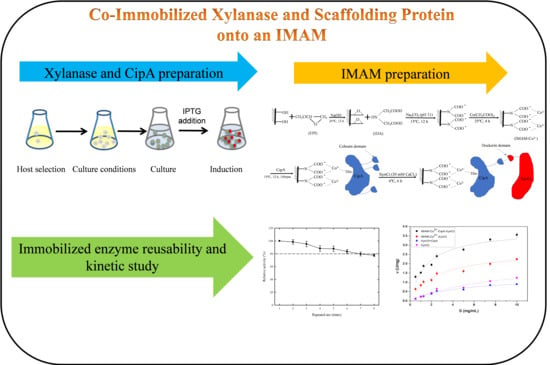
Co-Immobilization of Xylanase and Scaffolding Protein onto an Immobilized Metal Ion Affinity Membrane
Abstract
:1. Introduction
2. Results and Discussion
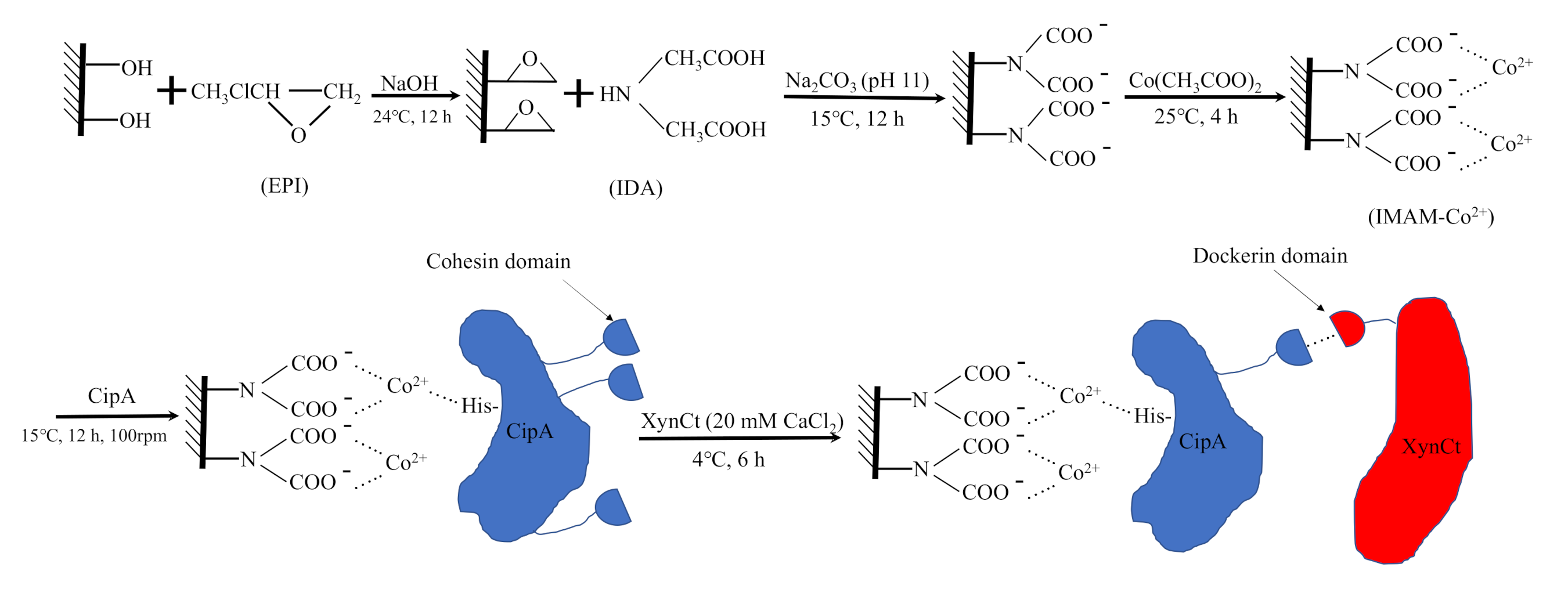
2.1. Constructs of CipA and XynCt
2.2. Expression of CipA and XynCt Proteins
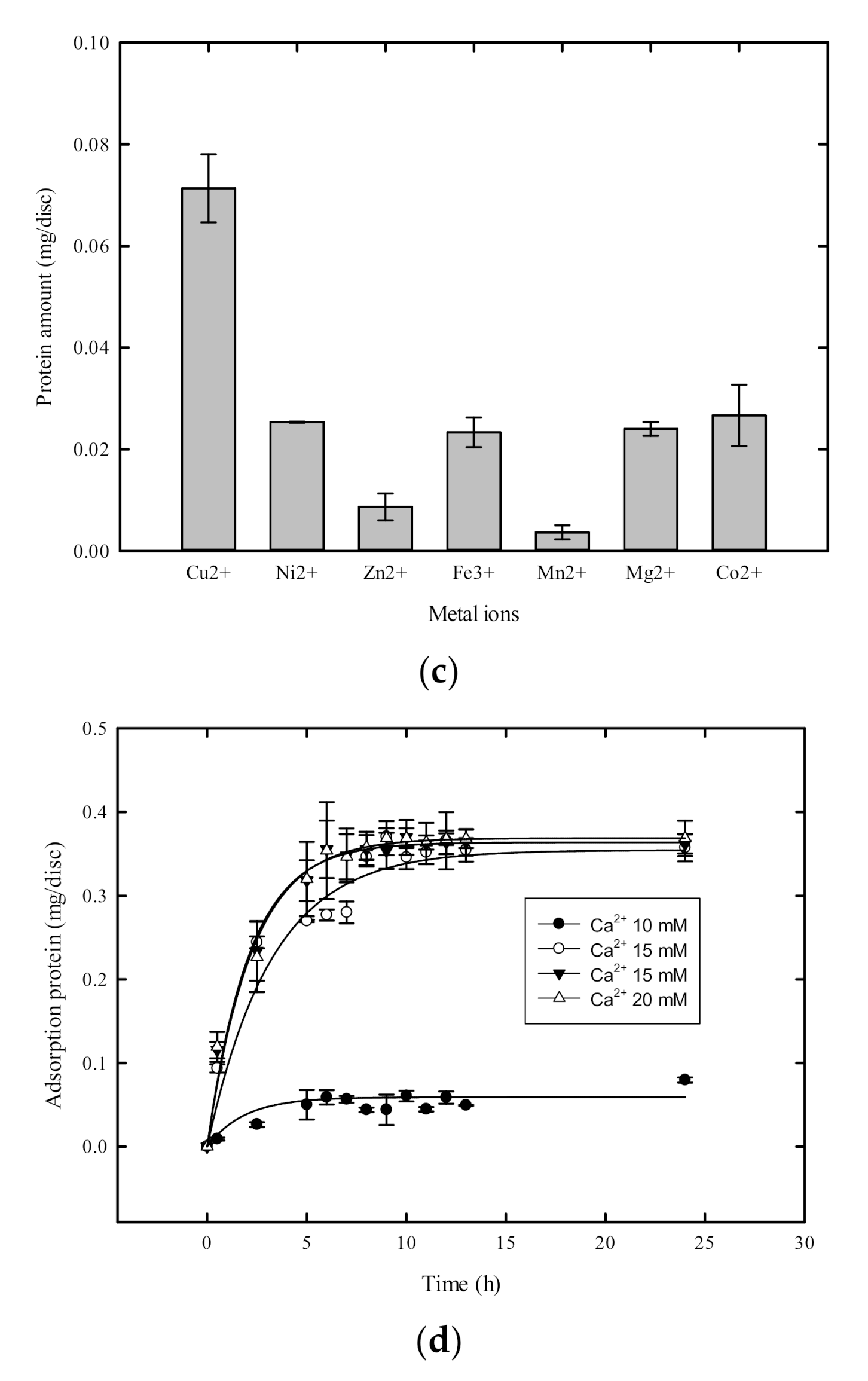
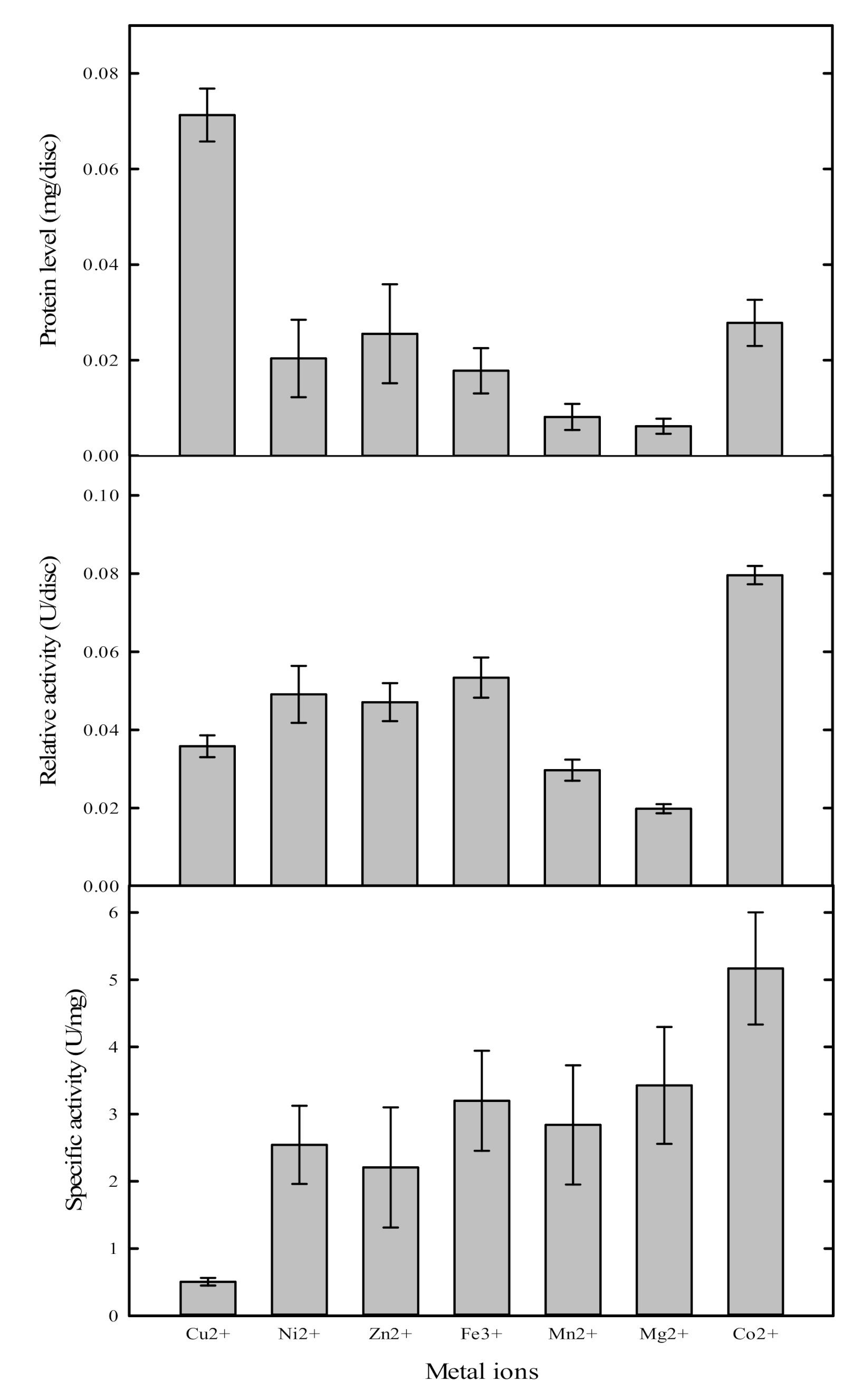
2.3. Metal Ion Selection
2.4. Characteristics of the Immobilized Enzyme
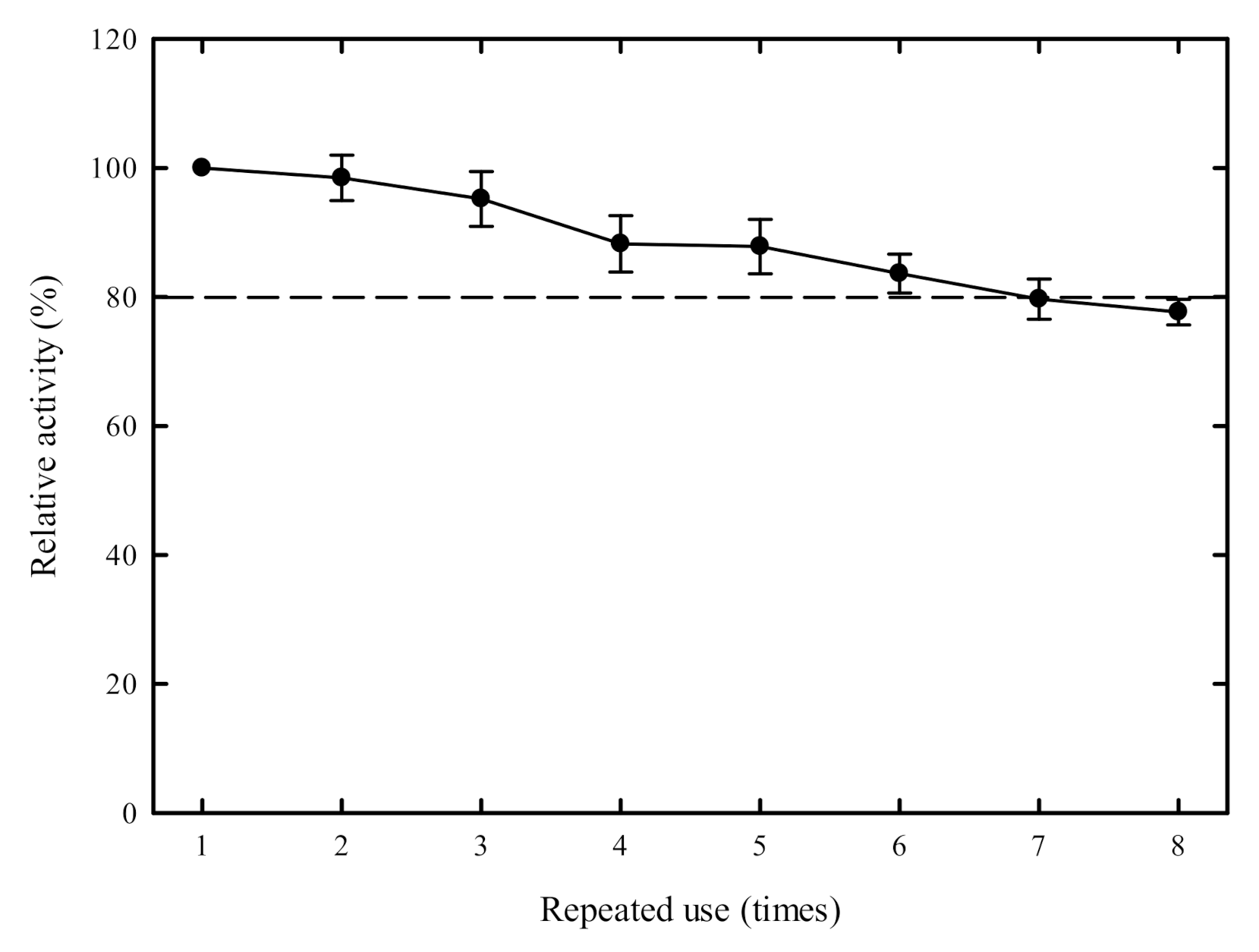
2.5. Reusability of the Immobilized Enzyme
2.6. Kinetics Analyses of Various XynC Enzymes
3. Materials and Methods
3.1. Chemicals, Strains, and Plasmids
3.2. Constructs of the XynCt and CipA
3.3. Strain Cultivation and Protein Expression
3.4. IMAM Preparation and Enzyme Immobilization
3.5. Assays
3.6. The pH and Temperature Effect
3.7. Kinetics Model
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, L.; Rowbotham, J.S.; Greenwell, C.H.; Dyer, P.W. An introduction to pyrolysis and catalytic pyrolysis: Versatile techniques for biomass conversion. In New and Future Developments in Catalysis: Catalytic Biomass Conversion; Elsevier: Amsterdam, The Netherlands, 2013; pp. 173–208. [Google Scholar]
- Wyman, C.E.; Decker, S.R.; Himmel, M.E.; Brady, J.W. Hydrolysis of Cellulose and Hemicellulose, in Polysaccharides: Structural Diversity and Functional Versatility, 2nd ed.; Dumitriu, S., Ed.; Marcel Dekker INC.: New York, NY, USA, 2005; pp. 995–1033. [Google Scholar]
- Dhiman, S.S.; Sharma, J.; Battan, B. Industrial applications and future prospects of microbial xylanases: A review. Bioresources 2008, 3, 1377–1402. [Google Scholar]
- Gírio, F.M.; Fonseca, C.; Carvalheiro, F.; Duarte, L.C.; Marques, S.; Bogel-Łukasik, R. Hemicelluloses for fuel ethanol: A review. Bioresour. Technol. 2010, 101, 4775–4800. [Google Scholar] [CrossRef]
- Peng, F.; Peng, P.; Xu, F.; Sun, R.-C. Fractional purification and bioconversion of hemicelluloses. Biotechnol. Adv. 2012, 30, 879–903. [Google Scholar] [CrossRef]
- Saha, B.C. a-L-Arabinofuranosidases: Biochemistry, molecular biology and application in biotechnology. Biotechnol. Adv. 2000, 18, 403–423. [Google Scholar] [CrossRef]
- Orita, T.; Sakka, M.; Kimura, T.; Sakka, K. Recombinant cellulolytic or xylanolytic complex comprising the full-length scaffolding protein RjCipA and cellulase RjCel5B or xylanase RjXyn10C of Ruminiclostridium josui. Enzym. Microb. Technol. 2017, 97, 63–70. [Google Scholar] [CrossRef]
- Kumar, P.; Gupta, A.; Dhakate, S.R.; Mathur, R.B.; Nagar, S.; Gupta, V.K. Covalent immobilization of xylanase produced from Bacillus pumilus SV-85S on electrospun polymethyl methacrylate nanofiber membrane. Biotechnol. Appl. Biochem. 2013, 60, 162–169. [Google Scholar] [CrossRef]
- Naidja, A.; Huang, P.M.; Bollag, J.-M. Activity of tyrosinase immobilized on hydroxyaluminum-montmorillonite complexes. J. Mol. Catal. A Chem. 1997, 115, 305–316. [Google Scholar] [CrossRef]
- He, J.; Li, X.; Evans, D.G.; Duan, X.; Li, C. A new support for the immobilization of penicillin acylase. J. Mol. Catal. B Enzym. 2000, 11, 45–53. [Google Scholar] [CrossRef]
- Kotha, A.; Selvaraj, L.; Rajan, C.R.; Ponrathnam, S.; Kumar, K.K.; Ambekar, G.R.; Shewale, J.G. Adsorption and expression of penicillin-G acylase immobilized onto methacrylate polymers generated with varying pore generating solvent volume. Appl. Biochem. Biotechnol. 1991, 30, 297–302. [Google Scholar] [CrossRef]
- Chaga, G.S. Twenty-five years of immobilized metal ion affinity chromatography: Past, present and future. J. Biochem. Biophys. Methods 2001, 49, 313–334. [Google Scholar] [CrossRef]
- Chaga, G.S.; Ersson, B.; Porath, J.O. Isolation of calcium-binding proteins on selective adsorbents application to purification of bovine calmodulin. J. Chromatogr. A 1996, 732, 261–269. [Google Scholar] [CrossRef]
- Boccù, E.; Gianferrara, T.; Gardossi, L.; Veronese, F.M. E. coli penicillin acylase: Purification by affinity chromatography and covalent binding to nylon. Farmaco 1990, 45, 203–214. [Google Scholar]
- Fitton, V.; Verdoni, N.; Sanchez, J.; Santarelli, X. Penicillin acylase purification with the aid of pseudo-affinity chromatography. J. Biochem. Biophys. Methods 2001, 49, 553–560. [Google Scholar] [CrossRef]
- Armisén, P.; Mateo, C.; Cortés, E.; Barredo, J.L.; Salto, F.; Diez, B.; Rodés, L.; García, J.L.; Fernández-Lafuente, R.; Guisán, J.M. Selective adsorption of poly-His tagged glutaryl acylase on tailor-made metal chelate supports. J. Chromatogr. A 1999, 848, 61–70. [Google Scholar] [CrossRef]
- Gibert, S.; Bakalara, N.; Santarelli, X. Three-step chromatographic purification procedure for the production of a His-tag recombinant kinesin overexpressed in E. coli. J. Chromatogr. B Biomed. Sci. Appl. 2000, 737, 143–150. [Google Scholar] [CrossRef]
- Bresolin, I.T.L.; Borsoi-Ribeiro, M.; Tamashiro, W.M.S.C.; Augusto, E.F.P.; Vijayalakshmi, M.A.; Bueno, S.M.A. Evaluation of Immobilized Metal-Ion Affinity Chromatography (IMAC) as a Technique for IgG1 Monoclonal Antibodies Purification: The Effect of Chelating Ligand and Support. Appl. Biochem. Biotechnol. 2010, 160, 2148–2165. [Google Scholar] [CrossRef]
- Hu, H.-L.; Wang, M.-Y.; Chung, C.-H.; Suen, S.-Y. Purification of VP3 protein of infectious bursal disease virus using nickel ion-immobilized regenerated cellulose-based membranes. J. Chromatogr. B 2006, 840, 76–84. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Changchien, C.-C.; Suen, S.-Y. Purification of penicillin G acylase using immobilized metal affinity membranes. J. Chromatogr. B 2003, 794, 67–76. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Suen, S.-Y.; Chen, S.-C.; Tzeng, J.-H. Analysis of protein adsorption on regenerated cellulose-based immobilized copper ion affinity membranes. J. Chromatogr. A 2003, 996, 53–70. [Google Scholar] [CrossRef]
- Chen, C.-S.; Suen, S.-Y.; Lai, S.-Y.; Chang, G.R.-L.; Lu, T.-C.; Lee, M.-S.; Wang, M.-Y. Purification of capsid-like particles of infectious bursal disease virus (IBDV) VP2 expressed in E. coli with a metal-ion affinity membrane system. J. Virol. Methods 2005, 130, 51–58. [Google Scholar] [CrossRef]
- Ke, Y.-M.; Chen, C.-I.; Kao, P.-M.; Chen, H.-B.; Huang, H.-C.; Yao, C.-J.; Liu, Y.-C. Preparation of the immobilized metal affinity membrane with high amount of metal ions and protein adsorption efficiencies. Process Biochem. 2010, 45, 500–506. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Suen, S.-Y.; Huang, C.-W.; Changchien, C.-C. Effects of spacer arm on penicillin G acylase purification using immobilized metal affinity membranes. J. Membr. Sci. 2005, 251, 201–207. [Google Scholar] [CrossRef]
- Pal, A.; Khanum, F. Covalent immobilization of xylanase on glutaraldehyde activated alginate beads using response surface methodology: Characterization of immobilized enzyme. Process Biochem. 2011, 46, 1315–1322. [Google Scholar] [CrossRef]
- Dos Santos, J.P.; da Rosa Zavareze, E.; Dias, A.R.G.; Vanier, N.L. Immobilization of xylanase and xylanase–β-cyclodextrin complex in polyvinyl alcohol via electrospinning improves enzyme activity at awide pH and temperature range. Int. J. Biol. Macromol. 2018, 118, 1676–1684. [Google Scholar] [CrossRef]
- Singh, V.; Kaul, S.; Singla, P.; Kumar, V.; Sandhir, R.; Chung, J.H.; Garg, P.; Singhal, N.K. Xylanase immobilization on magnetite and magnetite core/shell nanocomposites using two different flexible alkyl length organophosphonates: Linker length and shell effect on enzyme catalytic activity. Int. J. Biol. Macromol. 2018, 115, 590–599. [Google Scholar] [CrossRef]
- Chen, C.-I.; Ko, Y.-M.; Shieh, C.-J.; Liu, Y.-C. Direct penicillin G acylase immobilization by using the self-prepared immobilized metal affinity membrane. J. Membr. Sci. 2011, 380, 34–40. [Google Scholar] [CrossRef]
- Ko, Y.-M.; Chen, C.-I.; Shieh, C.-J.; Liu, Y.-C. Simultaneous purification and immobilization of d-hydantoinase on the immobilized metal affinity membrane via coordination bonds. Biochem. Eng. J. 2012, 61, 20–27. [Google Scholar] [CrossRef]
- Choi, S.K.; Ljungdahl, L.G. Dissociation of the cellulosome of Clostridium thermocellum in the presence of ethylenediaminetetraacetic acid occurs with the formation of trucated polypeptides. Biochemistry 1996, 35, 4897–4905. [Google Scholar] [CrossRef]
- Craig, S.J.; Foong, F.C.; Nordon, R. Engineered proteins containing the cohesin and dockerin domains from Clostridium thermocellum provides a reversible, high affinity interaction for biotechnology applications. J. Biotechnol. 2006, 121, 165–173. [Google Scholar] [CrossRef]
- Arora, R.; Behera, S.; Sharma, N.K.; Kumar, S. Bioprospecting thermostable cellulosomes for efficient biofuel production from lignocellulosic biomass. Bioresour. Bioprocess. 2015, 2, 38. [Google Scholar] [CrossRef] [Green Version]
- Mongay, C.; Cerdà, V. A Britton-Robinson buffer of known ionic strength. Ann. Chim. 1974, 64, 409–412. [Google Scholar]
- Amo, G.S.; Bezerra-Bussoli, C.; da Silva, R.R.; Kishi, L.T.; Ferreira, H.; Mariutti, R.B.; Arni, R.K.; Gomes, E.; Bonilla-Rodriguez, G.O. Heterologous expression, purification and biochemical characterization of a new xylanase from Myceliophthora heterothallica F.2.1.4. Int. J. Biol. Macromol. 2019, 131, 798–805. [Google Scholar] [CrossRef]
- Elgharbi, F.; Hlima, H.B.; Farhat-Khemakhem, A.; Ayadi-Zouari, D.; Bejar, S.; Hmida-Sayari, A. Expression of A. niger US368 xylanase in E. coli: Purification, characterization and copper activation. Int. J. Biol. Macromol. 2015, 74, 263–270. [Google Scholar] [CrossRef]
- Vitcosque, G.L.; Ribeiro, L.F.C.; de Lucas, R.C.; da Silva, T.M.; de Lima Damasio, A.R.; Farinas, C.S.; Gonçalves, A.Z.L.; Segato, F.; Buckeridge, M.S.; Jorge, J.A.; et al. The functional properties of a xyloglucanase (GH12) of Aspergillus terreus expressed in Aspergillus nidulans may increase performance of biomass degradation. Appl. Microbiol. Biotechnol. 2016, 100, 9133–9144. [Google Scholar] [CrossRef]
- Mehnati-Najafabadi, V.; Taheri-Kafrani, A.; Bordbar, A.-K. Xylanase immobilization on modified superparamagnetic graphene oxide nanocomposite: Effect of PEGylation on activity and stability. Int. J. Biol. Macromol. 2018, 107, 418–425. [Google Scholar] [CrossRef]
- Bailey, M.J.; Biely, P.; Poutanen, K. Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 1992, 23, 257–270. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to Image J: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]






| Enzyme | R2 | Km (mg/mL) | νmax (U/mg) |
|---|---|---|---|
| XynCt | 0.963 | 8.445 | 2.235 |
| XynCt + CipA | 0.972 | 4.736 | 1.330 |
| IMAM-Co2+-XynCt | 0.953 | 2.446 | 2.713 |
| IMAM-Co2+-CipA-XynCt | 0.929 | 1.513 | 3.831 |
| Strain or Plasmid | Genotype and Relevant Characteristics | Source |
|---|---|---|
| DH5α | F- endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG pΦ 80dlacZΔM15 Δ(lacZYA-argF) U169, hsdR17 (rk− mk+), λ– | Novagen, USA |
| BL21(DE3) | E. coli str. B F− ompT gal dcm lon hsdSB (rB− mB−) λ (DE3 [lacI lacUV5-T7gene ind1 sam7 nin5]) [malB+] K-12(λS) | Novagen, USA |
| ER2566(DE3) | F-λ-fhuA2 [lon] ompT lacZ::T7 gene 1gal sulA11 Δ(mcrC-mrr)114::IS10R(mcr-73::miniTn10-TetS)2 R(zgb210::Tn10) (TetS) endA1 [dcm] | New England Biolabs, USA |
| JM109(DE3) | endA1 glnV44 thi-1 relA1 lon::IS186 mcrB+_(lac-proAB) e14-[F’ traD36 proAB+ lacIq lacZ△M15] hsdR17(rK− mK+)λ−(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Promega, Fitchburg, WI, USA |
| pET21b and pET21d | bacterial vectors for inducible expression of N-terminally T7-tagged protein | Novagen, USA |
| T-vector | pBluescript IISK(–) with modified MCS | Yeastern Biotech Co. |
| CipA, DocT, XynC | Cellulosome scaffolding gene, Dockerin gene, Xylanase gene | Professor Li (Academia Sinica, Taiwan) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, H.-L.; Hu, N.-J.; Juang, T.-Y.; Liu, Y.-C. Co-Immobilization of Xylanase and Scaffolding Protein onto an Immobilized Metal Ion Affinity Membrane. Catalysts 2020, 10, 1408. https://doi.org/10.3390/catal10121408
Wong H-L, Hu N-J, Juang T-Y, Liu Y-C. Co-Immobilization of Xylanase and Scaffolding Protein onto an Immobilized Metal Ion Affinity Membrane. Catalysts. 2020; 10(12):1408. https://doi.org/10.3390/catal10121408
Chicago/Turabian StyleWong, Ho-Lam, Nien-Jen Hu, Tzong-Yuan Juang, and Yung-Chuan Liu. 2020. "Co-Immobilization of Xylanase and Scaffolding Protein onto an Immobilized Metal Ion Affinity Membrane" Catalysts 10, no. 12: 1408. https://doi.org/10.3390/catal10121408
APA StyleWong, H.-L., Hu, N.-J., Juang, T.-Y., & Liu, Y.-C. (2020). Co-Immobilization of Xylanase and Scaffolding Protein onto an Immobilized Metal Ion Affinity Membrane. Catalysts, 10(12), 1408. https://doi.org/10.3390/catal10121408