, S.; Tominaga, Y. Enzymatic synthesis of high-purity structured lipids with caprylic acid at 1, 3-positions and polyunsaturated fatty acid at 2-position. JAOCS, J. Am. Oil Chem. Soc. 2001, 78, 611–616. [Google Scholar] [CrossRef]
- Lo, S.K.; Tan, C.P.; Long, K.; Yusoff, M.S.A.; Lai, O.M. Diacylglycerol oil-properties, processes and products: A review. Food Bioprocess Technol. 2008, 1, 223–233. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Yamane, T. Enzymatic synthesis of structured lipids. Adv. Biochem. Eng. Biotechnol. 2004, 10, 129–140. [Google Scholar]
- Kim, B.H.; Akoh, C.C. Recent Research Trends on the Enzymatic Synthesis of Structured Lipids. J. Food Sci. 2015, 80, C1713–C1724. [Google Scholar] [CrossRef]
- Utama, Q.D.; Sitanggang, A.B.; Adawiyah, D.R.; Hariyadi, P. Lipase-catalyzed interesterification for the synthesis of medium-long-medium (MLM) structured lipids - A review. Food Technol. Biotechnol. 2019, 57, 305–318. [Google Scholar] [CrossRef]
- Nunes, P.A.; Pires-Cabral, P.; Guillén, M.; Valero, F.; Luna, D.; Ferreira-Dias, S. Production of MLM-type structured lipids catalyzed by immobilized heterologous Rhizopus oryzae lipase. JAOCS, J. Am. Oil Chem. Soc. 2011, 64, 57–68. [Google Scholar] [CrossRef]
- Mota, D.A.; Rajan, D.; Heinzl, G.C.; Osório, N.M.; Gominho, J.; Krause, L.C.; Soares, C.M.F.; Nampoothiri, K.M.; Sukumaran, R.K.; Ferreira-Dias, S. Production of low-calorie structured lipids from spent coffee grounds or olive pomace crude oils catalyzed by immobilized lipase in magnetic nanoparticles. Bioresour. Technol. 2020, 307, 123223–123232. [Google Scholar] [CrossRef]
- Costa, C.M.; Osório, N.M.; Canet, A.; Rivera, I.; Sandoval, G.; Valero, F.; Ferreira-Dias, S. Production of MLM Type Structured Lipids From Grapeseed Oil Catalyzed by Non-Commercial Lipases. Eur. J. Lipid Sci. Technol. 2018, 120, 1700320–1700328. [Google Scholar] [CrossRef]
- Nagao, T.; Kawashima, A.; Sumida, M.; Watanabe, Y.; Akimoto, K.; Fukami, H.; Sugihara, A.; Shimada, Y. Production of Structured TAG Rich in 1,3-Capryloyl-2-arachidonoyl Glycerol from Mortierella Single-Cell Oil. JAOCS, J. Am. Oil Chem. Soc. 2003, 80, 867–872. [Google Scholar] [CrossRef]
- Nunes, P.A.; Pires-Cabral, P.; Guillén, M.; Valero, F.; Ferreira-Dias, S. Batch operational stability of immobilized heterologous Rhizopus oryzae lipase during acidolysis of virgin olive oil with medium-chain fatty acids. Biochem. Eng. J. 2012, 67, 265–268. [Google Scholar] [CrossRef]
- Nunes, P.A.; Pires-Cabral, P.; Guillén, M.; Valero, F.; Ferreira-Dias, S. Optimized production of MLM triacylglycerols catalyzed by immobilized heterologous Rhizopus oryzae lipase. JAOCS, J. Am. Oil Chem. Soc. 2012, 89, 1287–1295. [Google Scholar] [CrossRef]
- Balieiro, A.L.; Osório, N.M.; Lima, Á.S.; Soares, C.M.F.; Valero, F.; Ferreira-Dias, S. Production of dietetic triacylglycerols from olive oil catalyzed by immobilized heterologous Rhizopus oryzae lipase. Chem. Eng. Trans. 2018, 64, 385–390. [Google Scholar]
- Esteban, L.; Jiménez, M.J.; Hita, E.; González, P.A.; Martín, L.; Robles, A. Production of structured triacylglycerols rich in palmitic acid at sn-2 position and oleic acid at sn-1,3 positions as human milk fat substitutes by enzymatic acidolysis. Biochem. Eng. J. 2011, 54, 62–69. [Google Scholar] [CrossRef]
- Simões, T.; Valero, F.; Tecelão, C.; Ferreira-Dias, S. Production of human milk fat substitutes catalyzed by a heterologous Rhizopus oryzae lipase and commercial lipases. JAOCS, J. Am. Oil Chem. Soc. 2014, 91, 411–419. [Google Scholar] [CrossRef]
- Faustino, A.R.; Osório, N.M.; Tecelão, C.; Canet, A.; Valero, F.; Ferreira-Dias, S. Camelina oil as a source of polyunsaturated fatty acids for the production of human milk fat substitutes catalyzed by a heterologous Rhizopus oryzae lipase. Eur. J. Lipid Sci. Technol. 2016, 118, 532–544. [Google Scholar] [CrossRef]
- del Muñío, M.M.; Robles, A.; Esteban, L.; González, P.A.; Molina, E. Synthesis of structured lipids by two enzymatic steps: Ethanolysis of fish oils and esterification of 2-monoacylglycerols. Process Biochem. 2009, 44, 723–730. [Google Scholar]
- Hita, E.; Robles, A.; Camacho, B.; Ramírez, A.; Esteban, L.; Jiménez, M.J.; Muñío, M.M.; González, P.A.; Molina, E. Production of structured triacylglycerols (STAG) rich in docosahexaenoic acid (DHA) in position 2 by acidolysis of tuna oil catalyzed by lipases. Process Biochem. 2007, 42, 415–422. [Google Scholar] [CrossRef]
- Rodríguez, A.; Esteban, L.; Martín, L.; Jiménez, M.J.; Hita, E.; Castillo, B.; González, P.A.; Robles, A. Synthesis of 2-monoacylglycerols and structured triacylglycerols rich in polyunsaturated fatty acids by enzyme catalyzed reactions. Enzyme Microb. Technol. 2012, 51, 148–155. [Google Scholar] [CrossRef]
- Zhou, D.; Xu, X.; Mu, H.; HØY, C.E.; Adler-Nissen, J. Lipase-catalyzed production of structured lipids via acidolysis of fish oil with caprylic acid. J. Food Lipids 2000, 7, 263–274. [Google Scholar] [CrossRef]
- Paula, A. V.; Nunes, G.F.M.; De Castro, H.F.; Santos, J.C. Synthesis of structured lipids by enzymatic interesterification of milkfat and soybean oil in a basket-type stirred tank reactor. Ind. Eng. Chem. Res. 2015, 54, 1731–1737. [Google Scholar] [CrossRef]
- Ray, J.; Nagy, Z.K.; Smith, K.W.; Bhaggan, K.; Stapley, A.G.F. Kinetic study of the acidolysis of high oleic sunflower oil with stearic-palmitic acid mixtures catalysed by immobilised Rhizopus oryzae lipase. Biochem. Eng. J. 2013, 73, 17–28. [Google Scholar] [CrossRef]
- Yang, T.; Fruekilde, M.B.; Xu, X. Suppression of acyl migration in enzymatic production of structured lipids through temperature programming. Food Chem. 2005, 92, 101–107. [Google Scholar] [CrossRef]
- SÁ, A.G.A.; Meneses, A.C. de; Araújo, P.H.H. de; Oliveira, D. de A review on enzymatic synthesis of aromatic esters used as flavor ingredients for food, cosmetics and pharmaceuticals industries. Trends Food Sci. Technol. 2017, 69, 95–105. [Google Scholar] [CrossRef]
- Gao, W.; Wu, K.; Chen, L.; Fan, H.; Zhao, Z.; Gao, B.; Wang, H.; Wei, D. A novel esterase from a marine mud metagenomic library for biocatalytic synthesis of short-chain flavor esters. Microb. Cell Fact. 2016, 15, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.-M.; Huang, H.-Y.; Chen, Y.-M.; Kuo, C.-H.; Shieh, C.-J. Continuous Production of 2-Phenylethyl Acetate in a Solvent-Free System Using a Packed-Bed Reactor with Novozym® 435. Catalysts 2020, 10, 714–727. [Google Scholar] [CrossRef]
- Bayout, I.; Bouzemi, N.; Guo, N.; Mao, X.; Serra, S.; Riva, S.; Secundo, F. Natural flavor ester synthesis catalyzed by lipases. Flavour Fragr. J. 2020, 35, 209–218. [Google Scholar] [CrossRef]
- Asmat, S.; Husain, Q. A robust nanobiocatalyst based on high performance lipase immobilized to novel synthesised poly(o-toluidine) functionalized magnetic nanocomposite: Sterling stability and application. Mater. Sci. Eng. C 2019, 99, 25–36. [Google Scholar] [CrossRef]
- Asmat, S.; Anwer, A.H.; Husain, Q. Immobilization of lipase onto novel constructed polydopamine grafted multiwalled carbon nanotube impregnated with magnetic cobalt and its application in synthesis of fruit flavours. Int. J. Biol. Macromol. 2019, 140, 484–495. [Google Scholar] [CrossRef]
- Moreira, W.C.; Elias, A.L.P.; Osório, W.R.; Padilha, G.S. Alternative method to improve the ethyl valerate yield using an immobilised Burkholderia cepacia lipase. J. Microencapsul. 2019, 36, 327–337. [Google Scholar] [CrossRef]
- Rodriguez-Nogales, J.M.; Roura, E.; Contreras, E. Biosynthesis of ethyl butyrate using immobilized lipase: A statistical approach. Process Biochem. 2005, 40, 63–68. [Google Scholar] [CrossRef]
- Guillén, M.; Benaiges, M.D.; Valero, F. Biosynthesis of ethyl butyrate by immobilized recombinant Rhizopus oryzae lipase expressed in Pichia pastoris. Biochem. Eng. J. 2012, 65, 1–9. [Google Scholar] [CrossRef]
- Guillén, M.; Benaiges, M.D.; Valero, F. Improved ethyl butyrate synthesis catalyzed by an immobilized recombinant Rhizopus oryzae lipase: A comprehensive statistical study by production, reaction rate and yield analysis. J. Mol. Catal. B Enzym. 2016, 133, S371–S376. [Google Scholar] [CrossRef]
- Grosso, C.; Ferreira-Dias, S.; Pires-Cabral, P. Modelling and optimization of ethyl butyrate production catalysed by Rhizopus oryzae lipase. J. Food Eng. 2013, 115, 475–480. [Google Scholar] [CrossRef]
- Ben Salah, R.; Ghamghui, H.; Miled, N.; Mejdoub, H.; Gargouri, Y. Production of butyl acetate ester by lipase from novel strain of Rhizopus oryzae. J. Biosci. Bioeng. 2007, 103, 368–372. [Google Scholar] [CrossRef]
- Kumari, A.; Mahapatra, P.; Garlapati, V.K.; Banerjee, R.; Dasgupta, S. Lipase mediated isoamyl acetate synthesis in solvent-free system using vinyl acetate as acyl donor. Food Technol. Biotechnol. 2009, 47, 13–18. [Google Scholar]
- Ghamgui, H.; Karra-Chaâbouni, M.; Bezzine, S.; Miled, N.; Gargouri, Y. Production of isoamyl acetate with immobilized Staphylococcus simulans lipase in a solvent-free system. Enzyme Microb. Technol. 2006, 38, 788–794. [Google Scholar] [CrossRef]
- Garlapati, V.K.; Banerjee, R. Solvent-free synthesis of flavour esters through immobilized lipase mediated transesterification. Enzyme Res. 2013, 2013. [Google Scholar] [CrossRef] [Green Version]
- Dhake, K.P.; Deshmukh, K.M.; Patil, Y.P.; Singhal, R.S.; Bhanage, B.M. Improved activity and stability of Rhizopus oryzae lipase via immobilization for citronellol ester synthesis in supercritical carbon dioxide. J. Biotechnol. 2011, 156, 46–51. [Google Scholar] [CrossRef]
- Rios, N.S.; Pinheiro, B.B.; Pinheiro, M.P.; Bezerra, R.M.; dos Santos, J.C.S.; Barros Gonçalves, L.R. Biotechnological potential of lipases from Pseudomonas: Sources, properties and applications. Process Biochem. 2018, 75, 99–120. [Google Scholar] [CrossRef]
- Kirchner, G.; Scollar, M.P.; Klibanov, A.M. Resolution of Racemic Mixtures via Lipase Catalysis in Organic Solvents. J. Am. Chem. Soc. 1985, 107, 7072–7076. [Google Scholar] [CrossRef]
- Palomo, J.M.; Segura, R.L.; Fernandez-Lorente, G.; Guisán, J.M.; Fernandez-Lafuente, R. Enzymatic resolution of (±)-glycidyl butyrate in aqueous media. Strong modulation of the properties of the lipase from Rhizopus oryzae via immobilization techniques. Tetrahedron Asymmetry 2004, 15, 1157–1161. [Google Scholar] [CrossRef]
- Songür, R.; Lurçi, B.; Bayraktar, E.; Mehmetoǧlu, Ü.; Demir, A.S. Enantioselective production of benzoin from benzoin acetate via kinetic resolution and deracemization using Rhizopus oryzae. Artif. Cells Blood Substit. Biotechnol. 2011, 39, 162–168. [Google Scholar]
- Cabrera, Z.; Palomo, J.M. Enantioselective desymmetrization of prochiral diesters catalyzed by immobilized Rhizopus oryzae lipase. Tetrahedron Asymmetry 2011, 22, 2080–2084. [Google Scholar] [CrossRef]
- Yousefi, M.; Mohammadi, M.; Habibi, Z. Enantioselective resolution of racemic ibuprofen esters using different lipases immobilized on octyl sepharose. J. Mol. Catal. B Enzym. 2014, 104, 87–94. [Google Scholar] [CrossRef]
- Matsumoto, T.; Ito, M.; Fukuda, H.; Kondo, A. Enantioselective transesterification using lipase-displaying yeast whole-cell biocatalyst. Appl. Microbiol. Biotechnol. 2004, 64, 481–485. [Google Scholar] [CrossRef]
- Nakamura, Y.; Matsumoto, T.; Nomoto, F.; Ueda, M.; Fukuda, H.; Kondo, A. Enhancement of activity of lipase-displaying yeast cells and their application to optical resolution of (R,S)-1-benzyloxy-3-chloro-2-propyl monosuccinate. Biotechnol. Prog. 2006, 22, 998–1002. [Google Scholar] [CrossRef]




| Supplier | Name | Application | Lipase Properties |
|---|---|---|---|
| Amano | Lipase DF “Amano” 15 | Oil and fats | Optimum pH range 6–7; stable pH range 4–7, optimum temperature range 35–40 °C, relatively specific to fatty acids |
| Sigma | Lipase from R. oryzae (no. 62305) | Oil and fats | Optimum pH 8, optimum temperature 40 °C |
| Sigma | Lipase, immobilised on Immobead 150 from R. oryzae (no. 89445) | Pharmaceutical and bioenergy | Optimum pH 7.5, optimum temperature 40 °C |
| Lipase Name 1 | MW (kDa) | Isoelectric Point | pH Optimum | T Optimum (°C) | Substrate Specificity | Ref. |
|---|---|---|---|---|---|---|
| rROL | 29 | 8/7.25 2 | 30/40 2 | C12 > C10 > C8 > C4 4 | [39] | |
| proROL | 32 | 7.25 | 40 | C8 > C12 > C10 > C4 4 | [39] | |
| rROL | 30 | 8.5 | [56] | |||
| entire-proROL | 40 | 8 | [56] | |||
| pre-entire-proROL 3 | 42 | 8 | [56] | |||
| rROL | 29 | 8 | 37 | [57] | ||
| rROL | 29 | [58] | ||||
| proROL | 32 | [58] | ||||
| proROL | 34 | 6–6.5 | 35 | [60] | ||
| rROL | 30 | 6 | 40 | [60] | ||
| proROL | 35 | 9 | 40 | C16 > C18 > C12 > C8 > C4 5 C16 > C12 > C8 > C18 > C4 6 | [68] | |
| proROL | 32 | 6.9 | [70] | |||
| rROL | 30 | 9.3 | 8.25 | 30 | C8 > C10 > C6 > C4 > C12 > C16,C14 > C2 6 | [73] |
| proROL | 35 | 5.2 | 30 | C12 > C10 > C8 > C6 > C16 > C5 > C4 > C3 > C2 4 | [74] | |
| proROL | 32 | 7.6 | 7.5 | 35 | C8 > C6 > C4 > C2 6 | [75] |
| rROL | 29 | C12 > C10 > C8 > C6 > C4 > C3 > C2 4 C8 > C10 > C18 > C4 > C6 6 | [90] | |||
| proROL | 34 | C2 > C3 > C8 > C6 > C12 > C10 > C4 4 C8 > C10 > C4 > C6 > C18 6 | [90] | |||
| proROL | 8 | 40 | [92] | |||
| rROL | 30.3 | 8.6 | 8–8.5 | 30 | [93] | |
| proROL | 8.5 | 30 | [94] | |||
| proROL | 37 | 8.5 | 40 | [95] | ||
| rROL | 29 | 8 | [96] | |||
| ROL | 17 | 4.2 | 7 | 40 | [97] | |
| ROL | 7 | 40 | [98] | |||
| ROL | 6 | 45 | C8 > C4 > C6 > C2 6 C8 > C12 > C14 > C16 > C18 5 | [99] | ||
| proROL | 32 | 7 | 35 | [100] | ||
| ROL | 6 | 30 | C7,C8,C12,C16 > C2,C3,C4,C18 5 | [101] | ||
| ROL | 7.5 | 50 | [102] | |||
| proROL | 32 | 7.5 | 30–40 | [103] | ||
| ROL | 14.45 | 6.5 | 9 | 30–40 | C16 > C18 > C12 > C8 > C4 > C2 4 | [104] |
| 8.3 | 35–37 | [105] | ||||
| proROL | 35 | C10 > C14 > C12 > C8 > C6 > C4 > C16 6 | [65] | |||
| entire-proROL | 46 | [65] |
| Cell Factory | Promotor/Vector | Lipase | Production | Lipolytic Activity | Reference |
|---|---|---|---|---|---|
| E. coli Origami DE3 | pET11 | proROL | Intracellular | 166 U mL−1 | [121] |
| pET22 | proROL | Intracellular | 82 U mL−1 | ||
| S. cerevisiae | UPR-ICL | rROL | Extracellular | 0.29 U flask−1 | [65] |
| UPR-ICL | proROL | Extracellular | 191 U flask−1 |
| Gene | Prom. | Co-expr. Δ Delet. | Phenot. | Operational Mode | Pmax AU mL−1 | Μ h−1 | qp AU gX−1 h−1 | YP/X AU gX−1 | Vol. Prod. AU L−1 h−1 | Spec. Vol. Prod. AU gX−1 h−1 | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rROL | AOX | Mut+ SC | Fed-batch MeOH added when CO2 decrease | 500 | 5435 | [73] | |||||
| rROL | AOX | Mut+ SC | Fed-batch MeOH added manual control | 1334 | 12,888 | 268 | [128] | ||||
| rROL | AOX | Mut+ SC | Fed-batch MeOH added manual control 2 g L−1 | 150 | 0.036 | 130 | 2470 | 3000 | 49 | [129] | |
| rROL | AOX | Mut+ MC | Fed-batch MeOH added manual control 2 g L−1 | 175 | 0.012 | 71 | 3578 | 1894 | 40 | [129] | |
| rROL | AOX | MutS His+ SC | Fed-batch MeOH added manual control 2 g L−1 | 205 | 0.005 | 83 | 5775 | 2246 | 63 | [129] | |
| rROL | AOX | MutS His- MC | Fed-batch MeOH added manual control 2 g L−1 | 270 | 0.004 | 59 | 7500 | 1500 | 46 | [129] | |
| rROL | AOX | Mut+ SC | Fed-batch MeOH controlled 3 g L−1 MNLFB + MLFB to maintain 25% DO | 644 | 0.03 | 277 | 7800 | 8110 | 98 | [130] | |
| rROL | AOX | Mut+ SC | Fed-batch Temperature limited | 534 | 0.13 | 130 | 5830 | 5200 | 57 | [130] | |
| rROL | AOX | Mut+ SC | Fed-batch MeOH controlled 3 g L−1 MNLFB + temperature limited | 713 | 0.02 | 161 | 6960 | 5980 | 58 | [130] | |
| rROL | AOX | Mut+ SC | Fed-batch MeOH controlled 3 g L−1 MNLFB | 280 | 0.046 | 322 | 5282 | 5406 | 102 | [131] | |
| rROL | AOX | Mut+ SC | Fed-batch MNLFB µ = 0.015 h−1 | 135 | 0.014 | 46 | 2644 | 1857 | 36 | [131] | |
| rROL | AOX | Mut+ SC | Fed-batch MeOH controlled 3 g L−1 MNLFB DO 25% | 368 | 0.034 | 256 | 7700 | 5490 | [132] | ||
| rROL | AOX | Mut+ SC | Continuous MeOH controlled 3 g L−1 | 0.02 | 0.91 | 45.7 | 1.6 | [133] | |||
| rROL | AOX | Mut+ SC | Continuous MeOH controlled 3 g L−1 | 0.05 | 0.98 | 19.6 | 2.7 | [133] | |||
| rROL | AOX | MutS SC | Fed-batch MeOH controlled 1 g L−1 | 490 | 0.004 | 148 | 11,236 | 4901 | 112 | [134] | |
| rROL | AOX | MutS SC | Fed-batch MeOH controlled 2 g L−1 Sorbitol exponential feeding rate 0.02 h−1 | 488 | 0.02 | 326 | 10,369 | 6346 | 135 | [135] | |
| rROL | AOX | MutS SC | Fed-batch MeOH controlled 2 g L−1 Glycerol exponential feeding rate 0.02 h−1 | 471 | 0.025 | 149 | 6373 | 5416 | 73 | [136] | |
| rROL | FLD1 | Mut+ SC | Fed-batch Manual SNLFB 8 g L−1 | 385 | 0.02 | 244 | 7634 | 4379 | 87 | [137] | |
| rROL | FLD1 | GAP HAC1 S. cerevisiae | Mut+ SC | Fed-batch MLFB µ = 0.005 h−1 | 73 | 0.005 | 52 | 3630 | 1147 | 57 | [138] |
| rROL | FLD1 | Δ GAS1 | Mut+ SC | Fed-batch MLFB µ = 0.005 h−1 | 240 | 0.006 | 145 | 7172 | 2790 | 82 | [138] |
| rROL | FLD1 | Δ GAS1 GAP HAC1 S. cerevisiae | Mut+ SC | Fed-batch MLFB µ = 0.005 h−1 | 206 | 0.005 | 206 | 13,186 | 3274 | 144 | [138] |
| proROL | AOX | Mut+ SC | Fed-batch Methanol controlled at 0.1 v/v | 12,019 | [139] | ||||||
| proROL | AOX | Mut+ MC | Fed-batch Methanol controlled at 3 g L−1 | 140 | 4375 | 1521 | [67] | ||||
| rROL | AOX | Mut+ MC | Fed-batch Methanol controlled at 3 g L−1 | 195 | 5417 | 2130 | [67] | ||||
| proROL | AOX | Mut+ MC | Fed-batch Methanol-sorbitol to maintain DO 20–50% | 20,500 | 175,213 | 178,261 | 1524 | [140] | |||
| proROL | AOX | Ubc1 | Mut+ MC | Fed-batch Methanol-sorbitol to maintain DO 20–50% | 28,600 | 223,883 | 223,833 | 1865 | [140] | ||
| proROL | AOX | Hrd1 | Mut+ MC | Fed-batch Methanol-sorbitol to maintain DO 20–50% | 29,600 | 229,922 | 233,543 | 1810 | [140] | ||
| proROL | AOX | Ubc1 + Hrd1 | Mut+ MC | Fed-batch Methanol-sorbitol to maintain DO 20–50% | 33,900 | 280,165 | 266,929 | 2206 | [140] | ||
| proROL | AOX | Ssa4 + Sso2 + Bmh2 | Mut+ MC | Fed-batch Methanol-sorbitol to maintain DO 20–50% | 36,578 | 302,000 | 290,000 | 2206 | [141] | ||
| proROL | AOX | Ssa4 + Sso2 + Bmh2 + Vhb9 | Mut+ MC | Fed-batch Methanol-sorbitol to maintain DO 20–50% | 41,700 | 345,000 | 331,000 | 2206 | [141] | ||
| proROL | AOX | Mut+ | Fed-batch Methanol to maintain DO over 20% | 21,000 | 107,000 | 125,000 | [68] | ||||
| proROL | GAP | Mut+ SC | Fed-batch Constant feeding rate to maintain DO 20–40% | 11 | 131 | 1 | [62] | ||||
| proROL | GAP | Glycosylation Mutant A + B | Mut+ SC | Fed-batch Constant feeding rate to maintain DO 20–40% | 2600 | 30,952 | 221 | [62] | |||
| 28proROL | AOX | Mut+ SC | Fed-batch MNLFB µ = 0.015 h−1 | 219 | 0.011 | 57 | 5264 | 2763 | 49 | [39] | |
| 28proROL | AOX | Mut+ SC | Fed-batch MNLFB µ = 0.045 h−1 | 147 | 0.038 | 69 | 1908 | 2782 | 44 | [39] | |
| 28proROL | AOX | Mut+ SC | Fed-batch MNLFB 3 g L−1 | 358 | 0.065 | 308 | 4972 | 7160 | 99 | [39] |
| Substrates | Lipase | Immobilisation Technique | Reactor Type | Stepwise Addition | Biodiesel Generation | Yield-Conversion/Operational Stability | Ref. |
|---|---|---|---|---|---|---|---|
| OO + MeOH | rROL | IA onto ReliZymeTM OD 403M | PBR | Yes | 1st | Y: PBR 49.1% OS: second batch 44.8% | [185] |
| OO + MeOH | rROL | IA onto ReliZymeTM OD 403M | STR | Yes | 1st | Y: STR 33.56% OS: second batch 7.7% | [185] |
| RO + MeOH | proROL | WCB over agar plate | SLLB | No | 1st | No biodiesel production | [186] |
| RO + EtOH | proROL | WCB over agar plate | SLLB | No | 1st | No biodiesel production | [186] |
| RO + MeOH | proROL | WCB over agar plate | SGLB | No | 1st | Y: 58% | [186] |
| RO + EtOH | proROL | WCB over agar plate | SGLB | No | 1st | Y: 72% | [186] |
| Crude CO + MeOH | proROL | Free enzymes | BR | Yes | 1st | Y: 68.56% | [187] |
| Crude CO + MeOH | proROL-CRL | Free enzymes | BR | Yes | 1st | Y: 84.25% | [187] |
| Crude CO + MeOH | proROL-CRL | CI onto functionalised silica gel | BR | Yes | 1st | Y: 88.9% | [187] |
| SYO + MeOH | proROL | WCB immobilised into BSPs | BR | Yes | 1st | Y: 82.2% OS: after 6 cycles almost all activity loss | [188] |
| SYO + MeOH | proROL | CI WCB immobilised onto BSPs | BR | Yes | 1st | Y: 92.2% OS: after 6 cycles no loss of activity | [188] |
| SYO + EtOH | proROL | IA onto microporous resin NKA (polystyrene) | BR | Yes | 1st | Y: 58.5% | [189] |
| SYO + EtOH | proROL-CRL | IA onto microporous resin NKA (polystyrene) | BR | Yes | 1st | Y: 80.8% | [189] |
| SYO + EtOH | proROL-Novozyme 435 | proROL: IA onto microporous resin NKA (polystyrene). Novozyme 435: IA onto Lewatit VP OC 1600 | BR | Yes | 1st | Y: 98.5% OS: after 20 cycles Y decreased to 78.3% | [189] |
| SYO + EtOH | proROL-PFL | IA onto microporous resin NKA (polystyrene) | BR | Yes | 1st | Y: 55.8% | [189] |
| SYO + MeOH | proROL | CI onto magnetic chitosan microspheres | MSFBR | Yes | 1st | Y: 91.3% OS: after 6 reaction cycles Y decreased to around 80% | [190] |
| SYO + MeoH | proROL | WCB immobilised into BSPs | BR | Yes | 1st | Y: over 90% OS: after 10 reaction cycles Y decreased to 10% | [191] |
| SYO + MeoH | proROL | WCB immobilised into BSPs | PBR | Yes | 1st | Y: over 90% OS: after 10 reaction cycles Y decreased to 80% | [191] |
| SO + EtOH | proROL | CI onto modified sepiolite with p-hydroxybenzaldehyde linker | BR | No | 1st | C: 84.3% OS: after 9 cycles C decreased to 21.4% | [192] |
| SO + EtOH | proROL | CI onto modified sepiolite with benzylamine-terephthalic aldehyde linker | BR | No | 1st | [192] | |
| SO + EtOH | proROL | IE onto demineralised sepiolite | BR | No | 1st | Y: 90.2% OS: proROL IE after 9 cycles C decreased to 18.1% | [192] |
| Pistacia chinensis bge seed oil + MeOH | rROL | CI onto Amberlite IRA-93 | BR | Yes | 2nd | Y: 92% OS: after 8 cycles Y decreased to 60% | [196] |
| Pistacia chinensis bge seed oil + MeOH | rROL | IA microporous resin HPD-400 | BR | Yes | 2nd | Y: 94% OS: after 8 cycles Y decreased to 50% | [196] |
| Calophyllum inophyllum linn oil + MeOH | proROL | WCB immobilised into BSPs | PBR | Yes | 2nd | Y: 92% OS: after 6 cycles Y decreased a 4.9% | [197] |
| Oil extracted from Nannochloropsis gaditana + MeOH | proROL | WCB | BR | Yes | 3rd | Y: 83% OS: after 3 cycles Y decreased to 71% | [198] |
| Oil extracted from Nannochloropsis gaditana + MeOH | proROL | WCB immobilised into BSPs | BR | Yes | 3rd | Y: 70% OS: second cycle Y decreased to 43% | [198] |
| Oil extracted from Nannochloropsis gaditana + MeOH | proROL | WCB immobilised into BSPs | BR | Yes | 3rd | Y: 83% OS: after 3 cycles Y decreased to 71% | [199] |
| Oil extracted from Nannochloropsis gaditana + MeOH | proROL | WCB | TPB | No | 3rd | Y: 58% | [200] |
| Oil extracted from Nannochloropsis gaditana + EtOH | proROL | WCB | TPB | No | 3rd | Y: 92% | [200] |
| Oil extracted from Botryococcus braunii + MeOH | proROL | WCB | TPB | No | 3rd | Y: 58% | [200] |
| Oil extracted from Botryococcus braunii + EtOH | proROL | WCB | TPB | No | 3rd | Y: 68% | [200] |
| Oil extracted from Chlorella vulgaris + MeOH | proROL | Free enzyme | BR | Yes | 3rd | C: 75% | [201] |
| Oil extracted from Chlorella vulgaris + MeOH | proROL | IA onto MNP | BR | Yes | 3rd | C: 46% OS: after 5 cycles decreased to 10% | [201] |
| Oil extracted from Chlorella vulgaris + MeOH | proROL | CI onto AP modified MNP | BR | Yes | 3rd | C: 53% OS: after 5 cycles C decreased to 25% | [201] |
| Oil extracted from Chlorella vulgaris + MeOH | proROL | CI onto AP-GA modified MNP | BR | Yes | 3rd | C: 69.8% OS: after 5 cycles C decreased to 45% | [201] |
| Sludge palm oil + MeOH | proROL | IE into alginate-polyvinyl alcohol beads | BR | No | 3rd | Y: 91.30% OS: no activity loss after 15 cycles | [202] |
| Oil extracted from SCG + MeOH | R. delemar (= oryzae) lipase | Free enzyme | BR | No | 3rd | Y: 18% | [203] |
| WCO + MeOH | proROL | Free enzyme | BR | 3rd | Y: 93% | [204] | |
| WCO + iso-propanol | proROL | Free enzyme | BR | 3rd | Y: 86.8% | [204] | |
| WCO + iso-butanol | proROL | Free enzyme | BR | 3rd | Y: 80.2% | [204] | |
| WCO + iso-amyl alcohol | proROL | Free enzyme | BR | 3rd | Y: 64% | [204] | |
| WCO + MeOH | proROL | WCB IE into calcium alginate beads | BR | 3rd | Y: 84% | [204] | |
| WCO + iso-propanol | proROL | WCB IE into calcium alginate beads | BR | 3rd | Y: 71% | [204] | |
| WCO + iso-butanol | proROL | WCB IE into calcium alginate beads | BR | 3rd | Y: 62% | [204] | |
| WCO+ iso-amyl alcohol | proROL | WCB IE into calcium alginate beads | BR | 3rd | Y: 43% | [204] | |
| JO + MeOH | proROL | WCB IE into sodium alginate beads | BR | No | 2nd | Y: 80.5% OS: after 6 cycles Y decreased to 61.5% | [205] |
| KO + MeOH | proROL | WCB IE into sodium alginate beads | BR | No | 2nd | Y: 78.3% OS: after 6 cycles Y decreased to 63.4% | [205] |
| SYO + MeOH | proROL | WCB | BR | Yes | 1st | Y: 80% OS: after 3 cycles Y decreased to 18% | [206] |
| SYO + MeOH | proROL | WCB immobilised into BSPs | BR | Yes | 1st | Y: 82% OS: after 10 cycles Y decreased to 10% | [206] |
| SYO + MeOH | proROL | CI WCB immobilised into BSPs | BR | Yes | 1st | Y: 74% OS: after 35 cycles Y decreased to 65% | [206] |
| SYO + MeOH | proROL | WCB immobilised into BSPs | BR | Yes | 1st | Y: 82% OS: after 6 cycles Y decreased to 48% | [207] |
| SYO + MeOH | proROL | CI WCB immobilised into BSPs | BR | Yes | 1st | Y: 80% OS: after 6 cycles Y decreased to 70% | [207] |
| ALO + MeOH | rROL | IA onto rice husk | BR | Yes | 2nd | [208] | |
| ALO + MeOH | rROL | IA onto ReliZymeTM OD403 | BR | Yes | 2nd | Y: 64.5% OS: after 7 cycles Y decreased to 41.3% | [208] |
| Crude microbial oil from Candida sp. LEB-M3 + MeOH | rROL | IA onto ReliZymeTM OD403 | BR | Yes | 3rd | Y: 38% OS: after 7 cycles Y decreased to 26.6% | [209] |
| Neutralised microbial oil from Candida sp. LEB-M3 + MeOH | rROL | IA onto ReliZymeTM OD403 | BR | Yes | 3rd | Y: 38% | [209] |
| OO + MeOH | rROL | IA onto ReliZymeTM OD403 | BR | Yes | 1st | Y: 54.3% OS: after 7 cycles Y decreased to 40% | [209] |
| OA + MeOH | rROL | IA onto ReliZymeTM OD403 | BR | Yes | 1st | Y: 68% | [209] |
| RO + EtOH | proROL | IA onto microporous resin NKA | BR | No | 1st | Y: above 98% OS: After 10 cycles Y decreased to 60% | [210] |
| JO + MeOH | proROL-CRL | WCB (proROL) and free enzyme (CRL) IE into sodium alginate beads | PBR | No | 2nd | Y: 84.2% | [211] |
| KO + MeOH | proROL-CRL | WCB (proROL) and free enzyme (CRL) IE into sodium alginate beads | PBR | No | 2nd | Y: 81% | [211] |
| WCO + MeOH | proROL | WCB IE into sodium alginate beads | BR | No | 3rd | Y: 94.01% | [212] |
| WCO + Methyl acetate | proROL | WCB IE into sodium alginate beads | BR | No | 3rd | Y: 91.11% | [212] |
| WCO + Ethyl acetate | proROL | WCB IE into sodium alginate beads | BR | No | 3rd | Y: 90.06 | [212] |
| WCO + MeOH | proROL | IE into sodium alginate beads | BR | No | 3rd | Y: 83% | [212] |
| WCO + Methyl acetate | proROL | IE into sodium alginate beads | BR | No | 3rd | Y: 80% | [212] |
| WCO + Ethyl acetate | proROL | IE into sodium alginate beads | BR | No | 3rd | Y: 78% | [212] |
| Oil extracted from Chlorella vulgaris + MeOH | proROL | IA into MNP | BR | Yes | 3rd | Y: 45% OS: after 5 cycles Y decreased to 10% | [213] |
| Oil extracted from Chlorella vulgaris + MeOH | proROL | IA into MGO | BR | Yes | 3rd | Y: 51% OS: after 5 cycles Y decreased to 16% | [213] |
| Oil extracted from Chlorella vulgaris + MeOH | proROL | IA into MGO-AP | BR | Yes | 3rd | Y: 54% OS: after 5 cycles Y decreased to 25% | [213] |
| Oil extracted from Chlorella vulgaris + MeOH | proROL | CI into MGO-AP-GA | BR | Yes | 3rd | Y: 68% OS: after 5 cycles Y decreased to 58.77% | [213] |
| Cottonseed oil + MeOH | proROL | WCB immobilised into BSPs | BR | Yes | 1st | Y: 27.9% | [214] |
| Rubber seed oil + MeOHe | proROL | Free enzyme | BR | Yes | 2nd | Y: 31% | [215] |
| Rubber seed oil + Ethyl acetate | proROL | Free enzyme | BR | No | 2nd | Y: 33.3% | |
| SYO + MeOH | proROL-CRL | CI onto silica gel pretreated with AP and GA | BR | Yes | 1st | Y: 99.99% OS: after 20 cycles Y decreased to 85% | [216] |
| RO deodoriser distillate + MeOH | proROL | Free enzyme | BR | Yes | 1st | Y: 93.07% | [217] |
| RO deodoriser distillate + MeOH | proROL-CRL | Free enzyme | BR | Yes | 1st | Y: 98.16% | [217] |
| ALO + MeOH | rROL | CI onto ET, AP and GA pretreated ReliZymeTM HFA403 | BR | Yes | 2nd | Y: 57.16% OS: after 5 cycles Y decreased a 12.31% | [181] |
| ALO + EtOH | rROL | CI onto ET, AP and GA pretreated ReliZymeTM HFA403 | BR | Yes | 2nd | Y: 60.25% OS: after 7 cycles Y decreased a 11.89% | [181] |
| Triolein + MeOH | rROL | Free enzyme | BR | No | 1st | Y: 71.2% | [117] |
| Triolein + EtOH | rROL | Free enzyme | BR | No | 1st | Y: 64.2% | [117] |
| Triolein + MeOH | rROL | IA onto RelyZymeTM OD403S | BR | No | 1st | Y: 82.6% | [117] |
| Triolein + EtOH | rROL | IA onto RelyZymeTM OD403S | BR | No | 1st | Y:100.7% | [117] |
| JO + MeOH | rROL | IA onto Lewatit VP OC 1600 | BR | Yes | 2nd | Y: 61% OS: after 10 cycles Y decreased a 40% | [218] |
| JO + MeOH | rROL | IA onto LifetechTM ECR1030M | BR | Yes | 2nd | Y: 63% OS: after 10 cycles Y decreased a 40% | [218] |
| JO + MeOH | rROL | IA onto LifetechTM AP1090M | BR | Yes | 2nd | Y: 55% OS: after 10 cycles Y decreased a 25% | [218] |
| JO + MeOH | rROL | CI onto LifetechTM ECR8285M | BR | Yes | 2nd | Y: 63% OS: after 10 cycles Y decreased a 60% | [218] |
| JO + MeOH | rROL | CI onto Amberlita IRA 96 | BR | Yes | 2nd | Y: 68% OS: after 10 cycles Y decreased a 20% | [218] |
| OO + MeOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 77% | [219] |
| OO + EtOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 62% | [219] |
| OO + Propanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 46% | [219] |
| OO + Butanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 18% | [219] |
| SYO + MeOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 50% | [219] |
| SYO + EtOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 46% | [219] |
| SYO + Propanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 35% | [219] |
| SYO + Butanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 10% | [219] |
| CO + MeOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 70% | [219] |
| CO + EtOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 56% | [219] |
| CO + Propanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 43% | [219] |
| CO + Butanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 16% | [219] |
| SO + MeOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 32% | [219] |
| SO + EtOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 28% | [219] |
| SO + Propanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 17% | [219] |
| SO + Butanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 1st | Y: 7% | [219] |
| Algal oil + MeOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 3rd | Y: 63% | [219] |
| Algal oil + EtOH | prorROL | IA onto Amberlite XAD 761 | BR | No | 3rd | Y: 55% | [219] |
| Algal oil + Propanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 3rd | Y: 40% | [219] |
| Algal oil + Butanol | prorROL | IA onto Amberlite XAD 761 | BR | No | 3rd | Y: 13% | [219] |
| ALO + MeOH | rROL | CI onto AP and GA treated ReliZymeTM HFA403 | BR | Yes | 2nd | Y: 28.62% OS: after 9 cycles, Y decreased a 43% | [170] |
| JO + MeOH | proROL | WCB immobilised into BSPs | BR | Yes | 2nd | Y: 88.6% OS: after 6 cycles Y decreased a 21% | [220] |
| OA + MeOH | proROL | WCB immobilised into BSPs | BR | No | 1st | Y: 80% OS: after 8 cycles, almost no activity loss. | [221] |
| Rice bran oil + MeOH | proROL | IA onto rod-like mesoporous silica | BR | No | 1st | Y: 81.7% OS: after 3 cycles Y decreased to 67.7% | [222] |
| JO + MeOH | proROL | IE into polyvinyl alcohol—alginate matrix | BR | No | 2nd | Yield: 87.1% | [223] |
| ALO + MeOH | rROL | IA Octadecyl-Sepabeads | BR | Yes | 2nd | Y: 58.31% OS: after 2 cycles Y decreased to 54.67% | [224] |
| Tung oil + MeOH | proROL | CI onto Amberlite IRA 93 | BR | Yes | 2nd | Y: 91.9% OS: after 6 cycles Y decreased to 85.1% | [95] |
| Babassu oil + EtOH | proROL | WCB immobilised into BSPs | BR | No | 1st | Y: 74.15% | [225] |
| SL Type | Definition | Properties | Ref. |
|---|---|---|---|
| Low caloric and dietetic TAGs |
|
| [246,247,248,249] |
| Human milk fat substitutes (HMFS) |
|
| [246,247,250,251] |
| Cocoa butter equivalents (CBE) |
|
| [246,247,252,253] |
| Trans-free plastic fats |
|
| [247,254,255] |
| TAGs rich in specific long-chain and polyunsaturated fatty acids (PUFAs) |
|
| [246,256] |
| MAGs and DAGs |
|
| [182,184,237,257] |
| Product | Substrates | Reaction Type | Lipase | Immobilisation Technique | ID/OS | Ref. |
|---|---|---|---|---|---|---|
| MLM | OO + CRA | Acidolysis | proROL/rROL | CI onto Eupergit®C/sepiolite (AlPO4-sepiolite) | ID: 21.6%. OS: half-life 159 h | [261] |
| MLM | OO + CA | Acidolysis | proROL/rROL | CI onto Eupergit®C/sepiolite (AlPO4-sepiolite) | ID: 34.82%. OS: half-life 136 h | [261] |
| MLM | SCG + CA | Acidolysis | proROL | CI onto GA treated MNP | ID: 50% | [262] |
| MLM | SCG + ethyl caprate | Interesterification | proROL | CI onto GA treated MNP | ID: 26% | [262] |
| MLM | OP + CA | Acidolysis | proROL | CI onto GA treated MNP | ID: 51% OS: 6.8 batches | [262] |
| MLM | OP + ethyl caprate | Interesterification | proROL | CI onto GA treated MNP | ID: 46%. OS: 9.1 batches | [262] |
| MLM | Grapeseed oil + CRA | Acidolysis | rROL | CI onto Amberlite IRA 96 | ID: 54%. OS: half-life 166 h | [263] |
| MLM | Grapeseed oil + CA | Acidolysis | rROL | CI onto Amberlite IRA 96 | ID: 69% OS: half-life 118 h | [263] |
| MLM | TGA58F + CA | Acidolysis | proROL | IA onto Dowex WBA | ID: 64.6% | [264] |
| MLM | TGA40 + CA | Acidolysis | proROL | IA onto Dowex WBA | ID: 62.8% | [264] |
| MLM | TGA55E + CA | Acidolysis | proROL | IA onto Dowex WBA | ID: 64.8% OS: 90 days in PBR1 dropped 10% | [264] |
| MLM | OO + CRA | Acidolysis | rROL | CI onto Eupergit® C/IA onto Lewatit VP OC 1600 | OS: half time 2.4 batches (54.3 h) with Eupergit®C | [265] |
| MLM | OO + CA | Acidolysis | rROL | CI onto Eupergit® C/IA onto Lewatit VP OC 1600 | OS: half time 10.2 batches (234 h) with Lewatit VP OC 1600 | [265] |
| MLM | OO + CRA | Acidolysis | rROL | CI onto Eupergit® C | ID: 15.5% | [266] |
| MLM | OO + CA | Acidolysis | rROL | CI onto Eupergit® C | ID: 33.3% | [266] |
| MLM | OO + CRA | Acidolysis | rROL | CI onto Amberlite IRA 96 | ID: 76.9 | [267] |
| MLM | OO + CA | Acidolysis | rROL | CI onto Amberlite IRA 96 | ID: 85.6% | [267] |
| HMFS | PA enriched TAGs + OA enriched mixtures | Acidolysis | proROL | IA onto Accurel® MP-1000 | ID: OA in sn-1,3 67.2% - PA in sn-2 67.8%. OS: no activity loss in 10 uses (190 h) | [268] |
| HMFS | Lard + FFA from EPAX 1050TG | Acidolysis | rROL | CI onto Accurel® MP-1000 | ID: 24 mol%. OS: after 4 batches, 55% of original activity | [269] |
| HMFS | Tripalmitin + FFA from camelina oil | Acidolysis | rROL | AI onto RelizymeTM OD403/S/CI onto Lewatit VP OC 1600 | ID: 52% | [270] |
| TAGs rich in PUFAs | cod liver + tuna oil + ethanol. | Alcoholysis | proROL | IA onto Accurel® MP-1000 | Alcoholysis ID: 72% OS: after 6 cycles, complete deactivation. | [271] |
| 2-MAG from alcoholysis + CRA | Esterification | proROL | IA onto Accurel® MP-1000 | ID: 95%. OS: after 5 cycles, no activity loss. | [271] | |
| TAGs rich in PUFAs | Tuna oil + CRA | Acidolysis | proROL | IA onto Accurel® MP-1000 | OS: over one week | [272] |
| TAGs rich in PUFAs | cod liver oil + ethanol 96% | Alcoholysis | proROL | IA onto Accurel® MP-1000 | Alcoholysis Y: 78%. OS: after 3 cycles, a 57% decrease | [273] |
| cod liver oil + 1-butanol | Alcoholysis | proROL | IA onto Accurel® MP-1000 | Alcoholysis Y: 78%. OS: after 3 cycles, no activity decrease | [273] | |
| Esterification: 2-MAG from alcoholysis + CRA | Esterification | proROL | IA onto Accurel® MP-1000 | Esterification Y: 71%. | [273] | |
| TAGs rich in PUFAs | Fish oil + CRA | Acidolysis | proROL | Non-immobilised | ID: 2.5% | [274] |
| HMFS | Milkfat + SYO | Interesterification | proROL | EI into polysiloxane-PVA | ID: 8.14%. OS: after 10 batches, no activity loss | [275] |
| CBE | SO + SA-PA mixtures | Acidolysis | proROL | IA onto Accurel® MP-1000 | [276] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Fernández, J.; Benaiges, M.D.; Valero, F. Rhizopus oryzae Lipase, a Promising Industrial Enzyme: Biochemical Characteristics, Production and Biocatalytic Applications. Catalysts 2020, 10, 1277. https://doi.org/10.3390/catal10111277
López-Fernández J, Benaiges MD, Valero F. Rhizopus oryzae Lipase, a Promising Industrial Enzyme: Biochemical Characteristics, Production and Biocatalytic Applications. Catalysts. 2020; 10(11):1277. https://doi.org/10.3390/catal10111277
Chicago/Turabian StyleLópez-Fernández, Josu, M. Dolors Benaiges, and Francisco Valero. 2020. "Rhizopus oryzae Lipase, a Promising Industrial Enzyme: Biochemical Characteristics, Production and Biocatalytic Applications" Catalysts 10, no. 11: 1277. https://doi.org/10.3390/catal10111277
APA StyleLópez-Fernández, J., Benaiges, M. D., & Valero, F. (2020). Rhizopus oryzae Lipase, a Promising Industrial Enzyme: Biochemical Characteristics, Production and Biocatalytic Applications. Catalysts, 10(11), 1277. https://doi.org/10.3390/catal10111277






