Glyoxyl-Activated Agarose as Support for Covalently Link Novo-Pro D: Biocatalysts Performance in the Hydrolysis of Casein
Abstract
:1. Introduction
2. Results
2.1. Effect of the Immobilization Time on the Immobilization Parameters of NPD on Agarose-Glyoxyl Support
2.2. Effect of Protein Loading on Immobilized Enzyme Performance
2.3. Effect of Temperature and pH on the NPD Activity
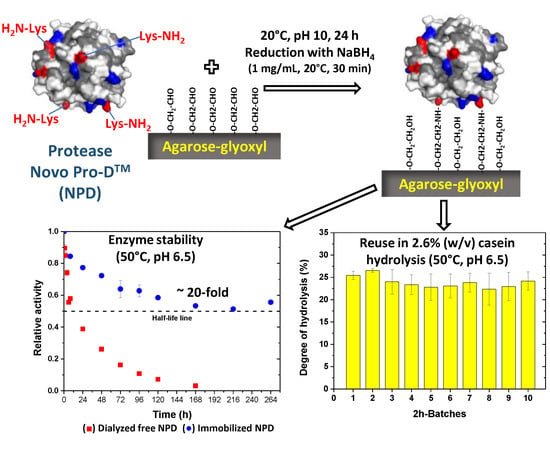
2.4. Thermal Stability at pH 6.5 of Soluble and Immobilized NPD
2.5. Performance of Soluble and Immobilized NPD in the Hydrolysis of Casein
3. Materials and Methods
3.1. Materials
3.2. Standard Enzyme Activity Assay Using a Small Substrate
3.3. NPD Immobilization Procedure
3.4. pH and Temperature Effects on Enzyme Activity
3.5. Thermal and pH Stabilities of Soluble and Immobilized NPD
3.6. Protein Hydrolysis Assays and Biocatalyst Reuse
3.7. Protein Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Datta, S.; Christena, L.R.; Rajaram, Y.R.S. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech 2012, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- DiCosimo, R.; McAuliffe, J.; Poulose, A.J.; Bohlmann, G. Industrial use of immobilized enzymes. Chem. Soc. Rev. 2013, 42, 6437–6474. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Zhao, X.S.; Bao, X.Y.; Guo, W.; Lee, F.Y. Immobilizing catalysts on porous materials. Mater. Today 2006, 9, 32–39. [Google Scholar] [CrossRef]
- Bilal, M.; Asgher, M.; Cheng, H.; Yan, Y.; Iqbal, H.M.N. Multi-point enzyme immobilization, surface chemistry, and novel platforms: A paradigm shift in biocatalyst design. Crit. Rev. Biotechnol. 2019, 39, 202–219. [Google Scholar] [CrossRef]
- Eş, I.; Vieira, J.D.G.; Amaral, A.C. Principles, techniques, and applications of biocatalyst immobilization for industrial application. Appl. Microbiol. Biotechnol. 2015, 99, 2065–2082. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- dos Santos Aguilar, J.G.; Sato, H.H. Microbial proteases: Production and application in obtaining protein hydrolysates. Food Res. Int. 2018, 103, 253–262. [Google Scholar] [CrossRef]
- Tavano, O.L.; Berenguer-Murcia, A.; Secundo, F.; Fernandez-Lafuente, R. Biotechnological Applications of Proteases in Food Technology. Compr. Rev. Food Sci. Food Saf. 2018, 17, 412–436. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, K.; Fernandez-Lafuente, R. Control of protein immobilization: Coupling immobilization and site-directed mutagenesis to improve biocatalyst or biosensor performance. Enzyme Microb. Technol. 2011, 48, 107–122. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fuentes, M.; Betancor, L.; Grazu, V.; López-Gallego, F.; Pessela, B.C.C.; Hidalgo, A.; Fernández-Lorente, G.; Fernández-Lafuente, R.; et al. Glyoxyl agarose: A fully inert and hydrophilic support for immobilization and high stabilization of proteins. Enzyme Microb. Technol. 2006, 39, 274–280. [Google Scholar] [CrossRef]
- Tardioli, P.W.; Pedroche, J.; Giordano, R.L.C.; Fernández-Lafuente, R.; Guisán, J.M. Hydrolysis of proteins by immobilized-stabilized alcalase-glyoxyl agarose. Biotechnol. Prog. 2003, 19, 352–360. [Google Scholar] [CrossRef]
- Blanco, R.M.; Guisan, J.M. Stabilization of enzymes by multipoint covalent attachment to agarose-aldehyde gels. Borohydride reduction of trypsin-agarose derivatives. Enzyme Microb. Technol. 1989, 11, 360–366. [Google Scholar] [CrossRef]
- López-Gallego, F.; Montes, T.; Fuentes, M.; Alonso, N.; Grazu, V.; Betancor, L.; Guisán, J.M.; Fernández-Lafuente, R. Improved stabilization of chemically aminated enzymes via multipoint covalent attachment on glyoxyl supports. J. Biotechnol. 2005, 116, 1–10. [Google Scholar] [CrossRef]
- Siar, E.H.; Zaak, H.; Kornecki, J.F.; Zidoune, M.N.; Barbosa, O.; Fernandez-Lafuente, R. Stabilization of ficin extract by immobilization on glyoxyl agarose. Preliminary characterization of the biocatalyst performance in hydrolysis of proteins. Process Biochem. 2017, 58, 98–104. [Google Scholar] [CrossRef]
- Pedroche, J.; del Mar Yust, M.; Mateo, C.; Fernández-Lafuente, R.; Girón-Calle, J.; Alaiz, M.; Vioque, J.; Guisán, J.M.; Millán, F. Effect of the support and experimental conditions in the intensity of the multipoint covalent attachment of proteins on glyoxyl-agarose supports: Correlation between enzyme–support linkages and thermal stability. Enzyme Microb. Technol. 2007, 40, 1160–1166. [Google Scholar] [CrossRef]
- Santos, J.C.S.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the support properties for immobilization or purification of enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef] [Green Version]
- Zucca, P.; Fernandez-Lafuente, R.; Sanjust, E. Agarose and its derivatives as supports for enzyme immobilization. Molecules 2016, 21, 1577. [Google Scholar] [CrossRef]
- Mateo, C.; Abian, O.; Bernedo, M.; Cuenca, E.; Fuentes, M.; Fernandez-Lorente, G.; Palomo, J.M.; Grazu, V.; Pessela, B.C.C.C.; Giacomini, C.; et al. Some special features of glyoxyl supports to immobilize proteins. Enzyme Microb. Technol. 2005, 37, 456–462. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Rocha-Martin, J.; Mateo, C.; Cava, F.; Berenguer, J.; Vega, D.; Fernandez-Lafuente, R.; Guisan, J.M. Purification and stabilization of a glutamate dehygrogenase from Thermus thermophilus via oriented multisubunit plus multipoint covalent immobilization. J. Mol. Catal. B Enzym. 2009, 58, 158–163. [Google Scholar] [CrossRef]
- Grazu, V.; Betancor, L.; Montes, T.; Lopez-gallego, F.; Guisan, J.M.; Fernandez-Lafuente, R. Glyoxyl agarose as a new chromatographic matrix. Enzyme Microb. Technol. 2006, 38, 960–966. [Google Scholar] [CrossRef]
- Lima, L.N.; Aragon, C.C.; Mateo, C.; Palomo, J.M.; Giordano, R.L.C.; Tardioli, P.W.; Guisan, J.M.; Fernandez-Lorente, G. Immobilization and stabilization of a bimolecular aggregate of the lipase from Pseudomonas fluorescens by multipoint covalent attachment. Process Biochem. 2013, 48, 118–123. [Google Scholar] [CrossRef]
- Bashir, F.; Asgher, M.; Hussain, F.; Randhawa, M.A. Development and characterization of cross-linked enzyme aggregates of thermotolerant alkaline protease from Bacillus licheniformis. Int. J. Biol. Macromol. 2018, 113, 944–951. [Google Scholar] [CrossRef]
- Tardioli, P.W.; Fernández-Lafuente, R.; Guisán, J.M.; Giordano, R.L.C. Design of new immobilized-stabilized carboxypeptidase a derivative for production of aromatic free hydrolysates of proteins. Biotechnol. Prog. 2003, 19, 565–574. [Google Scholar] [CrossRef]
- Atacan, K.; Çakıroğlu, B.; Özacar, M. Covalent immobilization of trypsin onto modified magnetite nanoparticles and its application for casein digestion. Int. J. Biol. Macromol. 2017, 97, 148–155. [Google Scholar] [CrossRef]
- Pedroche, J.; Yust, M.M.; Girón-Calle, J.; Vioque, J.; Alaiz, M.; Mateo, C.; Guisán, J.M.; Millán, F. Stabilization-immobilization of carboxypeptidase A to aldehyde-agarose gels: A practical example in the hydrolysis of casein. Enzyme Microb. Technol. 2002, 31, 711–718. [Google Scholar] [CrossRef]
- Kamnerdpetch, C.; Weiss, M.; Kasper, C.; Scheper, T. An improvement of potato pulp protein hydrolyzation process by the combination of protease enzyme systems. Enzyme Microb. Technol. 2007, 40, 508–514. [Google Scholar] [CrossRef]
- Waglay, A.; Karboune, S. Enzymatic generation of peptides from potato proteins by selected proteases and characterization of their structural properties. Biotechnol. Prog. 2016, 32, 420–429. [Google Scholar] [CrossRef]
- Maluf, J.U.; Fiorese, M.L.; Maestre, K.L.; Dos Passos, F.R.; Finkler, J.K.; Fleck, J.F.; Borba, C.E. Optimization of the porcine liver enzymatic hydrolysis conditions. J. Food Process Eng. 2020, 43, e13370. [Google Scholar] [CrossRef]
- Rojas, M.J.; Siqueira, P.F.; Miranda, L.C.; Tardioli, P.W.; Giordano, R.L.C. Sequential proteolysis and cellulolytic hydrolysis of soybean hulls for oligopeptides and ethanol production. Ind. Crops Prod. 2014, 61, 202–210. [Google Scholar] [CrossRef]
- Tan, Y.; Chang, K.-C.; Meng, S. Comparing the kinetics of the hydrolysis of by-product from channel catfish (Ictalurus punctatus) fillet processing by eight proteases. LWT 2019, 111, 809–820. [Google Scholar] [CrossRef]
- Žuža, M.G.; Milašinović, N.Z.; Jonović, M.M.; Jovanović, J.R.; Kalagasidis Krušić, M.T.; Bugarski, B.M.; Knežević-Jugović, Z.D. Design and characterization of alcalase–chitosan conjugates as potential biocatalysts. Bioprocess Biosyst. Eng. 2017, 40, 1713–1723. [Google Scholar] [CrossRef]
- Yang, A.; Long, C.; Xia, J.; Tong, P.; Cheng, Y.; Wang, Y.; Chen, H. Enzymatic characterisation of the immobilised Alcalase to hydrolyse egg white protein for potential allergenicity reduction. J. Sci. Food Agric. 2017, 97, 199–206. [Google Scholar] [CrossRef]
- Ferreira, L.; Ramos, M.A.; Dordick, J.S.; Gil, M.H. Influence of different silica derivatives in the immobilization and stabilization of a Bacillus licheniformis protease (Subtilisin Carlsberg). J. Mol. Catal. B Enzym. 2003, 21, 189–199. [Google Scholar] [CrossRef] [Green Version]
- Ait Braham, S.; Hussain, F.; Morellon-Sterling, R.; Kamal, S.; Kornecki, J.F.; Barbosa, O.; Kati, D.E.; Fernandez-Lafuente, R. Cooperativity of covalent attachment and ion exchange on alcalase immobilization using glutaraldehyde chemistry: Enzyme stabilization and improved proteolytic activity. Biotechnol. Prog. 2019, 35, e2768. [Google Scholar] [CrossRef]
- Hussain, F.; Arana-Peña, S.; Morellon-Sterling, R.; Barbosa, O.; Braham, S.A.; Kamal, S.; Fernandez-Lafuente, R. Further stabilization of alcalase immobilized on glyoxyl supports: Amination plus modification with glutaraldehyde. Molecules 2018, 23, 3188. [Google Scholar] [CrossRef] [Green Version]
- Tardioli, P.W.; Sousa, R.; Giordano, R.C.; Giordano, R.L.C. Kinetic model of the hydrolysis of polypeptides catalyzed by Alcalase® immobilized on 10% glyoxyl-agarose. Enzyme Microb. Technol. 2005, 36, 555–564. [Google Scholar] [CrossRef]
- Sousa, R.; Lopes, G.P.; Tardioli, P.W.; Giordano, R.L.C.; Almeida, P.I.F.; Giordano, R.C. Kinetic model for whey protein hydrolysis by alcalase multipoint-immobilized on agarose gel particles. Brazilian J. Chem. Eng. 2004, 21, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Novozymes A/S. Novo-Pro D, Product Data Sheet. 2007; 1–2. [Google Scholar]
- da Rosa, L.O.L.; Santana, M.C.; Azevedo, T.L.; Brígida, A.I.S.; Godoy, R.; Pacheco, S.; Mellinger-Silva, C.; Cabral, L.M.C. A comparison of dual-functional whey hydrolysates by the use of commercial proteases. Food Sci. Technol. 2018, 38, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Goettig, P. Effects of glycosylation on the enzymatic activity and mechanisms of proteases. Int. J. Mol. Sci. 2016, 17, 1969. [Google Scholar] [CrossRef] [Green Version]
- Bonzom, C.; Hüttner, S.; Mirgorodskaya, E.; Chong, S.L.; Uthoff, S.; Steinbüchel, A.; Verhaert, R.M.D.; Olsson, L. Glycosylation influences activity, stability and immobilization of the feruloyl esterase 1a from Myceliophthora thermophila. AMB Express 2019, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Boudrant, J.; Woodley, J.M.; Fernandez-Lafuente, R. Parameters necessary to define an immobilized enzyme preparation. Process Biochem. 2020, 90, 66–80. [Google Scholar] [CrossRef]
- Sadana, A.; Henley, J.P. Single-step unimolecular non-first-order enzyme deactivation kinetics. Biotechnol. Bioeng. 1987, 30, 717–723. [Google Scholar] [CrossRef]
- Pronk, S.; Lindahl, E.; Kasson, P.M. Dynamic heterogeneity controls diffusion and viscosity near biological interfaces. Nat. Commun. 2014, 5, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Regan, D.L.; Lilly, M.D.; Dunnill, P. Influence of intraparticle diffuisional limitation on the observed kinetics of immobilized enzymes and on catalyst design. Biotechnol. Bioeng. 1974, 16, 1081–1093. [Google Scholar] [CrossRef]
- Shen, L.; Chen, Z. Critical review of the impact of tortuosity on diffusion. Chem. Eng. Sci. 2007, 62, 3748–3755. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Consolati, T.; Mayr, T.; Nidetzky, B. Shine a light on immobilized enzymes: Real-time sensing in solid supported biocatalysts. Trends Biotechnol. 2013, 31, 194–203. [Google Scholar] [CrossRef]
- Boniello, C.; Mayr, T.; Klimant, I.; Koenig, B.; Riethorst, W.; Nidetzky, B. Intraparticle concentration gradients for substrate and acidic product in immobilized cephalosporin C amidase and their dependencies on carrier characteristics and reaction parameters. Biotechnol. Bioeng. 2010, 106, 528–540. [Google Scholar] [CrossRef]
- Milessi, T.S.S.; Kopp, W.; Rojas, M.J.; Manrich, A.; Baptista-Neto, A.; Tardioli, P.W.; Giordano, R.C.; Fernandez-Lafuente, R.; Guisan, J.M.; Giordano, R.L.C. Immobilization and stabilization of an endoxylanase from Bacillus subtilis (XynA) for xylooligosaccharides (XOs) production. Catal. Today 2016, 259, 130–139. [Google Scholar] [CrossRef] [Green Version]
- Dal Magro, L.; Kornecki, J.F.; Klein, M.P.; Rodrigues, R.C.; Fernandez-Lafuente, R. Optimized immobilization of polygalacturonase from Aspergillus niger following different protocols: Improved stability and activity under drastic conditions. Int. J. Biol. Macromol. 2019, 138, 234–243. [Google Scholar] [CrossRef]
- Dal Magro, L.; Kornecki, J.F.; Klein, M.P.; Rodrigues, R.C.; Fernandez-Lafuente, R. Pectin lyase immobilization using the glutaraldehyde chemistry increases the enzyme operation range. Enzyme Microb. Technol. 2020, 132, 109397. [Google Scholar] [CrossRef]
- Minton, A.P. How can biochemical reactions within cells differ from those in test tubes? J. Cell Sci. 2006, 119, 2863–2869. [Google Scholar] [CrossRef] [Green Version]
- Vieira, D.C.; Lima, L.N.; Mendes, A.A.; Adriano, W.S.; Giordano, R.L.C.R.C.R.L.C.; Giordano, R.L.C.R.C.R.L.C.; Tardioli, P.W. Hydrolysis of lactose in whole milk catalyzed by β-galactosidase from Kluyveromyces fragilis immobilized on chitosan-based matrix. Biochem. Eng. J. 2013, 81, 54–64. [Google Scholar] [CrossRef]
- Sgarbieri, V.C. Structural and physicochemical properties of milk proteins. Brazilian J. food Technol. 2005, 8, 43–56. [Google Scholar]
- Sanchez, A.; Cruz, J.; Rueda, N.; Dos Santos, J.C.S.; Torres, R.; Ortiz, C.; Villalonga, R.; Fernandez-Lafuente, R. Inactivation of immobilized trypsin under dissimilar conditions produces trypsin molecules with different structures. RSC Adv. 2016, 6, 27329–27334. [Google Scholar] [CrossRef]
- Siar, E.-H.; Arana-Peña, S.; Barbosa, O.; Zidoune, M.; Fernandez-Lafuente, R. Immobilization/stabilization of ficin extract on glutaraldehyde-activated agarose beads. Variables that control the final stability and activity in protein hydrolyses. Catalysts 2018, 8, 149. [Google Scholar] [CrossRef] [Green Version]
- Siar, E.-H.; Morellon-Sterling, R.; Zidoune, M.N.; Fernandez-Lafuente, R. Amination of ficin extract to improve its immobilization on glyoxyl-agarose: Improved stability and activity versus casein. Int. J. Biol. Macromol. 2019, 133, 412–419. [Google Scholar] [CrossRef]
- Siar, E.-H.; Morellon-Sterling, R.; Zidoune, M.N.; Fernandez-Lafuente, R. Use of glyoxyl-agarose immobilized ficin extract in milk coagulation: Unexpected importance of the ficin loading on the biocatalysts. Int. J. Biol. Macromol. 2020, 144, 419–426. [Google Scholar] [CrossRef]
- Guisán, J. Aldehyde-agarose gels as activated supports for immobilization-stabilization of enzymes. Enzyme Microb. Technol. 1988, 10, 375–382. [Google Scholar] [CrossRef]
- Hummel, B.C.W. A modified spectrophotometric determination of chymotrypsin, trypsin, and thrombin. Can. J. Biochem. Physiol. 1959, 37, 1393–1399. [Google Scholar] [CrossRef]
- Tsabouri, S.; Douros, K.; Priftis, K. Cow’s milk allergenicity. Endocr. Metab. Immune Disord. Targets 2014, 14, 16–26. [Google Scholar] [CrossRef]
- Miciński, J.; Kowalski, I.M.; Zwierzchowski, G.; Szarek, J.; Pierożyński, B.; Zabłocka, E. Characteristics of cow’s milk proteins including allergenic properties and methods for its reduction. Polish Ann. Med. 2013, 20, 69–76. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Enzymic Hydrolysis of Food Proteins; Elsevier Applied Science Publishers: New York, NY, USA, 1986. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Naito, K.; Iio, T.; Katagi, M.; Yoshikawa, Y.; Ohtsuka, H.; Orino, K. Binding analysis of bovine milk proteins, especially casein interactions and the interaction between α-casein and lactoferrin, using beads immobilised with zinc ion, poly-l-lysine or α-casein. Int. Dairy J. 2020, 105, 104690. [Google Scholar] [CrossRef]












| Protein Loaded (mg/g) | Theoretical Immobilized Activity (UBTEE/g) | Theoretical Immobilized Activity (Ucasein/g) | RA (%) Using Small Substrate (0.36 mM BTEE) | RA (%) Using Large Substrate (20 g/L casein) 1 |
|---|---|---|---|---|
| 1.4 | 20.6 ± 0.2 | 88.1 ± 7.0 | 63.3 ± 0.7 | 16.3 ± 0.9 |
| 18.9 | 275.6 ± 0.2 | 1166.4 ± 93.3 | 13.8 ± 1.2 | 14.5 ± 4.4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, L.A.; Novelli, P.K.; Fernandez-Lafuente, R.; Tardioli, P.W.; Giordano, R.L.C. Glyoxyl-Activated Agarose as Support for Covalently Link Novo-Pro D: Biocatalysts Performance in the Hydrolysis of Casein. Catalysts 2020, 10, 466. https://doi.org/10.3390/catal10050466
Lopes LA, Novelli PK, Fernandez-Lafuente R, Tardioli PW, Giordano RLC. Glyoxyl-Activated Agarose as Support for Covalently Link Novo-Pro D: Biocatalysts Performance in the Hydrolysis of Casein. Catalysts. 2020; 10(5):466. https://doi.org/10.3390/catal10050466
Chicago/Turabian StyleLopes, Laiane Antunes, Paula Kern Novelli, Roberto Fernandez-Lafuente, Paulo Waldir Tardioli, and Raquel Lima Camargo Giordano. 2020. "Glyoxyl-Activated Agarose as Support for Covalently Link Novo-Pro D: Biocatalysts Performance in the Hydrolysis of Casein" Catalysts 10, no. 5: 466. https://doi.org/10.3390/catal10050466